Structural and Functional Heat Stress Responses of Chloroplasts of Arabidopsis thaliana
Abstract
1. Introduction
2. Materials and Methods
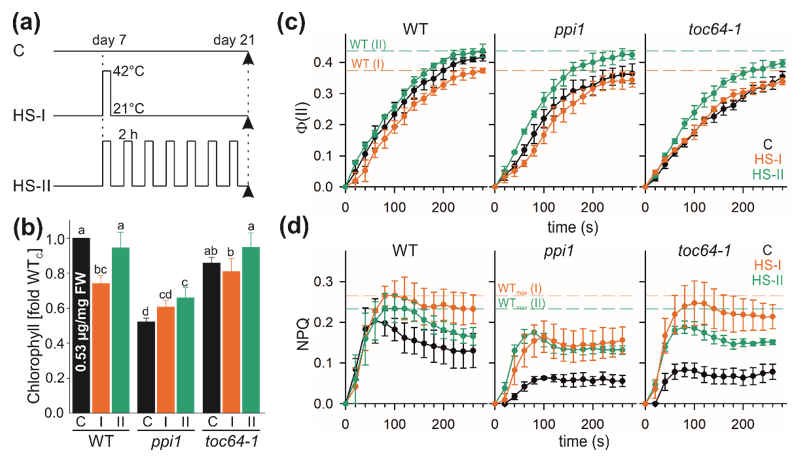
2.1. Plant Material and Stress Treatment
2.2. Measurement of Chlorophyll and Photosynthetic Activity
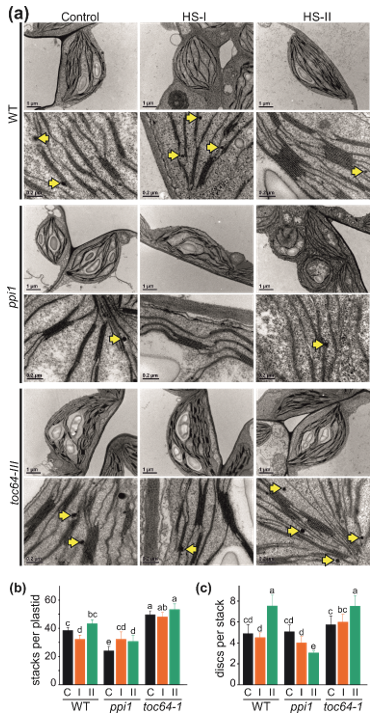
2.3. Analysis of Chloroplast Ultrastructure by Transmission Electron Microscopy
2.4. Isolation of Chloroplasts
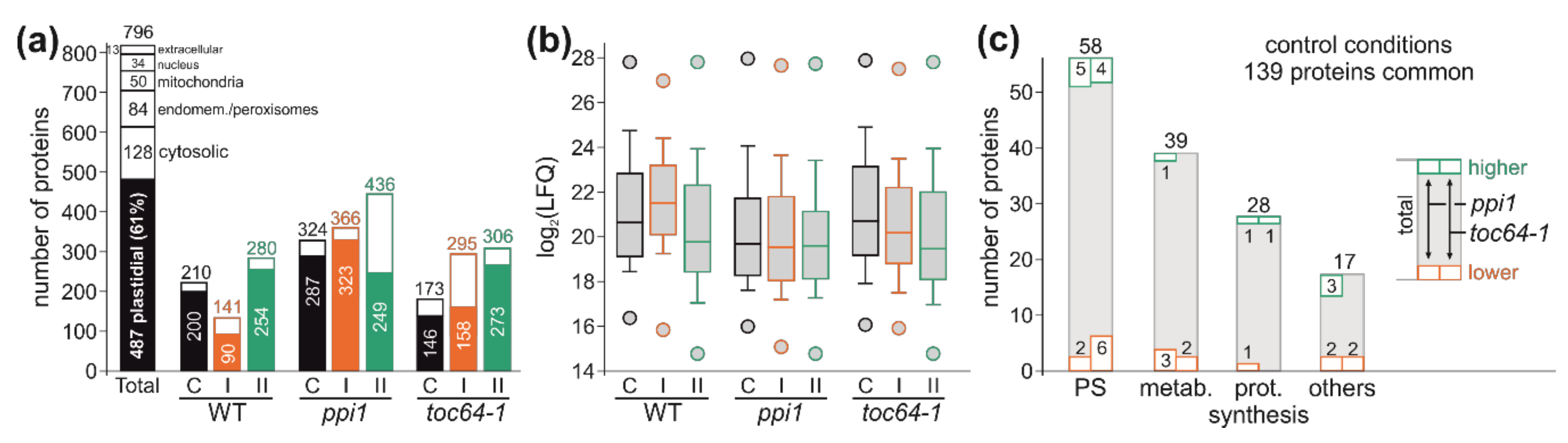
2.5. Shotgun Proteomics (GEL-LC-Orbitrap-MS)
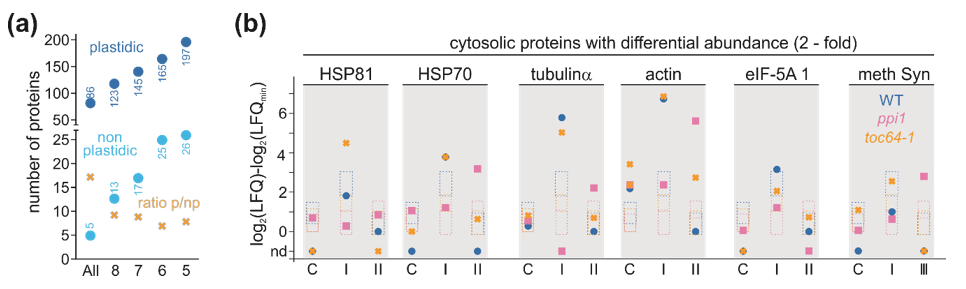
2.6. Quantitative Analysis of the Proteome
3. Results
3.1. Heat Stress Alters Chlorophyll Content and Photosynthetic Performance
3.2. The Impact of Heat Stress on Chloroplasts Structure
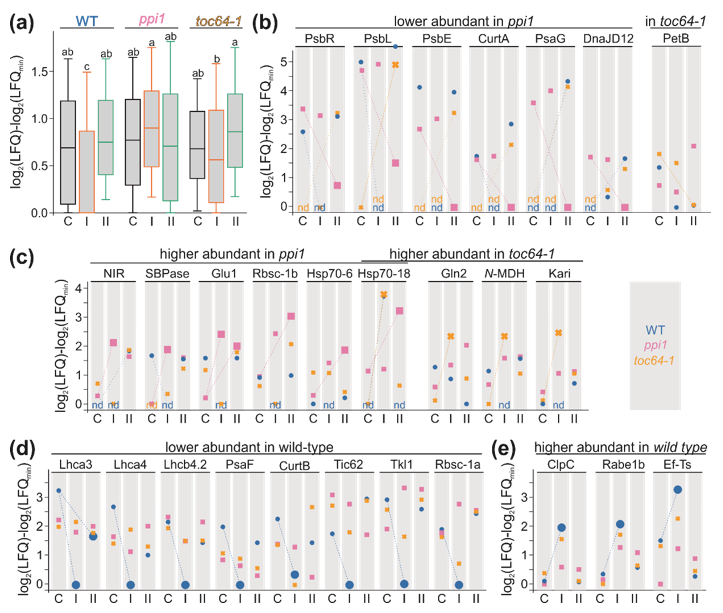
3.3. The Chloroplast Proteome of Wild-Type, ppi1, and toc64-1
3.4. Proteome Changes in Response to Heat Stress
4. Discussion
4.1. The Change of Chloroplast Proteome and Performance in Mutants of the TOC Translocon
4.2. The Response of Chloroplasts to Temperature Elevations
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Williams, B.; Verchot, J.; Dickman, M.B. When supply does not meet demand-ER stress and plant programmed cell death. Front. Plant Sci. 2014, 5, 211. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Hipp, M.S.; Kasturi, P.; Hartl, F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019, 20, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Morán Luengo, T.; Mayer, M.P.; Rüdiger, S.G.D. The Hsp70-Hsp90 Chaperone Cascade in Protein Folding. Trends Cell Biol. 2019, 29, 164–177. [Google Scholar] [CrossRef] [PubMed]
- Schleiff, E.; Becker, T. Common ground for protein translocation: Access control for mitochondria and chloroplasts. Nat. Rev. Mol. Cell Biol. 2011, 12, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Kunze, M.; Berger, J. The similarity between N-terminal targeting signals for protein import into different organelles and its evolutionary relevance. Front. Physiol. 2015, 6, 259. [Google Scholar] [CrossRef] [PubMed]
- Mikhaleva, S.; Lemke, E.A. Beyond the Transport Function of Import Receptors: What’s All the FUS about? Cell 2018, 173, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085. [Google Scholar] [CrossRef]
- Paul, P.; Simm, S.; Blaumeiser, A.; Scharf, K.-D.; Fragkostefanakis, S.; Mirus, O.; Schleiff, E. The protein translocation systems in plants—Composition and variability on the example of Solanum lycopersicum. BMC Genom. 2013, 14, 189. [Google Scholar] [CrossRef]
- Paul, P.; Simm, S.; Mirus, O.; Scharf, K.-D.; Fragkostefanakis, S.; Schleiff, E. The complexity of vesicle transport factors in plants examined by orthology search. PLoS ONE 2014, 9, e97745. [Google Scholar] [CrossRef]
- Shanmugabalaji, V.; Kessler, F. CHLORAD: Eradicating Translocon Components from the Outer Membrane of the Chloroplast. Mol. Plant. 2019, 12, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Oreb, M.; Tews, I.; Schleiff, E. Policing Tic ’n’ Toc, the doorway to chloroplasts. Trends Cell Biol. 2008, 18, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R. Protein Quality Control in the Endoplasmic Reticulum of Plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef] [PubMed]
- Vögtle, F.-N.; Meisinger, C. Sensing mitochondrial homeostasis: The protein import machinery takes control. Dev. Cell 2012, 23, 234–236. [Google Scholar] [CrossRef]
- Leister, D. Chloroplast research in the genomic age. Trends Genet. 2003, 19, 47–56. [Google Scholar] [CrossRef]
- Kosová, K.; Vítámvás, P.; Prášil, I.T. Proteomics of stress responses in wheat and barley-search for potential protein markers of stress tolerance. Front. Plant Sci. 2014, 5, 711. [Google Scholar] [CrossRef]
- Bokszczanin, K.L. Solanaceae Pollen Thermotolerance Initial Training Network (SPOT-ITN) Consortium; Fragkostefanakis, S. Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant Sci. 2013, 4, 315. [Google Scholar] [CrossRef]
- Bäurle, I. Plant Heat Adaptation: Priming in response to heat stress. F1000 Res. 2016, 5, F1000. [Google Scholar] [CrossRef]
- Hossain, Z.; Nouri, M.Z.; Komatsu, S. Plant cell organelle proteomics in response to abiotic stress. J. Proteome Res. 2012, 11, 37–48. [Google Scholar] [CrossRef]
- Rurek, M. Plant mitochondria under a variety of temperature stress conditions. Mitochondrion 2014, 19, 289–294. [Google Scholar] [CrossRef]
- Lee, D.-G.; Ahsan, N.; Lee, S.-H.; Kang, K.Y.; Bahk, J.D.; Lee, I.-J.; Lee, B.-H. A proteomic approach in analyzing heat-responsive proteins in rice leaves. Proteomics 2007, 7, 3369–3383. [Google Scholar] [CrossRef]
- Ferreira, S.; Hjernø, K.; Larsen, M.; Wingsle, G.; Larsen, P.; Fey, S.; Roepstorff, P.; Salomé Pais, M. Proteome profiling of Populus euphratica Oliv. upon heat stress. Ann. Bot. 2006, 98, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Majoul, T.; Bancel, E.; Triboï, E.; Ben Hamida, J.; Branlard, G. Proteomic analysis of the effect of heat stress on hexaploid wheat grain: Characterization of heat-responsive proteins from total endosperm. Proteomics 2003, 3, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Laino, P.; Shelton, D.; Finnie, C.; De Leonardis, A.M.; Mastrangelo, A.M.; Svensson, B.; Lafiandra, D.; Masci, S. Comparative proteome analysis of metabolic proteins from seeds of durum wheat (cv. Svevo) subjected to heat stress. Proteomics 2010, 10, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Huang, B. Root proteomic responses to heat stress in two Agrostis grass species contrasting in heat tolerance. J. Exp. Bot. 2008, 59, 4183–4194. [Google Scholar] [CrossRef]
- Taylor, N.L.; Tan, Y.-F.; Jacoby, R.P.; Millar, A.H. Abiotic environmental stress induced changes in the Arabidopsis thaliana chloroplast, mitochondria and peroxisome proteomes. J. Proteom. 2009, 72, 367–378. [Google Scholar] [CrossRef]
- Soll, J.; Schleiff, E. Protein import into chloroplasts. Nat. Rev. Mol. Cell Biol. 2004, 5, 198–208. [Google Scholar] [CrossRef]
- Yamori, W.; Hikosaka, K.; Way, D.A. Temperature response of photosynthesis in C3, C4, and CAM plants: Temperature acclimation and temperature adaptation. Photosynth. Res. 2014, 119, 101–117. [Google Scholar] [CrossRef]
- Sun, A.Z.; Guo, F.Q. Chloroplast Retrograde Regulation of Heat Stress Responses in Plants. Front. Plant Sci. 2016, 7, 398. [Google Scholar] [CrossRef]
- Wang, Q.L.; Chen, J.H.; He, N.Y.; Guo, F.Q. Metabolic Reprogramming in Chloroplasts under Heat Stress in Plants. Int. J. Mol. Sci. 2018, 19, E849. [Google Scholar] [CrossRef]
- Sharkey, T.D. Effects of moderate heat stress on photosynthesis: Importance of thylakoid reactions, rubisco deactivation, reactive oxygen species, and thermotolerance provided by isoprene. Plant Cell Environ. 2005, 28, 269–277. [Google Scholar] [CrossRef]
- Allakhverdiev, S.I.; Kreslavski, V.D.; Klimov, V.V.; Los, D.A.; Carpentier, R.; Mohanty, P. Heat stress: An overview of molecular responses in photosynthesis. Photosynth. Res. 2008, 98, 541–550. [Google Scholar] [CrossRef]
- Law, R.D.; Crafts-Brandner, S.J. Inhibition and acclimation of photosynthesis to heat stress is closely correlated with activation of ribulose-1,5-bisphosphate Carboxylase/Oxygenase. Plant Physiol. 1999, 120, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Semenova, G. Structural reorganization of thylakoid systems in response to heat treatment. Photosynthetica 2004, 42, 521–527. [Google Scholar] [CrossRef]
- Vani, B.; Pardha Saradhi, P.; Mohanty, P. Alteration in chloroplast structure and thylakoid membrane composition due to in vivo heat treatment of rice seedlings: Correlation with the functional changes. J. Plant Physiol. 2001, 158, 583–592. [Google Scholar] [CrossRef]
- Gounaris, K.; Brain, A.R.R.; Quinn, P.J.; Williams, W.P. Structural reorganisation of chloroplast thylakoid membranes in response to heat-stress. Biochim. Biophys. Acta 1984, 766, 198–208. [Google Scholar] [CrossRef]
- Dutta, S.; Mohanty, S.; Tripathy, B.C. Role of temperature stress on chloroplast biogenesis and protein import in pea. Plant Physiol. 2009, 150, 1050–1561. [Google Scholar] [CrossRef]
- Heckathorn, S.A.; Downs, C.A.; Coleman, J.S. Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol during Severe Heat StressNuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol during Severe Heat Stress. Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol During Severe Heat Stress Nuclear-Encoded Chloroplast Proteins Accumulate in the Cytosol During Severe Heat StressInt. J. Plant Sci. 1998, 159, 39–45. [Google Scholar]
- Flores-Pérez, Ú.; Jarvis, P. Molecular chaperone involvement in chloroplast protein import. Biochim. Biophys. Acta. 2013, 1833, 332–340. [Google Scholar] [CrossRef]
- Sohrt, K.; Soll, J. Toc64, a new component of the protein translocon of chloroplasts. J. Cell Biol. 2000, 148, 1213–1221. [Google Scholar] [CrossRef]
- Qbadou, S.; Becker, T.; Mirus, O.; Tews, I.; Soll, J.; Schleiff, E. The molecular chaperone Hsp90 delivers precursor proteins to the chloroplast import receptor Toc64. EMBO J. 2006, 25, 1836–1847. [Google Scholar] [CrossRef]
- Qbadou, S.; Becker, T.; Bionda, T.; Reger, K.; Ruprecht, M.; Soll, J.; Schleiff, E. Toc64—A preprotein-receptor at the outer membrane with bipartide function. J. Mol. Biol. 2007, 367, 1330–1346. [Google Scholar] [CrossRef] [PubMed]
- Sommer, M.; Rudolf, M.; Tillmann, B.; Tripp, J.; Sommer, M.S.; Schleiff, E. Toc33 and Toc64-III cooperate in precursor protein import into the chloroplasts of Arabidopsis thaliana. Plant Cell Environ. 2013, 36, 970–983. [Google Scholar] [CrossRef] [PubMed]
- Rosenbaum Hofmann, N.; Theg, S.M. Toc64 is not required for import of proteins into chloroplasts in the moss Physcomitrella patens. Plant J. 2005, 43, 675–687. [Google Scholar] [CrossRef]
- Aronsson, H.; Boij, P.; Patel, R.; Wardle, A.; Töpel, M.; Jarvis, P. Toc64/OEP64 is not essential for the efficient import of proteins into chloroplasts in Arabidopsis thaliana. Plant J. 2007, 52, 53–68. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Fragkostefanakis, S.; Röth, S.; Schleiff, E.; Scharf, K.D. Prospects of engineering thermotolerance in crops through modulation of heat stress transcription factor and heat shock protein networks. Plant Cell Environ. 2015, 38, 1881–1895. [Google Scholar] [CrossRef]
- Driedonks, N.; Xu, J.; Peters, J.L.; Park, S.; Rieu, I. Multi-Level Interactions between Heat Shock Factors, Heat Shock Proteins, and the Redox System Regulate Acclimation to Heat. Front. Plant Sci. 2015, 6, 999. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, T.; Mirus, O.; von Haeseler, A.; Schleiff, E. The tetratricopeptide repeats of receptors involved in protein translocation across membranes. Mol. Biol. Evol. 2007, 24, 2763–2774. [Google Scholar] [CrossRef] [PubMed]
- Mirus, O.; Bionda, T.; von Haeseler, A.; Schleiff, E. Evolutionarily evolved discriminators in the 3-TPR domain of the Toc64 family involved in protein translocation at the outer membrane of chloroplasts and mitochondria. J. Mol. Model. 2009, 15, 971–982. [Google Scholar] [CrossRef]
- Panigrahi, R.; Adina-Zada, A.; Whelan, J.; Vrielink, A. Ligand recognition by the TPR domain of the import factor Toc64 from Arabidopsis thaliana. PLoS ONE 2013, 8, e83461. [Google Scholar] [CrossRef] [PubMed]
- Schweiger, R.; Soll, J.; Jung, K.; Heermann, R.; Schwenkert, S. Quantification of interaction strengths between chaperones and tetratricopeptide repeat domain-containing membrane proteins. J. Biol. Chem. 2013, 288, 30614–30625. [Google Scholar] [CrossRef] [PubMed]
- Nickel, C.; Horneff, R.; Heermann, R.; Neumann, B.; Jung, K.; Soll, J.; Schwenkert, S. Phosphorylation of the outer membrane mitochondrial protein OM64 influences protein import into mitochondria. Mitochondrion 2019, 44, 93–102. [Google Scholar] [CrossRef]
- Jarvis, P.; Chen, L.J.; Li, H.; Peto, C.A.; Fankhauser, C.; Chory, J. An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 1998, 282, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef]
- Oreb, M.; Zoryan, M.; Vojta, A.; Maier, U.G.; Eichacker, L.A.; Schleiff, E. Phospho-mimicry mutant of atToc33 affects early development of Arabidopsis thaliana. FEBS Lett. 2007, 581, 5945–5951. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, E. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 1963, 17, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Simm, S.; Papasotiriou, D.G.; Ibrahim, M.; Leisegang, M.S.; Müller, B.; Schorge, T.; Karas, M.; Mirus, O.; Sommer, M.S.; Schleiff, E. Defining the core proteome of the chloroplast envelope membranes. Front. Plant Sci. 2013, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Popov, N.; Schmitt, M.; Schulzeck, S.; Matthies, H. Reliable micromethod for determination of the protein content in tissue homogenates. Acta Biol. Med. Ger. 1975, 34, 1441–1446. [Google Scholar]
- Chaturvedi, P.; Ischebeck, T.; Egelhofer, V.; Lichtscheidl, I.; Weckwerth, W. Cell-specific analysis of the tomato pollen proteome from pollen mother cell to mature pollen provides evidence for developmental priming. J. Proteome Res. 2013, 12, 4892–4903. [Google Scholar] [CrossRef]
- Paul, P.; Chaturvedi, P.; Selymesi, M.; Ghatak, A.; Mesihovic, A.; Scharf, K.-D.; Weckwerth, W.; Simm, S.; Schleiff, E. The membrane proteome of male gametophyte in Solanum lycopersicum. J. Proteom. 2016, 131, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Keller, M.; Simm, S.; SPOT-ITN Consortium. The coupling of transcriptome and proteome adaptation during development and heat stress response of tomato pollen. BMC Genom. 2018, 19, 447. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.P.B.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stit, T.M. Mapman: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef] [PubMed]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M.; et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef] [PubMed]
- Hooper, C.M.; Castleden, I.R.; Tanz, S.K.; Aryamanesh, N.; Millar, A.H. SUBA4: The interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Res. 2017, 45, D1064–D1074. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.S.; Semedo, J.; Ramalho, J.C.; Lidon, F.C. Bread and Durum Wheat under Heat Stress: A Comparative Study on the Photosynthetic Performance. J. Agron. Crop Sci. 2011, 197, 50–56. [Google Scholar] [CrossRef]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef]
- König, J.; Baier, M.; Horling, F.; Kahmann, U.; Harris, G.; Schürmann, P.; Dietz, K.J. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proc. Natl. Acad. Sci. USA 2002, 99, 5738–5743. [Google Scholar] [CrossRef]
- Dangoor, I.; Peled-Zehavi, H.; Wittenberg, G.; Danon, A. A chloroplast light-regulated oxidative sensor for moderate light intensity in Arabidopsis. Plant Cell. 2012, 24, 1894–1906. [Google Scholar] [CrossRef]
- König, J.; Muthuramalingam, M.; Dietz, K.J. Mechanisms and dynamics in the thiol/disulfide redox regulatory network: Transmitters, sensors and targets. Curr. Opin. Plant Biol. 2012, 15, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liu, J.; Liu, B.; Feng, D.; Da, Q.; Wang, P.; Shu, S.; Su, J.; Zhang, Y.; Wang, J.; et al. Evidence for a role of chloroplastic m-type thioredoxins in the biogenesis of photosystem II in Arabidopsis. Plant Physiol. 2013, 163, 1710–1728. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Liu, B.; Luo, L.; Feng, D.; Wang, P.; Liu, J.; Da, Q.; He, Y.; Qi, K.; Wang, J.; et al. HYPERSENSITIVE TO HIGH LIGHT1 interacts with LOW QUANTUM YIELD OF PHOTOSYSTEM II1 and functions in protection of photosystem II from photodamage in Arabidopsis. Plant Cell. 2014, 26, 1213–1229. [Google Scholar] [CrossRef] [PubMed]
- Calderon, R.H.; García-Cerdán, J.G.; Malnoë, A.; Cook, R.; Russell, J.J.; Gaw, C.; Dent, R.M.; de Vitry, C.; Niyogi, K.K. A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs. J. Biol. Chem. 2013, 288, 26688–26696. [Google Scholar] [CrossRef]
- Hey, D.; Rothbart, M.; Herbst, J.; Wang, P.; Müller, J.; Wittmann, D.; Gruhl, K.; Grimm, B. LIL3, a Light-Harvesting Complex Protein, Links Terpenoid and Tetrapyrrole Biosynthesis in Arabidopsis thaliana. Plant Physiol. 2017, 174, 1037–1050. [Google Scholar] [CrossRef]
- Armbruster, U.; Labs, M.; Pribil, M.; Viola, S.; Xu, W.; Scharfenberg, M.; Hertle, A.P.; Rojahn, U.; Jensen, P.E.; Rappaport, F.; et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 2013, 25, 2661–2678. [Google Scholar] [CrossRef]
- Latijnhouwers, M.; Xu, X.M.; Møller, S.G. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 2010, 232, 567–578. [Google Scholar] [CrossRef]
- Taira, M.; Valtersson, U.; Burkhardt, B.; Ludwig, R.A. Arabidopsis thaliana GLN2-encoded glutamine synthetase is dual targeted to leaf mitochondria and chloroplasts. Plant Cell. 2004, 16, 2048–2058. [Google Scholar] [CrossRef]
- Scheibe, R. Malate valves to balance cellular energy supply. Physiol. Plant. 2004, 120, 21–26. [Google Scholar] [CrossRef]
- Khozaei, M.; Fisk, S.; Lawson, T.; Gibon, Y.; Sulpice, R.; Stitt, M.; Lefebvre, S.C.; Raines, C.A. Overexpression of plastid transketolase in tobacco results in a thiamine auxotrophic phenotype. Plant Cell. 2015, 27, 432–447. [Google Scholar] [CrossRef]
- Sjögren, L.L.; Tanabe, N.; Lymperopoulos, P.; Khan, N.Z.; Rodermel, S.R.; Aronsson, H.; Clarke, A.K. Quantitative analysis of the chloroplast molecular chaperone ClpC/Hsp93 in Arabidopsis reveals new insights into its localization, interaction with the Clp proteolytic core, and functional importance. J. Biol. Chem. 2014, 289, 11318–11330. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Cai, C.; Wang, Z.; Fan, B.; Zhu, C.; Chen, Z. Plastid Translation Elongation Factor Tu Is Prone to Heat-Induced Aggregation Despite Its Critical Role in Plant Heat Tolerance. Plant Physiol. 2018, 176, 3027–3045. [Google Scholar] [CrossRef] [PubMed]
- Tillmann, B.; Röth, S.; Bublak, D.; Sommer, M.; Stelzer, E.H.; Scharf, K.D.; Schleiff, E. Hsp90 is involved in the regulation of cytosolic precursor protein abundance in tomato. Mol. Plant 2015, 8, 228–241. [Google Scholar] [CrossRef]
- Kubis, S.; Baldwin, A.; Patel, R.; Razzaq, A.; Dupree, P.; Lilley, K.; Kurth, J.; Leister, D.; Jarvis, P. The Arabidopsis ppi1 mutant is specifically defective in the expression, chloroplast import, and accumulation of photosynthetic proteins. Plant Cell 2003, 15, 1859–1871. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Petersson, U.A.; Haas, B.J.; Funk, C.; Schröder, W.P.; Kieselbach, T. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 8354–8365. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Toporik, H.; Borovikova, A.; Nelson, N. Structure determination and improved model of plant photosystem I. J. Biol. Chem. 2010, 285, 3478–3486. [Google Scholar] [CrossRef] [PubMed]
- Michelet, L.; Zaffagnini, M.; Morisse, S.; Sparla, F.; Pérez-Pérez, M.E.; Francia, F.; Danon, A.; Marchand, C.H.; Fermani, S.; Trost, P.; et al. Redox regulation of the Calvin-Benson cycle: Something old, something new. Front. Plant Sci. 2013, 4, 470. [Google Scholar] [CrossRef]
- Taniguchi, M.; Miyake, H. Redox-shuttling between chloroplast and cytosol: Integration of intra-chloroplast and extra-chloroplast metabolism. Curr. Opin. Plant Biol. 2012, 15, 252–260. [Google Scholar] [CrossRef]
- Fu, A.; He, Z.; Cho, H.S.; Lima, A.; Buchanan, B.B.; Luan, S. A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 15947–15952. [Google Scholar] [CrossRef]
- Jurić, S.; Hazler-Pilepić, K.; Tomasić, A.; Lepedus, H.; Jelicić, B.; Puthiyaveetil, S.; Bionda, T.; Vojta, L.; Allen, J.F.; Schleiff, E.; et al. Tethering of ferredoxin:NADP+ oxidoreductase to thylakoid membranes is mediated by novel chloroplast protein TROL. Plant J. 2009, 60, 783–794. [Google Scholar] [CrossRef]
- Järvi, S.; Suorsa, M.; Tadini, L.; Ivanauskaite, A.; Rantala, S.; Allahverdiyeva, Y.; Leister, D.; Aro, E.M. Thylakoid-Bound FtsH Proteins Facilitate Proper Biosynthesis of Photosystem I. Plant Physiol. 2016, 171, 1333–1343. [Google Scholar] [PubMed]
- Kato, Y.; Hyodo, K.; Sakamoto, W. The Photosystem II Repair Cycle Requires FtsH Turnover through the EngA GTPase. Plant Physiol. 2018, 178, 596–611. [Google Scholar] [CrossRef] [PubMed]
- Wada, M.; Kong, S.G. Actin-mediated movement of chloroplasts. J. Cell Sci. 2018, 131, jcs210310. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.S.; Park, E.; Nedo, A.; Alqarni, A.; Ren, L.; Hoban, K.; Modla, S.; McDonald, J.H.; Kambhamettu, C.; Dinesh-Kumar, S.P.; et al. Stromule extension along microtubules coordinated with actin-mediated anchoring guides perinuclear chloroplast movement during innate immunity. Elife 2018, 7, e23625. [Google Scholar] [CrossRef] [PubMed]
- Suorsa, M.; Regel, R.E.; Paakkarinen, V.; Battchikova, N.; Herrmann, R.G.; Aro, E.M. Protein assembly of photosystem II and accumulation of subcomplexes in the absence of low molecular mass subunits PsbL and PsbJ. Eur. J. Biochem. 2004, 271, 96–107. [Google Scholar] [CrossRef]
- Allahverdiyeva, Y.; Suorsa, M.; Rossi, F.; Pavesi, A.; Kater, M.M.; Antonacci, A.; Tadini, L.; Pribil, M.; Schneider, A.; Wanner, G.; et al. Arabidopsis plants lacking PsbQ and PsbR subunits of the oxygen-evolving complex show altered PSII super-complex organization and short-term adaptive mechanisms. Plant J. 2013, 75, 671–784. [Google Scholar] [CrossRef]
- Ido, K.; Kakiuchi, S.; Uno, C.; Nishimura, T.; Fukao, Y.; Noguchi, T.; Sato, F.; Ifuku, K. The conserved His-144 in the PsbP protein is important for the interaction between the PsbP N-terminus and the Cyt b559 subunit of photosystem II. J. Biol. Chem. 2012, 287, 26377–26387. [Google Scholar] [CrossRef]
- Morais, F.; Barber, J.; Nixon, P.J. The chloroplast-encoded alpha subunit of cytochrome b-559 is required for assembly of the photosystem two complex in both the light and the dark in Chlamydomonas reinhardtii. J. Biol. Chem. 1998, 273, 29315–29320. [Google Scholar] [CrossRef]

| Name | Process | Accession | log2(LFQ) | FC (LFQ) | |||
|---|---|---|---|---|---|---|---|
| wt | toc64-1 | ppi1 | toc64-1 | ppi1 | |||
| CYP38 | photosynthesis | AT3G01480 | 20.44 | 21.96 | 21.10 | 2.87 | 1.59 |
| FNR2 | photosynthesis | AT1G20020 | 21.98 | 23.13 | 22.91 | 2.22 | 1.91 |
| cpHSP70-1 | protein synth. | AT4G24280 | 19.71 | 20.82 | 19.92 | 2.16 | 1.16 |
| TROL | photosynthesis | AT4G01050 | 21.57 | 22.67 | 22.45 | 2.14 | 1.84 |
| Rpl15 | protein synth. | AT3G25920 | 18.01 | 19.08 | 19.07 | 2.10 | 2.08 |
| FTSH5 | photosynthesis | AT5G42270 | 22.11 | 23.12 | 22.93 | 2.02 | 1.77 |
| LHCb1.5 | photosynthesis | AT2G34420 | 21.34 | 22.11 | 23.95 | 1.71 | 6.11 |
| Tic62 | TIC | AT3G18890 | 21.27 | 22.22 | 22.58 | 1.93 | 2.48 |
| LOX2 | JA synth. | AT3G45140 | 21.09 | 21.20 | 22.24 | 1.07 | 2.22 |
| unknown | AT3G61870 | 19.60 | 20.08 | 20.75 | 1.40 | 2.22 | |
| PetE2 | photosynthesis | AT1G20340 | 22.12 | 22.11 | 23.27 | 0.99 | 2.21 |
| PsaE1 | photosynthesis | AT4G28750 | 23.61 | 24.14 | 24.70 | 1.45 | 2.14 |
| cpATPδ | photosynthesis | AT4G09650 | 21.75 | 22.17 | 22.84 | 1.34 | 2.14 |
| OE33 | photosynthesis | AT3G50820 | 22.15 | 23.10 | 23.21 | 1.92 | 2.08 |
| unknown | AT5G37360 | 18.59 | 18.78 | 19.61 | 1.14 | 2.03 | |
| LHCa4 | photosynthesis | AT3G47470 | 24.35 | 23.16 | 23.33 | 0.44 | 0.49 |
| GAPb | metabolism | AT1G42970 | 21.99 | 21.46 | 20.89 | 0.69 | 0.47 |
| PrxB | regulation | AT5G06290 | 21.53 | 20.19 | 20.40 | 0.39 | 0.46 |
| PsaF | photosynthesis | AT1G31330 | 25.84 | 24.89 | 24.68 | 0.52 | 0.45 |
| Ef-Ts | protein synth. | AT4G29060 | 19.02 | 19.13 | 17.82 | 1.08 | 0.44 |
| Trx-M1 | regulation | AT1G03680 | 19.54 | 18.58 | 18.30 | 0.52 | 0.43 |
| GAPa | metabolism | AT3G26650 | 23.48 | 22.94 | 22.23 | 0.68 | 0.42 |
| GLU1 | metabolism | AT5G04140 | 20.81 | 20.40 | 19.38 | 0.75 | 0.37 |
| Psb27 | photosynthesis | AT1G03600 | 22.30 | 21.26 | 22.32 | 0.49 | 1.01 |
| PsbS | photosynthesis | AT1G44575 | 24.27 | 23.22 | 23.53 | 0.48 | 0.60 |
| PsaL | photosynthesis | AT4G12800 | 24.04 | 22.92 | 23.32 | 0.46 | 0.61 |
| GLN2 | metabolism | AT5G35630 | 22.26 | 21.14 | 21.62 | 0.46 | 0.64 |
| NADP-MDH | metabolism | AT5G58330 | 18.95 | 17.75 | 18.47 | 0.44 | 0.72 |
| LHCa3 | photosynthesis | AT1G61520 | 25.19 | 23.94 | 24.19 | 0.42 | 0.50 |
| PsbL | photosynthesis | ATCG00560 | 21.12 | 16.75 | 20.91 | 0.05 | 0.86 |
| Name | Accession | FC [84] | log2(LFQ) | FC (LFQ) | |||
|---|---|---|---|---|---|---|---|
| wt | toc64-1 | ppi1 | toc64-1 | ppi1 | |||
| HSP90 homolog | At2g04030 | 2.4/1.8 | 19.96 | n.d. | 19.97 | n.d. | 1.00 |
| Chaperonin 60β | At1g55490 | 1.9/1.6 | 21.92 | 22.56 | 22.09 | 1.57 | 1.13 |
| At3g13470 | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| HSP70 homolog | At4g24280 | 1.8 | 19.71 | 20.82 | 19.92 | 2.16 | 1.16 |
| At5g49910 | n.d. | n.d. | n.d. | n.d. | n.d. | ||
| PPIase | At3g62030 | 1.8 | 21.06 | 20.67 | 20.51 | 0.77 | 0.68 |
| EF-Tu homolog | At4g20360 | 1.7 | 22.74 | 22.33 | 22.58 | 0.75 | 0.90 |
| Chaperonin 60α | At2g28000 | 1.6 | 21.59 | 22.03 | 21.89 | 1.36 | 1.23 |
| R-5-P isomerase | At3g04790 | 0.6/0.5 | 19.37 | n.d. | 19.91 | n.d. | 1.45 |
| LHCb5 | At4g10340 | 0.6 | 25.82 | 25.65 | 26.30 | 0.89 | 1.39 |
| LHCII type 1 | At1g29910 | 0.6 | 25.85 | 25.58 | 26.66 | 0.83 | 1.74 |
| At1g29920 | |||||||
| At1g29930 | |||||||
| At2g34420 | 21.34 | 22.12 | 23.95 | 1.72 | 6.11 | ||
| OE23 | At1g06680 | 0.6 | 24.82 | 25.27 | 25.19 | 1.37 | 1.29 |
| OE33 | At3g50820 | 0.6/0.5 | 22.15 | 23.11 | 23.21 | 1.93 | 2.08 |
| At5g66570 | 25.03 | 25.66 | 25.88 | 1.55 | 1.79 | ||
| SSU | At1g67090 | 0.4 | 23.62 | 23.37 | 23.48 | 0.85 | 0.91 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paul, P.; Mesihovic, A.; Chaturvedi, P.; Ghatak, A.; Weckwerth, W.; Böhmer, M.; Schleiff, E. Structural and Functional Heat Stress Responses of Chloroplasts of Arabidopsis thaliana. Genes 2020, 11, 650. https://doi.org/10.3390/genes11060650
Paul P, Mesihovic A, Chaturvedi P, Ghatak A, Weckwerth W, Böhmer M, Schleiff E. Structural and Functional Heat Stress Responses of Chloroplasts of Arabidopsis thaliana. Genes. 2020; 11(6):650. https://doi.org/10.3390/genes11060650
Chicago/Turabian StylePaul, Puneet, Anida Mesihovic, Palak Chaturvedi, Arindam Ghatak, Wolfram Weckwerth, Maik Böhmer, and Enrico Schleiff. 2020. "Structural and Functional Heat Stress Responses of Chloroplasts of Arabidopsis thaliana" Genes 11, no. 6: 650. https://doi.org/10.3390/genes11060650
APA StylePaul, P., Mesihovic, A., Chaturvedi, P., Ghatak, A., Weckwerth, W., Böhmer, M., & Schleiff, E. (2020). Structural and Functional Heat Stress Responses of Chloroplasts of Arabidopsis thaliana. Genes, 11(6), 650. https://doi.org/10.3390/genes11060650






