A Deletion in GDF7 is Associated with a Heritable Forebrain Commissural Malformation Concurrent with Ventriculomegaly and Interhemispheric Cysts in Cats
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Pedigree
2.2. DNA Array Genotyping
2.3. Genome-Wide Association Studies
2.4. Haplotype Analysis
2.5. Homozygosity Analysis
2.6. Whole Genome Sequencing
2.7. Variant Validation and Genotyping
3. Results
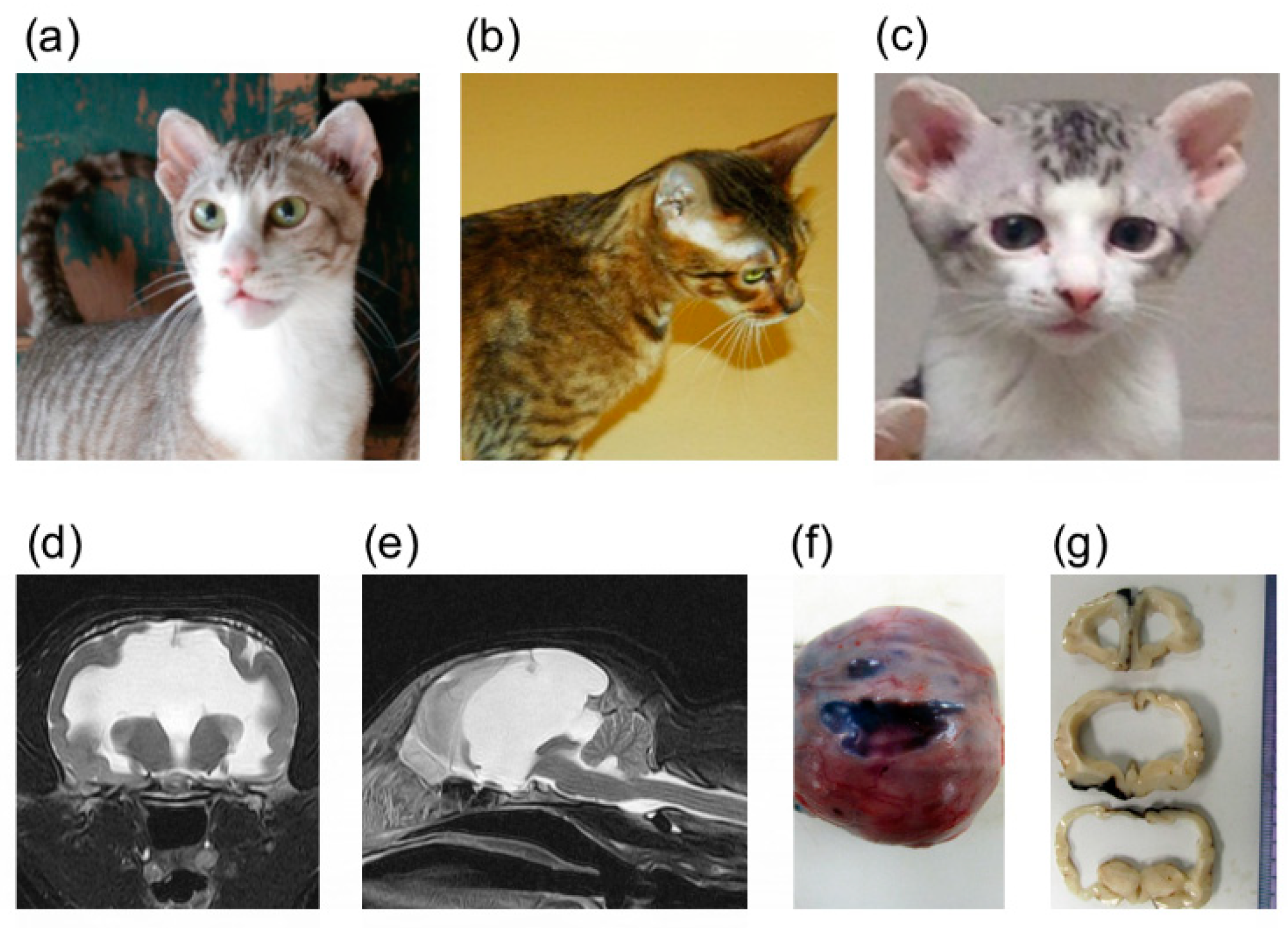
3.1. Pedigree and Genotyping
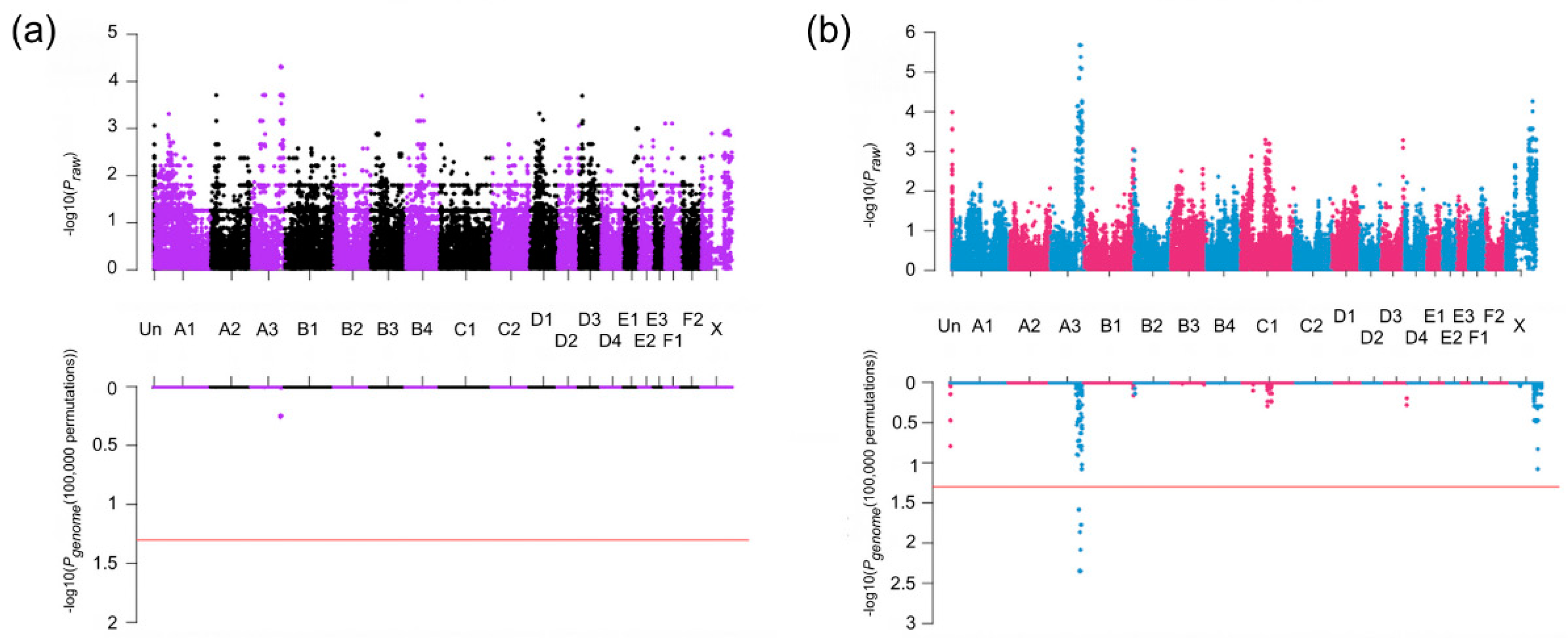
3.2. Association Studies
3.3. Haplotype Analysis
3.4. Homozygosity Analysis
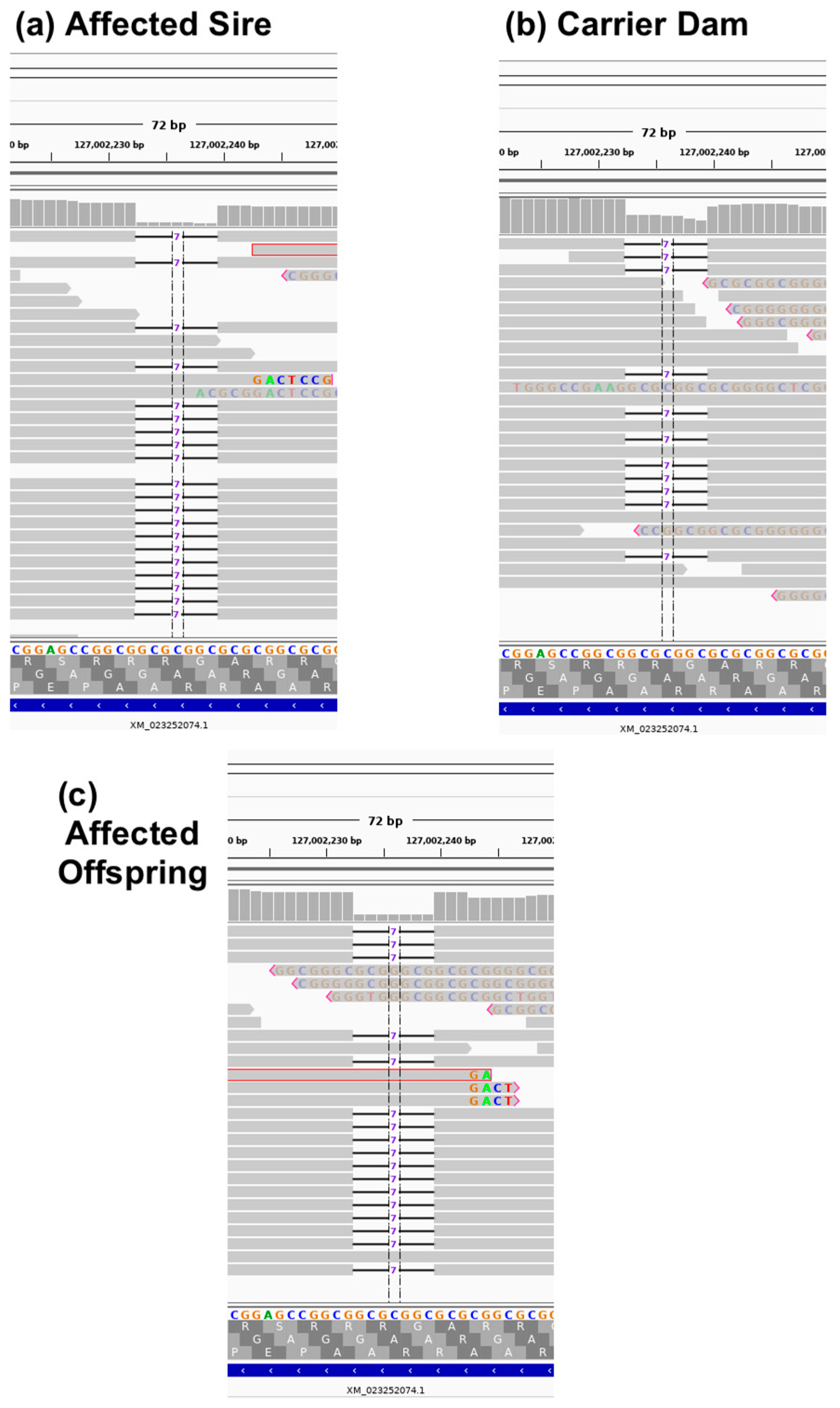
3.5. Whole Genome Sequencing
3.6. Variant Validation and Genotyping
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- MacKillop, E. Magnetic resonance imaging of intracranial malformations in dogs and cats. Vet. Radiol. Ultrasound. 2011, 52, S42–S51. [Google Scholar] [CrossRef] [PubMed]
- Jurney, C.; Haddad, J.; Crawford, N.; Miller, A.D.; Van Winkle, T.J.; Vite, C.H.; Sponenberg, P.; Inzana, K.D.; Cook, C.R.; Britt, L.; et al. Polymicrogyria in standard poodles. J. Vet. Intern. Med. 2009, 23, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, R.; Volk, H.; Smith, P.M.; Penderis, J.; Garosi, L.; MacKillop, E.; de Stefani, A.; Cherubini, G.; McConnell, J.F. Corpus callosal abnormalities in dogs. J. Vet. Intern. Med. 2014, 28, 1275–1279. [Google Scholar] [CrossRef] [PubMed]
- Bernardino, F.; Rentmeister, K.; Schmidt, M.J.; Bruehschwein, A.; Matiasek, K.; Matiasek, L.A.; Lauda, A.; Schoon, H.A.; Fischer, A. Inferior cerebellar hypoplasia resembling a Dandy-Walker-like malformation in purebred Eurasier dogs with familial non-progressive ataxia: A retrospective and prospective clinical cohort study. PLoS ONE 2015, 10, e0117670. [Google Scholar] [CrossRef] [PubMed]
- Gerber, M.; Fischer, A.; Jagannathan, V.; Drogemuller, M.; Drogemuller, C.; Schmidt, M.J.; Bernardino, F.; Manz, E.; Matiasek, K.; Rentmeister, K.; et al. A deletion in the VLDLR gene in Eurasier dogs with cerebellar hypoplasia resembling a Dandy-Walker-like malformation (DWLM). PLoS ONE 2015, 10, e0108917. [Google Scholar] [CrossRef]
- Estey, C.M. Congenital Hydrocephalus. Vet. Clin. North. Am. Small Anim. Pract. 2016, 46, 217–229. [Google Scholar] [CrossRef]
- Selby, L.A.; Hayes, H.M., Jr.; Becker, S.V. Epizootiologic features of canine hydrocephalus. Am. J. Vet. Res. 1979, 40, 411–413. [Google Scholar]
- Knowler, S.P.; Galea, G.L.; Rusbridge, C. Morphogenesis of Canine Chiari Malformation and Secondary Syringomyelia: Disorders of Cerebrospinal Fluid Circulation. Front. Vet. Sci. 2018, 5, 171. [Google Scholar] [CrossRef]
- Schmidt, M.J.; Kampschulte, M.; Enderlein, S.; Gorgas, D.; Lang, J.; Ludewig, E.; Fischer, A.; Meyer-Lindenberg, A.; Schaubmar, A.R.; Failing, K.; et al. The Relationship between Brachycephalic Head Features in Modern Persian Cats and Dysmorphologies of the Skull and Internal Hydrocephalus. J. Vet. Intern. Med. 2017, 31, 1487–1501. [Google Scholar] [CrossRef]
- Schlueter, C.; Budras, K.D.; Ludewig, E.; Mayrhofer, E.; Koenig, H.E.; Walter, A.; Oechtering, G.U. Brachycephalic feline noses: CT and anatomical study of the relationship between head conformation and the nasolacrimal drainage system. J. Feline Med. Surg. 2009, 11, 891–900. [Google Scholar] [CrossRef]
- Farnworth, M.J.; Chen, R.; Packer, R.M.; Caney, S.M.; Gunn-Moore, D.A. Flat feline faces: Is brachycephaly associated with respiratory abnormalities in the domestic cat (Felis catus)? PLoS ONE 2016, 11, e0161777. [Google Scholar] [CrossRef] [PubMed]
- Mestrinho, L.A.; Louro, J.M.; Gordo, I.S.; Niza, M.; Requicha, J.F.; Force, J.G.; Gawor, J.P. Oral and dental anomalies in purebred, brachycephalic Persian and exotic cats. J. Am. Vet. Med. Assoc. 2018, 253, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Sponenberg, D.P.; Graf-Webster, E. Hereditary meningoencephalocele in Burmese cats. J. Hered. 1986, 77, 60. [Google Scholar] [CrossRef] [PubMed]
- Lyons, L.A.; Erdman, C.A.; Grahn, R.A.; Hamilton, M.J.; Carter, M.J.; Helps, C.R.; Alhaddad, H.; Gandolfi, B. Aristaless-Like Homeobox protein 1 (ALX1) variant associated with craniofacial structure and frontonasal dysplasia in Burmese cats. Dev. Biol. 2016, 409, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Lowrie, M.; Wessmann, A.; Gunn-Moore, D.; Penderis, J. Quadrigeminal cyst management by cystoperitoneal shunt in a 4-year-old Persian cat. J. Feline Med. Surg. 2009, 11, 711–713. [Google Scholar] [CrossRef]
- Reed, S.; Cho, D.Y.; Paulsen, D. Quadrigeminal arachnoid cysts in a kitten and a dog. J. Vet. Diagn. Invest. 2009, 21, 707–710. [Google Scholar] [CrossRef]
- Herrmann, A.; Hecht, W.; Herden, C. Lissencephaly and microencephaly combined with hypoplasia of corpus callosum and cerebellum in a domestic cat. Tierarztl Prax Ausg K Kleintiere Heimtiere 2011, 39, 116–120. [Google Scholar]
- Shimbo, G.; Tagawa, M.; Yanagawa, M.; Miyahara, K. MRI of lobar holoprosencephaly in a cat with hypodipsic hypernatraemia. JFMS Open Rep. 2018, 4. [Google Scholar] [CrossRef]
- Boccanera, C.; Stabile, F.; Corvi, R.; Mariscoli, M.; Mandara, M.T. Hydrocephalus, supratentorial diverticulum and agenesis of the interthalamic adhesion and corpus callosum in a cat: MRI findings, treatment and follow-up. Vet. Record Case Rep. 2018, 6, e000416. [Google Scholar] [CrossRef]
- Woerde, D.J.; Hoffmann, K.L.; Brown, N.L. Frontoethmoidal encephalocele in a cat. JFMS Open Rep. 2018, 4. [Google Scholar] [CrossRef]
- Keating, M.K.; Sturges, B.K.; Siso, S.; Wisner, E.R.; Creighton, E.K.; Lyons, L.A. Characterization of an Inherited Neurologic Syndrome in Toyger Cats with Forebrain Commissural Malformations, Ventriculomegaly and Interhemispheric Cysts. J. Vet. Intern. Med. 2016, 30, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Alhaddad, H.; Abdi, M.; Bach, L.H.; Creighton, E.K.; Davis, B.W.; Decker, J.E.; Dodman, N.H.; Ginns, E.I.; Grahn, J.C.; et al. Applications and efficiencies of the first cat 63K DNA array. Sci Rep. 2018, 8, 7024. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, M.J.; Amigues, Y.; Blasi, M.; Broad, T.E.; Cherbonnel, C.; Cho, G.J.; Corley, S.; Daftari, P.; Delattre, D.R.; Dileanis, S.; et al. An international parentage and identification panel for the domestic cat (Felis catus). Anim Genet. 2007, 38, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Jones, O.R.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef]
- Wang, J. A simulation module in the computer program COLONY for sibship and parentage analysis. Mol. Ecol. Resour. 2013, 13, 734–739. [Google Scholar] [CrossRef]
- Mullikin, J.C.; Hansen, N.F.; Shen, L.; Ebling, H.; Donahue, W.F.; Tao, W.; Saranga, D.J.; Brand, A.; Rubenfield, M.J.; Young, A.C.; et al. Light whole genome sequence for SNP discovery across domestic cat breeds. BMC Genom. 2010, 11, 406. [Google Scholar] [CrossRef][Green Version]
- Buckley, R.M.; Davis, B.W.; Brashear, W.A.; Farias, F.H.G.; Kuroki, K.; Graves, T.; Hillier, L.W.; Kremitzki, M.; Li, G.; Middleton, R.; et al. A new domestic cat genome assembly based on long sequence reads empowers feline genomic medicine and identifies a novel gene for dwarfism. bioRxiv 2020. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Spielman, R.S.; Ewens, W.J. A sibship test for linkage in the presence of association: The sib transmission/disequilibrium test. Am. J. Hum. Genet. 1998, 62, 450–458. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Lyons, L.A.; Creighton, E.K.; Alhaddad, H.; Beale, H.C.; Grahn, R.A.; Rah, H.; Maggs, D.J.; Helps, C.R.; Gandolfi, B. Whole genome sequencing in cats, identifies new models for blindness in AIPL1 and somite segmentation in HES7. BMC Genom. 2016, 17, 265. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Thorvaldsdottir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Toonen, R.J.; Hughes, S. Increased throughput for fragment analysis on an ABI PRISM 377 automated sequencer using a membrane comb and STRand software. Biotechniques 2001, 31, 1320–1324. [Google Scholar] [PubMed]
- Buckley, R.M.; Gandolfi, B.; Creighton, E.K.; Pyne, C.A.; Leroy, M.L.; Senter, D.A.; Bouhan, D.M.; Gobble, J.R.; Abitbol, M.; Lyons, L.A.; et al. Werewolf, there wolf: Variants in Hairless associated wih hypotrichia and roaning in the lykoi cat breed. bioRxiv 2020. [Google Scholar] [CrossRef]
- Ancot, F.; Lemay, P.; Knowler, S.P.; Kennedy, K.; Griffiths, S.; Cherubini, G.B.; Sykes, J.; Mandigers, P.J.J.; Rouleau, G.A.; Rusbridge, C.; et al. A genome-wide association study identifies candidate loci associated to syringomyelia secondary to Chiari-like malformation in Cavalier King Charles Spaniels. BMC Genet. 2018, 19, 16. [Google Scholar] [CrossRef] [PubMed]
- Marchant, T.W.; Dietschi, E.; Rytz, U.; Schawalder, P.; Jagannathan, V.; Hadji Rasouliha, S.; Gurtner, C.; Waldvogel, A.S.; Harrington, R.S.; Drogemuller, M.; et al. An ADAMTS3 missense variant is associated with Norwich Terrier upper airway syndrome. PLoS Genet. 2019, 15, e1008102. [Google Scholar] [CrossRef] [PubMed]
- Aberdein, D.; Munday, J.S.; Gandolfi, B.; Dittmer, K.E.; Malik, R.; Garrick, D.J.; Lyons, L.A.; Lives, C. A FAS-ligand variant associated with autoimmune lymphoproliferative syndrome in cats. Mamm Genome 2017, 28, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, B.; Grahn, R.A.; Creighton, E.K.; Williams, D.C.; Dickinson, P.J.; Sturges, B.K.; Guo, L.T.; Shelton, G.D.; Leegwater, P.A.; Longeri, M.; et al. COLQ variant associated with Devon Rex and Sphynx feline hereditary myopathy. Anim Genet. 2015, 46, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Mauler, D.A.; Gandolfi, B.; Reinero, C.R.; O’Brien, D.P.; Spooner, J.L.; Lyons, L.A.; 99 Lives Consortium. Precision Medicine in Cats: Novel Niemann-Pick Type C1 Diagnosed by Whole-Genome Sequencing. J. Vet. Intern. Med. 2017, 31, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Oh, A.; Pearce, J.W.; Gandolfi, B.; Creighton, E.K.; Suedmeyer, W.K.; Selig, M.; Bosiack, A.P.; Castaner, L.J.; Whiting, R.E.; Belknap, E.B.; et al. Early-Onset Progressive Retinal Atrophy Associated with an IQCB1 Variant in African Black-Footed Cats (Felis nigripes). Sci. Rep. 2017, 7, 43918. [Google Scholar] [CrossRef] [PubMed]
- Ontiveros, E.S.; Ueda, Y.; Harris, S.P.; Stern, J.A.; Lives, C. Precision medicine validation: Identifying the MYBPC3 A31P variant with whole-genome sequencing in two Maine Coon cats with hypertrophic cardiomyopathy. J. Feline Med. Surg. 2019, 21, 1086–1093. [Google Scholar] [CrossRef]
- Jaffey, J.A.; Reading, N.S.; Giger, U.; Abdulmalik, O.; Buckley, R.M.; Johnstone, S.; Lyons, L.A.; Lives Cat Genome, C. Clinical, metabolic, and genetic characterization of hereditary methemoglobinemia caused by cytochrome b5 reductase deficiency in cats. J. Vet. Intern. Med. 2019, 33, 2725–2731. [Google Scholar] [CrossRef]
- Buckley, R.M.; Grahn, R.A.; Gandolfi, B.; Herrick, J.R.; Kittleson, M.D.; Bateman, H.L.; Newsom, J.; Swanson, W.F.; Prieur, D.J.; Lyons, L.A. Assisted reproduction mediated resurrection of a feline model for Chediak-Higashi syndrome caused by a large duplication in LYST. Sci. Rep. 2020, 10, 64. [Google Scholar] [CrossRef]
- Summers, A.D.; Reefhuis, J.; Taliano, J.; Rasmussen, S.A. Nongenetic risk factors for holoprosencephaly: An updated review of the epidemiologic literature. Am. J. Med. Genet. C Semin Med. Genet. 2018, 178, 151–164. [Google Scholar] [CrossRef]
- Dubourg, C.; Kim, A.; Watrin, E.; de Tayrac, M.; Odent, S.; David, V.; Dupe, V. Recent advances in understanding inheritance of holoprosencephaly. Am. J. Med. Genet. C Semin Med. Genet. 2018, 178, 258–269. [Google Scholar] [CrossRef]
- Kruszka, P.; Martinez, A.F.; Muenke, M. Molecular testing in holoprosencephaly. Am. J. Med. Genet. C Semin Med. Genet. 2018, 178, 187–193. [Google Scholar] [CrossRef]
- Roessler, E.; Hu, P.; Muenke, M. Holoprosencephaly in the genomics era. Am. J. Med. Genet. C Semin Med. Genet. 2018, 178, 165–174. [Google Scholar] [CrossRef]
- Lee, K.J.; Mendelsohn, M.; Jessell, T.M. Neuronal patterning by BMPs: A requirement for GDF7 in the generation of a discrete class of commissural interneurons in the mouse spinal cord. Genes Dev. 1998, 12, 3394–3407. [Google Scholar] [CrossRef] [PubMed]
- Heussler, H.S.; Suri, M.; Young, I.D.; Muenke, M. Extreme variability of expression of a Sonic Hedgehog mutation: Attention difficulties and holoprosencephaly. Arch. Dis Child. 2002, 86, 293–296. [Google Scholar] [CrossRef] [PubMed]
- Marini, M.; Cusano, R.; De Biasio, P.; Caroli, F.; Lerone, M.; Silengo, M.; Ravazzolo, R.; Seri, M.; Camera, G. Previously undescribed nonsense mutation in SHH caused autosomal dominant holoprosencephaly with wide intrafamilial variability. Am. J. Med. Genet. A 2003, 117A, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Hehr, U.; Gross, C.; Diebold, U.; Wahl, D.; Beudt, U.; Heidemann, P.; Hehr, A.; Mueller, D. Wide phenotypic variability in families with holoprosencephaly and a sonic hedgehog mutation. Eur. J. Pediatr. 2004, 163, 347–352. [Google Scholar] [CrossRef]
- Hogan, B.L. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 1996, 10, 1580–1594. [Google Scholar] [CrossRef]
- Mehler, M.F.; Mabie, P.C.; Zhang, D.; Kessler, J.A. Bone morphogenetic proteins in the nervous system. Trends Neurosci. 1997, 20, 309–317. [Google Scholar] [CrossRef]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis 2014, 1, 87–105. [Google Scholar] [CrossRef]
- Mikic, B.; Bierwert, L.; Tsou, D. Achilles tendon characterization in GDF-7 deficient mice. J. Orthop Res. 2006, 24, 831–841. [Google Scholar] [CrossRef]
- Mikic, B.; Ferreira, M.P.; Battaglia, T.C.; Hunziker, E.B. Accelerated hypertrophic chondrocyte kinetics in GDF-7 deficient murine tibial growth plates. J. Orthop Res. 2008, 26, 986–990. [Google Scholar] [CrossRef]
- Settle, S.; Marker, P.; Gurley, K.; Sinha, A.; Thacker, A.; Wang, Y.; Higgins, K.; Cunha, G.; Kingsley, D.M. The BMP family member Gdf7 is required for seminal vesicle growth, branching morphogenesis, and cytodifferentiation. Dev. Biol. 2001, 234, 138–150. [Google Scholar] [CrossRef][Green Version]
- Maloul, A.; Rossmeier, K.; Mikic, B.; Pogue, V.; Battaglia, T. Geometric and material contributions to whole bone structural behavior in GDF-7-deficient mice. Connect. Tissue Res. 2006, 47, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Palles, C.; Chegwidden, L.; Li, X.; Findlay, J.M.; Farnham, G.; Castro Giner, F.; Peppelenbosch, M.P.; Kovac, M.; Adams, C.L.; Prenen, H.; et al. Polymorphisms near TBX5 and GDF7 are associated with increased risk for Barrett’s esophagus. Gastroenterology 2015, 148, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Becker, J.; May, A.; Gerges, C.; Anders, M.; Schmidt, C.; Veits, L.; Noder, T.; Mayershofer, R.; Kreuser, N.; Manner, H.; et al. The Barrett-associated variants at GDF7 and TBX5 also increase esophageal adenocarcinoma risk. Cancer Med. 2016, 5, 888–891. [Google Scholar] [CrossRef] [PubMed]
| SNP * | Chr | Position † | Sib-TDT | Case-Control (Initial) | Case-Control (2nd) |
|---|---|---|---|---|---|
| chrA3.164724433 | A3 | 123353491 | >0.6 | 0.0001 | 0.0045 |
| chrA3.164567500 | A3 | 122318611 | >0.6 | 0.0001 | 0.0045 |
| chrA3.164340161 | A3 | 122513677 | >0.6 | 0.0001 | 0.0045 |
| chrA3.164113252 | A3 | 122698750 | >0.6 | 0.0001 | 0.0045 |
| chrA3.163320257 | A3 | 123353491 | 0.5639 | 0.0001 | 0.0045 |
| chrA3.162970354 | A3 | 123644765 | >0.6 | 0.0001 | 0.0045 |
| chrA3.162343840 | A3 | 124176474 | >0.6 | 0.0004 | 0.0045 |
| chrA3.158624618 | A3 | 127189752 | >0.6 | 0.0001 | 0.0045 |
| chrA3.159621145 | A3 | 126377299 | 0.5639 | 0.0001 | 0.0082 |
| chrA3.162413594 | A3 | 124100380 | >0.6 | 0.0014 | 0.0137 |
| chrA3.156826206 | A3 | 128667138 | 0.5639 | 0.0001 | 0.0169 |
| chrA3.156620632 | A3 | 128837125 | >0.6 | 0.0001 | 0.0169 |
| chrA3.155936886 | A3 | 129372537 | >0.6 | 0.0004 | 0.0169 |
| chrA3.168960567 | A3 | 119105247 | >0.6 | 0.0128 | 0.0261 |
| chrA3.168031908 | A3 | 119810207 | >0.6 | 0.0264 | 0.0261 |
| chrA3.167492986 | A3 | 120088757 | >0.6 | 0.0061 | 0.0261 |
| chrA3.167322483 | A3 | 120215597 | >0.6 | 0.0292 | 0.0261 |
| chrA3.162621987 | A3 | 123934341 | 0.5521 | 0.0014 | >0.05 |
| chrA3.163679766 | A3 | 123055238 | 0.5639 | 0.0004 | >0.05 |
| chrA3.161984351 | A3 | 124475589 | 0.5639 | 0.0002 | >0.05 |
| chrA3.161943004 | A3 | 124509146 | 0.5639 | 0.0002 | >0.05 |
| chrA3.161399869 | A3 | 124945294 | 0.5639 | 0.0020 | >0.05 |
| chrA3.160673309 | A3 | 125511595 | 0.5639 | 0.0002 | >0.05 |
| Chr:Pos | Ref/Alt | No. Het * | No. Homo | Gene Name | Sequence Ontology | Effect | HGVS c. (Clinically Relevant) |
|---|---|---|---|---|---|---|---|
| A3:127002233 | GCGCGGC/- | 2 | 1 | GDF7 | frameshift | LoF | ENSFCAT00000063603: c.221_227delGCCGCGC |
| C1:96095693 | C/T | 2 | 1 | intergenic | Other | ||
| C1:96839645 | C/T | 2 | 1 | intergenic | Other | ||
| D2:33368378 | C/A | 2 | 1 | intergenic | Other | ||
| C1:106990675 | C/A | 1 | 2 | intergenic | Other | ||
| E1:9973078 | C/T | 1 | 2 | SPECC1 | Intron | Other | ENSFCAT00000005195: c.2850 + 13572G > A |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Creighton, E.K.; Buckley, R.M.; Lyons, L.A.; 99 Lives Consortium. A Deletion in GDF7 is Associated with a Heritable Forebrain Commissural Malformation Concurrent with Ventriculomegaly and Interhemispheric Cysts in Cats. Genes 2020, 11, 672. https://doi.org/10.3390/genes11060672
Yu Y, Creighton EK, Buckley RM, Lyons LA, 99 Lives Consortium. A Deletion in GDF7 is Associated with a Heritable Forebrain Commissural Malformation Concurrent with Ventriculomegaly and Interhemispheric Cysts in Cats. Genes. 2020; 11(6):672. https://doi.org/10.3390/genes11060672
Chicago/Turabian StyleYu, Yoshihiko, Erica K. Creighton, Reuben M. Buckley, Leslie A. Lyons, and 99 Lives Consortium. 2020. "A Deletion in GDF7 is Associated with a Heritable Forebrain Commissural Malformation Concurrent with Ventriculomegaly and Interhemispheric Cysts in Cats" Genes 11, no. 6: 672. https://doi.org/10.3390/genes11060672
APA StyleYu, Y., Creighton, E. K., Buckley, R. M., Lyons, L. A., & 99 Lives Consortium. (2020). A Deletion in GDF7 is Associated with a Heritable Forebrain Commissural Malformation Concurrent with Ventriculomegaly and Interhemispheric Cysts in Cats. Genes, 11(6), 672. https://doi.org/10.3390/genes11060672




