Genetic Diversity of C4 Photosynthesis Pathway Genes in Sorghum bicolor (L.)
Abstract
1. Introduction
2. Materials and Methods
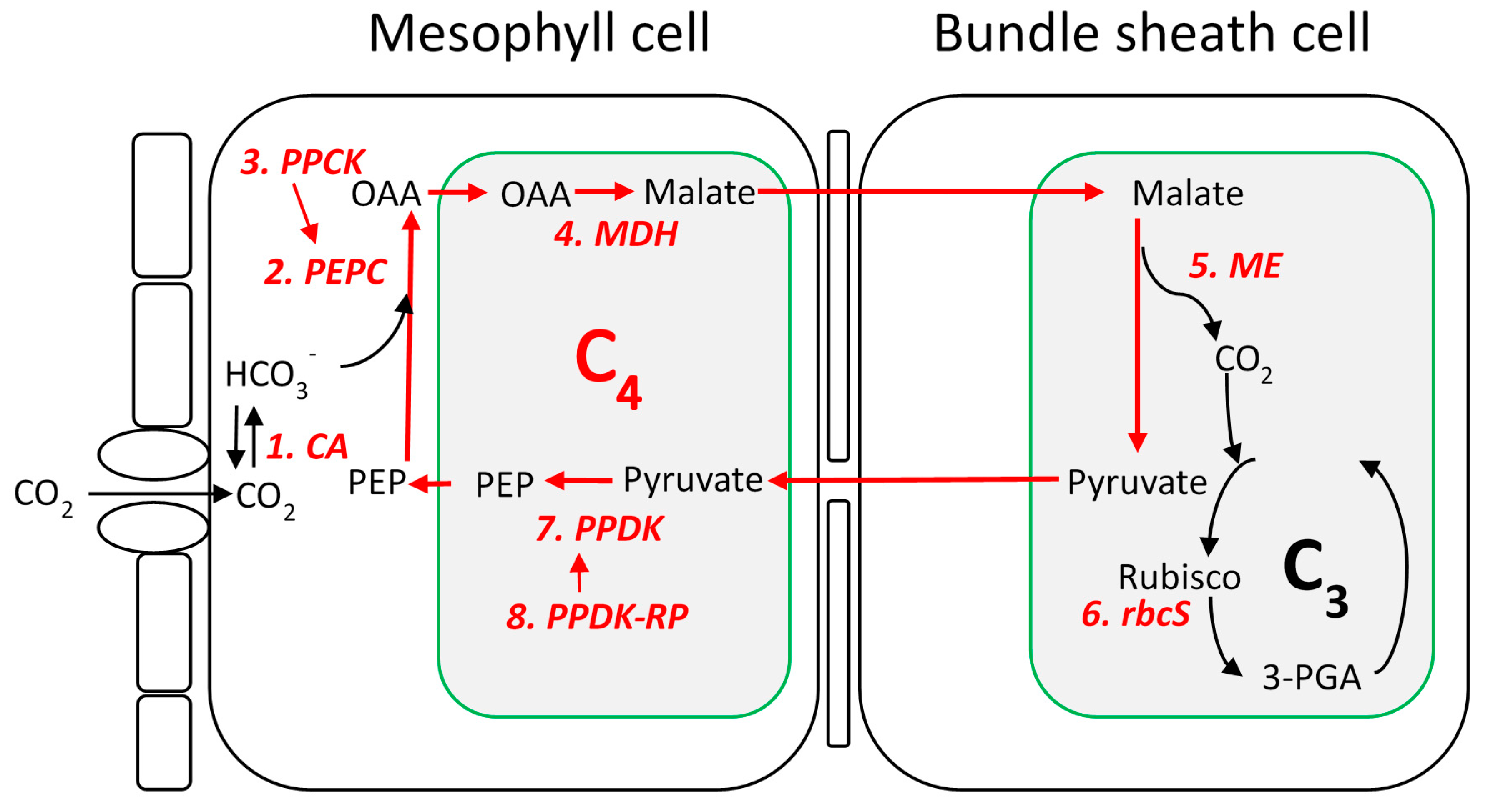
Identification of C4 Gene Families
3. Plant Material and Genomic Data
4. Gene-Level Population Genetic Analyses
5. SNP-Level Identification of Selection Signature
6. Phylogenetic and Haplotype Analysis
7. Results
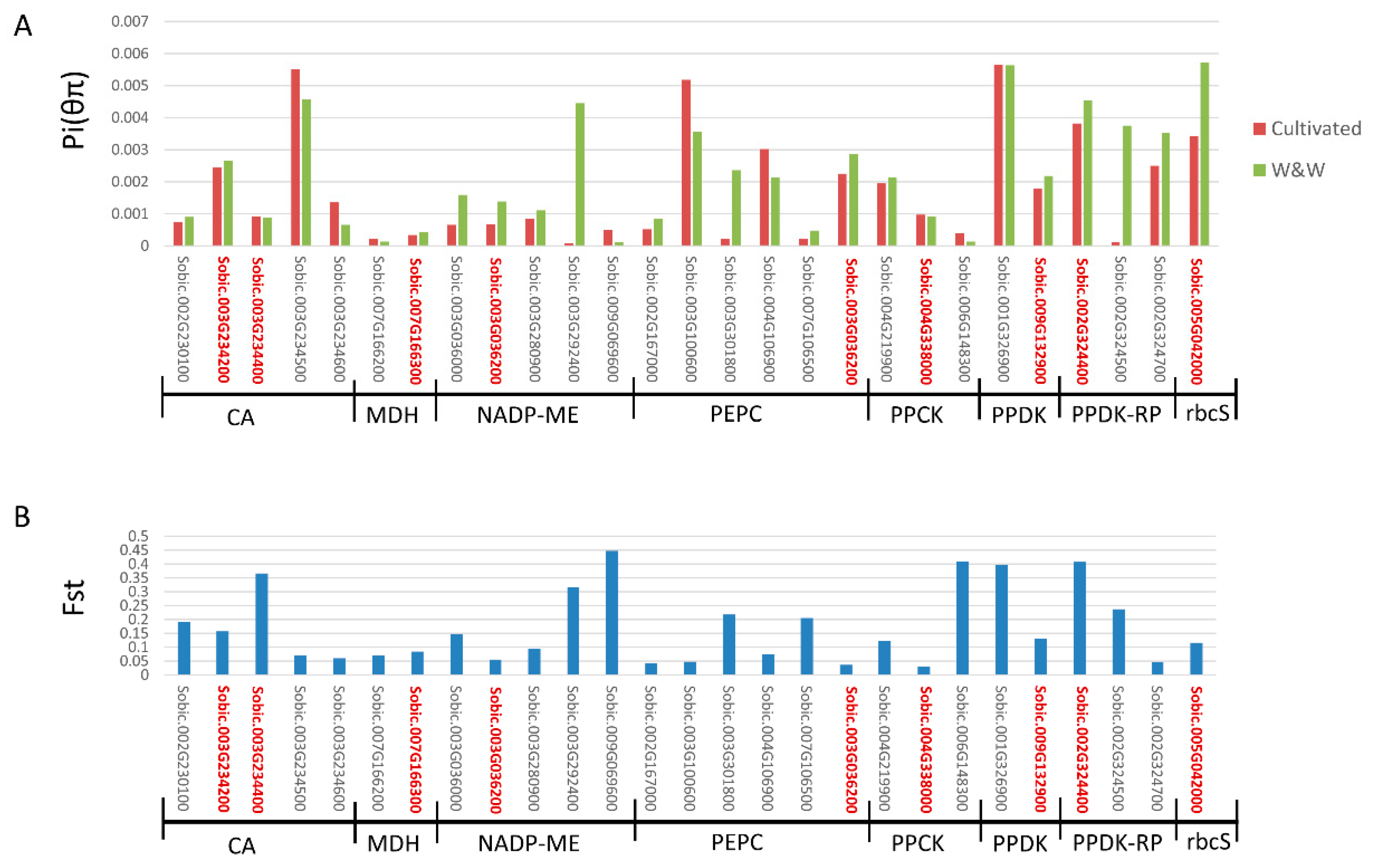
Nucleotide Diversity of Core C4 Gene Families in Sorghum
8. Identification of Selection Signals during Domestication across the 27 Genes
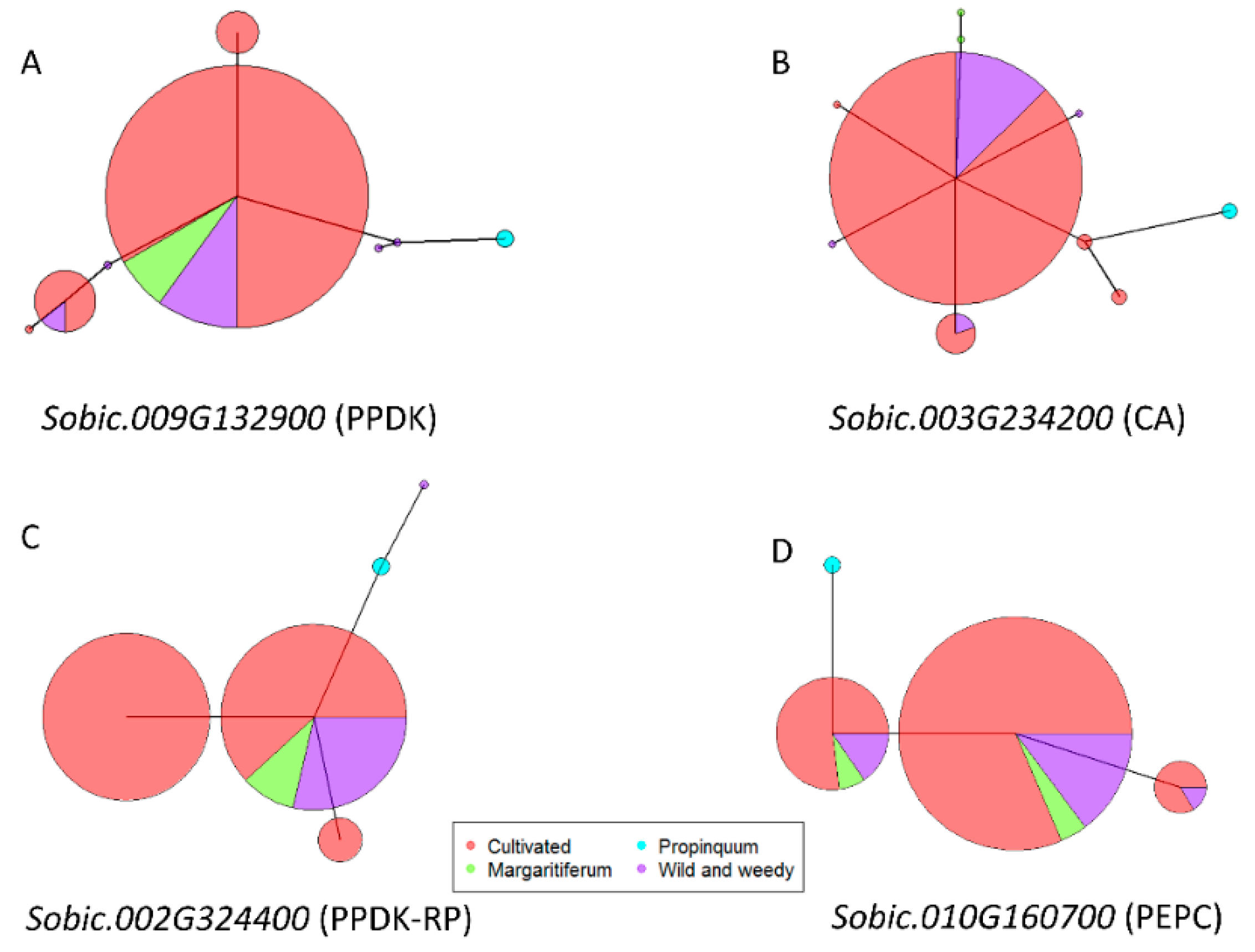
9. Allelic Variation of Core C4 Genes under Selection in Sorghum
10. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Sage, R.F.; Sage, T.L.; Kocacinar, F. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 2012, 63, 19–47. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F. The evolution of C4 photosynthesis. New Phytol. 2004, 161, 341–370. [Google Scholar] [CrossRef]
- Zelitch, I.; Schultes, N.P.; Peterson, R.B.; Brown, P.; Brutnell, T.P. High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 2009, 149, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Edwards, E.J.; Smith, S.A. Phylogenetic analyses reveal the shady history of C4 grasses. Proc. Natl. Acad. Sci. USA 2010, 107, 2532–2537. [Google Scholar] [CrossRef]
- Hibberd, J.M.; Covshoff, S. The regulation of gene expression required for C4 photosynthesis. Annu. Rev. Plant Biol. 2010, 61, 181–207. [Google Scholar] [CrossRef]
- von Caemmerer, S.; Furbank, R.T. The C4 pathway: An efficient CO2 pump. Photosynth. Res. 2003, 77, 191. [Google Scholar] [CrossRef] [PubMed]
- Hibberd, J.M.; Sheehy, J.E.; Langdale, J.A. Using C4 photosynthesis to increase the yield of rice—Rationale and feasibility. Curr. Opin. Plant Biol. 2008, 11, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Langdale, J.A. C4 cycles: Past, present, and future research on C4 photosynthesis. Plant Cell 2011, 23, 3879–3892. [Google Scholar] [CrossRef] [PubMed]
- von Caemmerer, S.; Furbank, R.T. Strategies for improving C4 photosynthesis. Curr. Opin. Plant Biol. 2016, 31, 125–134. [Google Scholar] [CrossRef]
- Still, C.J.; Berry, J.A.; Collatz, G.J.; DeFries, R.S. Global distribution of C3 and C4 vegetation: Carbon cycle implications. Glob. Biogeochem. Cy. 2003, 17, 1006. [Google Scholar] [CrossRef]
- Long, S.P.; Zhu, X.G.; Naidu, S.L.; Ort, D.R. Can improvement in photosynthesis increase crop yields? Plant Cell Environ. 2006, 29, 315–330. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.G.; Shan, L.L.; Wang, Y.; Quick, W.P. C4 Rice—An ideal arena for systems biology research. J. Integr. Plant Biol. 2010, 52, 762–770. [Google Scholar] [CrossRef] [PubMed]
- von Caemmerer, S.; Quick, W.P.; Furbank, R.T. The development of C4 rice: Current progress and future challenges. Science 2012, 336, 1671–1672. [Google Scholar] [CrossRef] [PubMed]
- Covshoff, S.; Szecowka, M.; Hughes, T.E.; Smith-Unna, R.; Kelly, S.; Bailey, K.J.; Sage, T.L.; Pachebat, J.A.; Leegood, R.; Hibberd, J.M. C4 Photosynthesis in the Rice Paddy: Insights from the Noxious Weed Echinochloa glabrescens. Plant Physiol. 2016, 170, 57–73. [Google Scholar] [CrossRef] [PubMed]
- Hatch, M.D.; Kagawa, T.; Craig, S. Subdivision of C4-pathway species based on differing C4 acid decarboxylating systems and ultrastructural features. Funct. Plant Biol. 1975, 2, 111–128. [Google Scholar] [CrossRef]
- Bräutigam, A.; Schliesky, S.; Külahoglu, C.; Osborne, C.P.; Weber, A.P.M. Towards an integrative model of C4 photosynthetic subtypes: Insights from comparative transcriptome analysis of NAD-ME, NADP-ME, and PEP-CK C4 species. J. Exp. Bot. 2014, 65, 3579–3593. [Google Scholar] [CrossRef]
- Sonawane, B.V.; Sharwood, R.E.; Whitney, S.; Ghannoum, O. Shade compromises the photosynthetic efficiency of NADP-ME less than PEP-CK and NAD-ME C4 Grasses. J. Exp. Bot. 2018, 69, 3053–3068. [Google Scholar] [CrossRef] [PubMed]
- Grass Phylogeny Working Group II. New grass phylogeny resolves deep evolutionary relationships and discovers C4 origins. New Phytol. 2012, 193, 304–312. [Google Scholar] [CrossRef]
- Kidambi, S.P.; Krieg, D.R.; Rosenow, D.T. Genetic variation for gas-exchange rates in grain sorghum. Plant Physiol. 1990, 92, 1211–1214. [Google Scholar] [CrossRef]
- Peng, S.B.; Krieg, D.R. Gas-exchange traits and their relationship to water-use efficiency of grain sorghum. Crop Sci. 1992, 32, 386–391. [Google Scholar] [CrossRef]
- Henderson, S.; von Caemmerer, S.; Farquhar, G.D.; Wade, L.J.; Hammer, G. Correlation between carbon isotope discrimination and transpiration efficiency in lines of the C4 species Sorghum bicolor in the glasshouse and the field. Aust. J. Plant Physiol. 1998, 25, 111–123. [Google Scholar] [CrossRef]
- Balota, M.; Payne, W.A.; Rooney, W.; Rosenow, D. Gas exchange and transpiration ratio in sorghum. Crop Sci. 2008, 48, 2361–2371. [Google Scholar] [CrossRef]
- Fernandez, M.G.S.; Strand, K.; Hamblin, M.T.; Westgate, M.; Heaton, E.; Kresovich, S. Genetic analysis and phenotypic characterization of leaf photosynthetic capacity in a sorghum (Sorghum spp.) diversity panel. Genet. Resour. Crop Ev. 2015, 62, 939–950. [Google Scholar] [CrossRef]
- Zheng, L.Y.; Guo, X.S.; He, B.; Sun, L.J.; Peng, Y.; Dong, S.S.; Liu, T.F.; Jiang, S.Y.; Ramachandran, S.; Liu, C.M.; et al. Genome-wide patterns of genetic variation in sweet and grain sorghum (Sorghum bicolor). Genome Biol. 2011, 12, R114. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Bowers, J.E.; Bruggmann, R.; Dubchak, I.; Grimwood, J.; Gundlach, H.; Haberer, G.; Hellsten, U.; Mitros, T.; Poliakov, A.; et al. The Sorghum bicolor genome and the diversification of grasses. Nature 2009, 457, 551–556. [Google Scholar] [CrossRef] [PubMed]
- Mace, E.S.; Tai, S.S.; Gilding, E.K.; Li, Y.H.; Prentis, P.J.; Bian, L.L.; Campbell, B.C.; Hu, W.S.; Innes, D.J.; Han, X.L.; et al. Whole-genome sequencing reveals untapped genetic potential in Africa’s indigenous cereal crop sorghum. Nat. Commun. 2013, 4, 2320. [Google Scholar] [CrossRef]
- Fankhauser, N.; Aubry, S. Post-transcriptional regulation of photosynthetic genes is a key driver of C4 leaf ontogeny. J. Exp. Bot. 2017, 68, 137–146. [Google Scholar] [CrossRef]
- Huang, P.; Studer, A.J.; Schnable, J.C.; Kellogg, E.A.; Brutnell, T.P. Cross species selection scans identify components of C4 photosynthesis in the grasses. J. Exp. Bot. 2017, 68, 127–135. [Google Scholar] [CrossRef]
- Burgess, S.J.; Hibberd, J.M. Insights into C4 metabolism from comparative deep sequencing. Curr. Opin. Plant Biol. 2015, 25, 138–144. [Google Scholar] [CrossRef]
- Reeves, G.; Grangé-Guermente, M.J.; Hibberd, J.M. Regulatory gateways for cell-specific gene expression in C4 leaves with Kranz anatomy. J. Exp. Bot. 2017, 68, 107–116. [Google Scholar] [CrossRef]
- Christin, P.-A.; Osborne, C.P.; Chatelet, D.S.; Columbus, J.T.; Besnard, G.; Hodkinson, T.R.; Garrison, L.M.; Vorontsova, M.S.; Edwards, E.J. Anatomical enablers and the evolution of C4 photosynthesis in grasses. Proc. Natl. Acad. Sci. USA 2013, 110, 1381–1386. [Google Scholar] [CrossRef]
- Moreno-Villena, J.J.; Dunning, L.T.; Osborne, C.P.; Christin, P.-A. Highly expressed genes are preferentially co-opted for C4 photosynthesis. Mol. Biol. Evol. 2018, 35, 94–106. [Google Scholar] [CrossRef]
- Borba, A.R.; Serra, T.S.; Gorska, A.; Gouveia, P.; Cordeiro, A.M.; Reyna-Llorens, I.; Knerova, J.; Barros, P.M.; Abreu, I.A.; Oliveira, M.M.O.; et al. Synergistic binding of bHLH transcription factors to the promoter of the maize NADP-ME gene used in C4 photosynthesis is based on an ancient code found in the ancestral C3 state. Mol. Biol. Evol. 2018, 35, 1690–1705. [Google Scholar] [CrossRef]
- Christin, P.-A.; Petitpierre, B.; Salamin, N.; Büchi, L.; Besnard, G. Evolution of C4 phosphoenolpyruvate carboxykinase in grasses, from genotype to phenotype. Mol. Biol. Evol. 2008, 26, 357–365. [Google Scholar] [CrossRef]
- Christin, P.-A.; Salamin, N.; Muasya, A.M.; Roalson, E.H.; Russier, F.; Besnard, G. Evolutionary switch and genetic convergence on rbcL following the evolution of C4 Photosynthesis. Mol. Biol. Evol. 2008, 25, 2361–2368. [Google Scholar] [CrossRef]
- Gowik, U.; Bräutigam, A.; Weber, K.L.; Weber, A.P.; Westhoff, P. Evolution of C4 photosynthesis in the genus Flaveria: How many and which genes does it take to make C4? Plant Cell 2011, 23, 2087–2105. [Google Scholar] [CrossRef]
- Mallmann, J.; Heckmann, D.; Bräutigam, A.; Lercher, M.J.; Weber, A.P.; Westhoff, P.; Gowik, U. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. eLife 2014, 3, e02478. [Google Scholar] [CrossRef]
- Williams, B.P.; Aubry, S.; Hibberd, J.M. Molecular evolution of genes recruited into C4 photosynthesis. Trends Plant Sci. 2012, 17, 213–220. [Google Scholar] [CrossRef]
- Wang, X.; Gowik, U.; Tang, H.; Bowers, J.E.; Westhoff, P.; Paterson, A.H. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 2009, 10, R68. [Google Scholar] [CrossRef]
- Ermakova, M.; Danila, F.R.; Furbank, R.T.; von Caemmerer, S. On the road to C4 rice: Advances and perspectives. Plant J. 2020, 101, 940–950. [Google Scholar] [CrossRef]
- Nei, M.; Li, W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [PubMed]
- Watterson, G.A. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Hudson, R.; Boos, D.D.; Kaplan, N. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 1992, 9, 138–151. [Google Scholar] [PubMed]
- Pfeifer, B.; Wittelsbürger, U.; Onsins, S.E.R.; Lercher, M.J. PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014, 31, 1929–1936. [Google Scholar] [CrossRef]
- Paradis, E.; Claude, J.; Strimmer, K.J.B. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Paradis, E. Pegas: An R package for population genetics with an integrated–modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef]
- Ehleringer, J.R.; Sage, R.F.; Flanagan, L.B.; Pearcy, R.W. Climate change and the evolution of C4 photosynthesis. Trends Ecol. Evol. 1991, 6, 95–99. [Google Scholar] [CrossRef]
- Christin, P.A.; Salamin, N.; Savolainen, V.; Duvall, M.R.; Besnard, G. C4 Photosynthesis evolved in grasses via parallel adaptive genetic changes. Curr. Biol. CB 2007, 17, 1241–1247. [Google Scholar] [CrossRef]
- Christin, P.A.; Samaritani, E.; Petitpierre, B.; Salamin, N.; Besnard, G. Evolutionary insights on C4 photosynthetic subtypes in grasses from genomics and phylogenetics. Genome Biol. Evol. 2009, 1, 221–230. [Google Scholar] [CrossRef][Green Version]
- Clark, J.D.; Stemler, A. Early domesticated sorghum from central Sudan. Nature 1975, 254, 588–591. [Google Scholar] [CrossRef]
- Mann, J.A.; Kimber, C.T.; Miller, F.R. The Origin and Early Cultivation of Sorghums in Africa; Bulletin 1454; Texas Agricultural Experiment Station: College Station, TX, USA, 1983. [Google Scholar]
- Wendorf, F.; Close, A.E.; Schild, R.; Wasylikowa, K.; Housley, R.A.; Harlan, J.R.; Królik, H. Saharan exploitation of plants 8,000 years BP. Nature 1992, 359, 721–724. [Google Scholar] [CrossRef]
- Tao, Y.; Mace, E.S.; Tai, S.; Cruickshank, A.; Campbell, B.C.; Zhao, X.; Van Oosterom, E.J.; Godwin, I.D.; Botella, J.R.; Jordan, D.R. Whole-genome analysis of candidate genes associated with seed size and weight in Sorghum bicolor reveals signatures of artificial selection and insights into parallel domestication in cereal crops. Front. Plant Sci. 2017, 8, 1237. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, X.; Mace, E.; Henry, R.; Jordan, D. Exploring and exploiting pan-genomics for crop improvement. Mol. Plant 2019, 12, 156–169. [Google Scholar] [CrossRef]
- Kümpers, B.M.C.; Burgess, S.J.; Reyna-Llorens, I.; Smith-Unna, R.; Boursnell, C.; Hibberd, J.M. Shared characteristics underpinning C4 leaf maturation derived from analysis of multiple C3 and C4 species of Flaveria. J. Exp. Bot. 2017, 68, 177–189. [Google Scholar] [CrossRef]
- Külahoglu, C.; Denton, A.K.; Sommer, M.; Maß, J.; Schliesky, S.; Wrobel, T.J.; Berckmans, B.; Gongora-Castillo, E.; Buell, C.R.; Simon, R.; et al. Comparative transcriptome atlases reveal altered gene expression modules between two Cleomaceae C3 and C4 plant species. Plant Cell 2014, 26, 3243–3260. [Google Scholar] [CrossRef]
- Massel, K.; Campbell, B.C.; Mace, E.S.; Tai, S.; Tao, Y.; Worland, B.G.; Jordan, D.R.; Botella, J.R.; Godwin, I.D. Whole genome sequencing reveals potential new targets for improving nitrogen uptake and utilization in Sorghum bicolor. Front. Plant Sci. 2016, 7, 1544. [Google Scholar] [CrossRef]
- Riley, R.M.; Jin, W.; Gibson, G. Contrasting selection pressures on components of the Ras-mediated signal transduction pathway in Drosophila. Mol. Ecol. 2003, 12, 1315–1323. [Google Scholar] [CrossRef]
- Campbell, B.C.; Gilding, E.K.; Mace, E.S.; Tai, S.; Tao, Y.; Prentis, P.J.; Thomelin, P.; Jordan, D.R.; Godwin, I.D. Domestication and the storage starch biosynthesis pathway: Signatures of selection from a whole sorghum genome sequencing strategy. Plant Biotechnol. J. 2016, 14, 2240–2253. [Google Scholar] [CrossRef]
- Wang, Y.-M.; Xu, W.-G.; Hu, L.; Zhang, L.; Li, Y.; Du, X.-H. Expression of maize gene encoding C4-pyruvate orthophosphate dikinase (PPDK) and C4-phosphoenolpyruvate carboxylase (PEPC) in transgenic Arabidopsis. Plant Mol. Biol. Rep. 2012, 30, 1367–1374. [Google Scholar] [CrossRef]
- Wang, D.; Portis, A.R.; Moose, S.P.; Long, S.P. Cool C4 photosynthesis: Pyruvate Pi dikinase expression and activity corresponds to the exceptional cold tolerance of carbon assimilation in Miscanthus × giganteus. Plant Physiol. 2008, 148, 557–567. [Google Scholar] [CrossRef]
- Naidu, S.L.; Moose, S.P.; AL-Shoaibi, A.K.; Raines, C.A.; Long, S.P. Cold Tolerance of C4 photosynthesis in Miscanthus × giganteus: Adaptation in amounts and sequence of C4 photosynthetic enzymes. Plant Physiol. 2003, 132, 1688–1697. [Google Scholar] [CrossRef]
- Chastain, C.J.; Failing, C.J.; Manandhar, L.; Zimmerman, M.A.; Lakner, M.M.; Nguyen, T.H.T. Functional evolution of C4 pyruvate, orthophosphate dikinase. J. Exp. Bot. 2011, 62, 3083–3091. [Google Scholar] [CrossRef]
- Bauwe, H.; Chollet, R. Kinetic properties of phosphoenolpyruvate carboxylase from C3, C4, and C3-C4 intermediate species of Flaveria (Asteraceae). Plant Physiol. 1986, 82, 695–699. [Google Scholar] [CrossRef]
- von Caemmerer, S.; Edwards, G.E.; Koteyeva, N.; Cousins, A.B. Single cell C4 photosynthesis in aquatic and terrestrial plants: A gas exchange perspective. Aquat. Bot. 2014, 118, 71–80. [Google Scholar] [CrossRef]
- Boyd, R.A.; Gandin, A.; Cousins, A.B. Temperature responses of C4 photosynthesis: Biochemical analysis of rubisco, phosphoenolpyruvate carboxylase, and carbonic anhydrase in Setaria viridis. Plant Physiol. 2015, 169, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Hatch, M.D.; Burnell, J.N. Carbonic anhydrase activity in leaves and its role in the first step of C4 photosynthesis. Plant Physiol. 1990, 93, 825–828. [Google Scholar] [CrossRef]
- von Caemmerer, S.; Quinn, V.; Hancock, N.; Price, G.D.; Furbank, R.T.; Ludwig, M. Carbonic anhydrase and C4 photosynthesis: A transgenic analysis. Plant Cell Environ. 2004, 27, 697–703. [Google Scholar] [CrossRef]
- Cousins, A.B.; Badger, M.R.; von Caemmerer, S. Carbonic anhydrase and its influence on carbon isotope discrimination during C4 photosynthesis. Insights from antisense RNA in Flaveria bidentis. Plant Physiol. 2006, 141, 232–242. [Google Scholar] [CrossRef]
- Osborn, H.L.; Alonso-Cantabrana, H.; Sharwood, R.E.; Covshoff, S.; Evans, J.R.; Furbank, R.T.; von Caemmerer, S. Effects of reduced carbonic anhydrase activity on CO2 assimilation rates in Setaria viridis: A transgenic analysis. J. Exp. Bot. 2016, 68, 299–310. [Google Scholar] [CrossRef]
- Watanabe, N.; Evans, J.R.; Chow, W.S. Changes in the Photosynthetic Properties of Australian wheat cultivars over the last century. Aust. J. Plant Physiol. 1994, 21, 169–183. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Zhang, C.; Dai, C.; Zhou, J.; Ren, C.; Zhang, J. Phosphoenolpyruvate carboxylase regulation in C4-PEPC-expressing transgenic rice during early responses to drought stress. Physiol. Plant. 2017, 159, 178–200. [Google Scholar] [CrossRef] [PubMed]
- Jeanneau, M.; Gerentes, D.; Foueillassar, X.; Zivy, M.; Vidal, J.; Toppan, A.; Perez, P. Improvement of drought tolerance in maize: Towards the functional validation of the Zm-Asr1 gene and increase of water use efficiency by over-expressing C4–PEPC. Biochimie 2002, 84, 1127–1135. [Google Scholar] [CrossRef]
- Tao, Y.; Zhao, X.; Wang, X.; Hathorn, A.; Hunt, C.; Cruickshank, A.W.; van Oosterom, E.J.; Godwin, I.D.; Mace, E.S.; Jordan, D.R. Large-scale GWAS in sorghum reveals common genetic control of grain size among cereals. Plant Biotechnol. J. 2020, 18, 1093–1105. [Google Scholar] [CrossRef]
| Gene ID | Enzyme | GL | CDSL | NoS | NoSiC | NoNS | NoSS | UPSGL | UBSGL | NoSUPS | NoNSUPS | NoSUBS | NoNSUBS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sobic.002G230100 | CA | 4823 | 1014 | 115 | 14 | 4 | 10 | No | No | 0 | 0 | 1 | 0 |
| Sobic.003G234200 | CA | 10440 | 1371 | 475 | 33 | 7 | 26 | No | No | 1 | 1 | 0 | 0 |
| Sobic.003G234400 | CA | 4749 | 615 | 138 | 13 | 3 | 10 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G234500 | CA | 2986 | 609 | 173 | 11 | 5 | 6 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G234600 | CA | 4750 | 771 | 210 | 18 | 10 | 8 | No | No | 0 | 0 | 0 | 0 |
| Sobic.007G166200 | NADP-MDH | 3354 | 1308 | 53 | 11 | 6 | 5 | No | No | 0 | 0 | 0 | 0 |
| Sobic.007G166300 | NADP-MDH | 3816 | 1290 | 108 | 12 | 3 | 9 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G036000 | NADP-ME | 6107 | 1941 | 111 | 11 | 4 | 7 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G036200 | NADP-ME | 5447 | 1911 | 141 | 12 | 3 | 9 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G280900 | NADP-ME | 5691 | 1782 | 175 | 22 | 13 | 9 | No | No | 1 | 1 | 0 | 0 |
| Sobic.003G292400 | NADP-ME | 4527 | 1782 | 95 | 22 | 8 | 14 | No | No | 10 | 2 | 0 | 0 |
| Sobic.009G069600 | NADP-ME | 3624 | 1713 | 118 | 34 | 10 | 24 | No | No | 3 | 1 | 0 | 0 |
| Sobic.002G167000 | PEPC | 5632 | 2904 | 41 | 11 | 6 | 5 | No | No | 0 | 0 | 0 | 0 |
| Sobic.003G100600 | PEPC | 8881 | 3117 | 371 | 43 | 9 | 34 | No | No | 0 | 0 | 21 | 2 |
| Sobic.003G301800 | PEPC | 7610 | 2901 | 138 | 19 | 3 | 17 | No | No | 0 | 0 | 0 | 0 |
| Sobic.004G106900 | PEPC | 6977 | 2883 | 146 | 34 | 5 | 29 | No | No | 0 | 0 | 7 | 0 |
| Sobic.007G106500 | PEPC | 5616 | 2895 | 64 | 12 | 8 | 4 | No | No | 1 | 1 | 0 | 0 |
| Sobic.010G160700 | PEPC | 6647 | 3087 | 193 | 28 | 9 | 19 | No | No | 0 | 0 | 2 | 0 |
| Sobic.004G219900 | PPCK | 1612 | 924 | 40 | 9 | 1 | 8 | No | No | 0 | 0 | 2 | 0 |
| Sobic.004G338000 | PPCK | 1749 | 855 | 37 | 9 | 4 | 4 | No | No | 0 | 0 | 0 | 0 |
| Sobic.006G148300 | PPCK | 1997 | 900 | 64 | 4 | 1 | 3 | No | No | 0 | 0 | 0 | 0 |
| Sobic.001G326900 | PPDK | 8494 | 2730 | 321 | 46 | 18 | 28 | No | Yes | 0 | 0 | 24 | 5 |
| Sobic.009G132900 | PPDK | 12748 | 2847 | 441 | 16 | 0 | 16 | No | No | 3 | 0 | 0 | 0 |
| Sobic.002G324400 | PPDK-RP | 2507 | 1290 | 79 | 22 | 8 | 14 | No | No | 0 | 0 | 3 | 0 |
| Sobic.002G324500 | PPDK-RP | 3072 | 1260 | 69 | 20 | 5 | 15 | No | No | 4 | 0 | 0 | 0 |
| Sobic.002G324700 | PPDK-RP | 4662 | 1587 | 222 | 28 | 19 | 9 | No | No | 1 | 1 | 2 | 2 |
| Sobic.005G042000 | RbcS | 1556 | 510 | 45 | 7 | 4 | 3 | No | No | 0 | 0 | 0 | 0 |
| GeneID | Enzyme | θπ–All | θπ-Cultivated | θπ-W&W | FST |
|---|---|---|---|---|---|
| Sobic.002G230100 | CA | 0.80 | 0.74 | 0.90 | 0.19 |
| Sobic.003G234200 | CA | 2.65 | 2.46 | 2.66 | 0.16 |
| Sobic.003G234400 | CA | 1.01 | 0.91 | 0.88 | 0.37 |
| Sobic.003G234500 | CA | 5.55 | 5.51 | 4.56 | 0.07 |
| Sobic.003G234600 | CA | 1.27 | 1.35 | 0.65 | 0.06 |
| Sobic.007G166200 | NADP-MDH | 0.18 | 0.21 | 0.13 | 0.07 |
| Sobic.007G166300 | NADP-MDH | 0.33 | 0.33 | 0.42 | 0.08 |
| Sobic.003G036000 | NADP-ME | 0.88 | 0.65 | 1.59 | 0.15 |
| Sobic.003G036200 | NADP-ME | 0.89 | 0.67 | 1.39 | 0.06 |
| Sobic.003G280900 | NADP-ME | 0.93 | 0.85 | 1.11 | 0.09 |
| Sobic.003G292400 | NADP-ME | 1.43 | 0.08 | 4.44 | 0.32 |
| Sobic.009G069600 | NADP-ME | 0.52 | 0.49 | 0.10 | 0.45 |
| Sobic.002G167000 | PEPC | 0.58 | 0.51 | 0.85 | 0.04 |
| Sobic.003G100600 | PEPC | 5.36 | 5.18 | 3.56 | 0.05 |
| Sobic.003G301800 | PEPC | 0.64 | 0.22 | 2.37 | 0.22 |
| Sobic.004G106900 | PEPC | 3.18 | 3.02 | 2.14 | 0.07 |
| Sobic.007G106500 | PEPC | 0.44 | 0.22 | 0.47 | 0.21 |
| Sobic.010G160700 | PEPC | 2.49 | 2.25 | 2.86 | 0.04 |
| Sobic.004G219900 | PPCK | 2.08 | 1.94 | 2.12 | 0.12 |
| Sobic.004G338000 | PPCK | 1.03 | 0.96 | 0.91 | 0.03 |
| Sobic.006G148300 | PPCK | 0.48 | 0.39 | 0.13 | 0.41 |
| Sobic.001G326900 | PPDK | 8.34 | 5.64 | 5.64 | 0.40 |
| Sobic.009G132900 | PPDK | 2.07 | 1.79 | 2.19 | 0.13 |
| Sobic.002G324400 | PPDK-RP | 5.04 | 3.82 | 4.55 | 0.41 |
| Sobic.002G324500 | PPDK-RP | 1.27 | 0.10 | 3.75 | 0.24 |
| Sobic.002G324700 | PPDK-RP | 2.58 | 2.50 | 3.51 | 0.05 |
| Sobic.005G042000 | rbcS | 4.32 | 3.41 | 5.72 | 0.12 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tao, Y.; George-Jaeggli, B.; Bouteillé-Pallas, M.; Tai, S.; Cruickshank, A.; Jordan, D.; Mace, E. Genetic Diversity of C4 Photosynthesis Pathway Genes in Sorghum bicolor (L.). Genes 2020, 11, 806. https://doi.org/10.3390/genes11070806
Tao Y, George-Jaeggli B, Bouteillé-Pallas M, Tai S, Cruickshank A, Jordan D, Mace E. Genetic Diversity of C4 Photosynthesis Pathway Genes in Sorghum bicolor (L.). Genes. 2020; 11(7):806. https://doi.org/10.3390/genes11070806
Chicago/Turabian StyleTao, Yongfu, Barbara George-Jaeggli, Marie Bouteillé-Pallas, Shuaishuai Tai, Alan Cruickshank, David Jordan, and Emma Mace. 2020. "Genetic Diversity of C4 Photosynthesis Pathway Genes in Sorghum bicolor (L.)" Genes 11, no. 7: 806. https://doi.org/10.3390/genes11070806
APA StyleTao, Y., George-Jaeggli, B., Bouteillé-Pallas, M., Tai, S., Cruickshank, A., Jordan, D., & Mace, E. (2020). Genetic Diversity of C4 Photosynthesis Pathway Genes in Sorghum bicolor (L.). Genes, 11(7), 806. https://doi.org/10.3390/genes11070806






