Abstract
Genome integrity is essential to maintain cellular function and viability. Consequently, genome instability is frequently associated with dysfunction in cells and associated with plant, animal, and human diseases. One consequence of relaxed genome maintenance that may be less appreciated is an increased potential for rapid adaptation to changing environments in all organisms. Here, we discuss evidence for the control and function of facultative heterochromatin, which is delineated by methylation of histone H3 lysine 27 (H3K27me) in many fungi. Aside from its relatively well understood role in transcriptional repression, accumulating evidence suggests that H3K27 methylation has an important role in controlling the balance between maintenance and generation of novelty in fungal genomes. We present a working model for a minimal repressive network mediated by H3K27 methylation in fungi and outline challenges for future research.
Keywords:
Cryptococcus; fungi; Fusarium; histones; lysine methylation; Neurospora; polycomb repressive complex; PRC2; Zymoseptoria 1. Introduction
Even though the first description of different chromatin types was cytological [1], transcriptional activity is frequently used to divide chromatin into three, sometimes four, categories that are correlated with post-translational modifications on histone tails [2]. Euchromatin is characterized by active transcription, and some examples of tell-tale histone marks are high levels of acetylation, e.g., on histone H3 lysine residues 9 and 27 (H3K9ac, H3K27ac), and di- or trimethylation of histone H3 lysine 4 or 36 (H3K4me2/3, H3K36me2/3). Constitutive heterochromatin lacks transcription of protein-coding genes, but non-coding RNA, either short siRNA or various types of long non-coding RNA (lncRNA) are produced from these regions, which are enriched in di- or trimethylation of histone H3 lysine 9 (H3K9me2/3) and methylated cytosines. A subtype of constitutive heterochromatin is “centrochromatin”, regions that contain the centromere and on which kinetochores form (reviewed in [3]). In many species, a histone H3 variant, CENPA (Cse4, Cid, CenH3), generates special nucleosomes that co-occur with canonical nucleosomes in centrochromatin [4]. The last type of chromatin is facultative heterochromatin, which typifies regions where transcription is reversibly turned on and off, either during responses to the environment, or during cell differentiation and development. The most intensely studied histone mark correlated with facultative heterochromatin is trimethylation of histone H3 lysine 27 (H3K27me3) [5,6]. This review is on facultative heterochromatin in fungi; we will largely focus on the past five years as several reviews have summarized earlier studies [7,8,9,10].
Compared to the breadth of information amassed in plants and animals (see recent reviews for summaries, e.g., [11,12,13,14,15,16,17,18,19]), the study of facultative heterochromatin in fungi can still be considered an immature field. Even though this form of heterochromatin had been recognized as important for differentiation and development for some time, it took until 2008 for the first demonstration of the presence of H3K27me3 in a fungus, the ascomycete Neurospora crassa [20]. Since then, our understanding has grown considerably, driven by comparative biology, and utilizing several key model organisms among the large and varied kingdom fungi, namely the saprobe N. crassa, the plant pathogens Dothistroma septosporum, several Fusarium species, Magnaporthe oryzae, Verticillium dahliae, Zymoseptoria tritici, the plant endosymbiont Epichloë festucae, and the facultative animal pathogen, Cryptococcus neoformans.
Here, we will briefly summarize what has been found in studies on the single histone H3K27 methyltransferase complex, polycomb repressive complex 2 (PRC2), review effects on development and pathogenicity in the absence of fungal PRC2, and discuss how the interplay of various histone marks affects genome plasticity and stability in fungi. We propose a testable working model predicting the minimal components of a reversible H3K27me3-mediated gene silencing system in fungi, touch on ongoing forward genetic screens that promise to discover novel components of the system, and present an outlook describing several key challenges in the area of facultative heterochromatin in fungi for the immediate future.
2. Setting the Stage: Reverse Genetics to Define Facultative Heterochromatin in Fungi
Polycomb group (PcG) proteins were discovered by their impact on Drosophila development [21], and it took ~25 years before the molecular basis for their importance became known (reviewed in [5,17]). The realization that the PcG proteins, E(z), Su(z)12, and Esc, were required for gene silencing came early and resulted in the development of “chromatin immunoprecipitation” (ChIP) [22]. The dominant Drosophila Pc mutant, first isolated in 1947 by P. Lewis, received its name because it altered the number of sex combs, a row of thick bristles, on the legs of male flies (reviewed in [23]). Usually sex combs are found only on the first pair of legs, but Pc mutants have sex combs on all three pairs. E. Lewis realized that this body plan transformation resulted from a requirement of Pc to repress homeotic genes of the Bithorax complex [21]. Biochemical studies showed that PcG proteins fall into two main complexes, polycomb repressive complex 1 and 2 (PRC1 and PRC2), the latter of which included E(z), Su(z)12, and Esc ([12,23]. Subsequent studies identified human homologs (EZH2, SUZ12, and EED, respectively) that catalyzed H3K27 methylation [24,25] and additional subunits, including RbAp48/Nurf55 (called Msi1, Cac3, Msl1, or NPF in different fungi) that co-purified with the core PRC2 ([12,23]. Interest among researchers studying fungi increased when H3K27me3 was found in Neurospora [20]; notably, this important histone mark is absent in both budding and fission yeast [10,23].
Although not easily discerned by overall sequence comparisons, homologs of core PRC2 components found in fungi have well-conserved domain architectures when compared to their plant and animal counterparts [8,10,26] (Table 1). Crystal structures of a partial core PRC2 of the ascomycete, Chaetomium thermophilum, helped define functional regions of E(z)/EZH2 and Esc/EED homologs, and revealed structural similarity between fungal and animal PRC2s [26,27,28,29,30,31], while biochemical studies by a combination of western analyses, ChIP-seq, and mass spectrometry have demonstrated that PRC2 in fungi methylates H3K27 [32,33]. The most striking dissimilarities are the presence of a large “insertion domain” in fungal EED proteins that is missing from plant and animal proteins; the domain structure and boundary is conserved but the primary amino acid sequence is taxon-specific [10]. SUZ12 homologs are largely different, except for the VEFS motif, whose presence is difficult to detect in some fungi; SUZ12 may be supplanted by functional homologs in some fungi, e.g., in C. neoformans [34].

Table 1.
Components of polycomb repressive complexes (PRC) in fungi. Known components from Drosophila, human, Fusarium graminearum, Neurospora crassa, Cryptococcus neoformans, and Arabidopsis thaliana. All fungi examined to date have single homologues of at least two core PRC2 subunits (KMT6 and EED). SUZ12 and PRC2-associated subunits, such as MSL1/NPF or PAS appear more specialized and may have impacts only on certain chromatin regions. While KDMs with similarity to JARID2 homologs, and RING proteins are present in fungi, PRC1 subunits or other PRC2-targeting complexes found in animals are not conserved in fungi.
Reverse genetics studies delineated the PRC2 of fungi in the ascomycetes, Neurospora [35] and Fusarium [32], and the basidiomycete, Cryptococcus [34]. Neurospora PRC2 was characterized by deletion of homologous genes for all three core subunits (SET-7/EZH2, EED, SUZ12), which abrogated all H3K27me3, while deletion of the gene encoding an associated subunit (NPF/CAC-3/MSL1) resulted in lower H3K27me3 levels in many interstitial regions and complete absence of H3K27me3 from subtelomeric regions [35]. In Fusarium graminearum, deletion of kmt6/EZH2 resulted in complete loss of H3K27me3 [32], and the same is true for deletion of eed or suz12, while deletion of msl1/cac3 had no effect on H3K27me3 levels (Connolly and Freitag, unpublished results). Loss of the C. neoformans PRC2 core components (Ezh2 and Eed1) or the non-canonical subunit, Bnd1, removed all H3K27me3, while loss of Msl1 or the novel accessory component Ccc1 resulted in regional reduction and relocalization of H3K27me3 [34]. This was the first study that delineated fungal PRC2 components by biochemical means, and it identified two novel fungal PRC2 subunits that may be functional homologs for SUZ12 (e.g., Bnd1) or connect PRC2 to histone deacetylase (HDAC) complexes (e.g., Ccc1). In contrast to the findings with Neurospora, Cryptococcus msl1 mutants showed only depletion of H3K27me3 in subtelomeric regions; ccc1 mutants showed a more pronounced reduction of H3K27me3 in these regions.
Studies in these three models, and more recently the wheat pathogen, Zymoseptoria tritici [36,37], defined the extent of H3K27me3 in fungal genomes by ChIP-seq, and revealed upregulation of genes covered by H3K27me3 in PRC2 mutants (Figure 1). Together, these studies demonstrated that fungi have a canonical or largely conserved PRC2 that appears to act as a general repressor of gene transcription, but they also suggested taxon-specific roles played by H3K27 methylation.
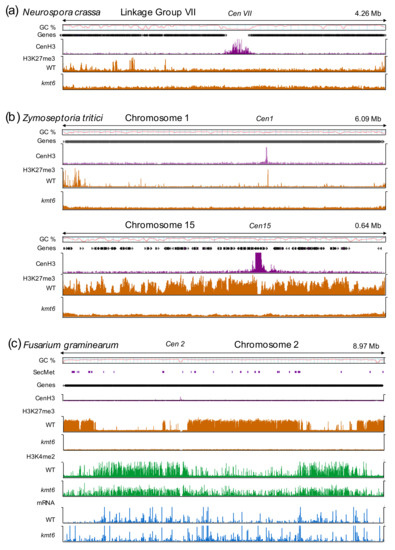
Figure 1.
Chromatin landscapes in selected filamentous fungi. (a) In N. crassa, H3K27me3 covers subtelomeric regions and discrete interstitial sequences along the chromosome arms. Deletion of kmt6 results in complete loss of H3K27me3. (b) H3K27me3 enrichment in Zymoseptoria tritici differs between core and accessory chromosomes. Core chromosomes (e.g., chromosome 1) show a similar distribution to N. crassa with enrichment in subtelomeric regions and local peaks. Conversely, accessory chromosomes (e.g., chromosome 15) are enriched with H3K27me3 over the entire length of the chromosome. Again, all H3K27me3 is lost upon deletion of kmt6. (c) Fusarium graminearum chromosomes are divided into large blocks of H3K27me3-enriched facultative heterochromatin and euchromatic regions, marked by H3K4me2. Loss of H3K27me3 increases both H3K4me2 enrichment and transcription (mRNA; blue) in many, if not all, former facultative heterochromatic blocks. H3K27me3 regions contain secondary metabolite (SecMet) clusters and pathogenicity genes that are activated in the absence of H3K27me3. ChIP-seq experiments with the centromere-specific histone H3, CenH3 (CENPA), indicate centromeric regions, which are largely free of H3K27me3, except small amounts in some Z. tritici centromeres [36]. Publicly available data were used to construct this figure [32,33,37,38].
3. Maintenance or De Novo H3K27 Methylation—or Both?
DNA replication requires disassembly of nucleosomes and eviction of DNA- and chromatin-interacting proteins. While strand-specific mechanisms of histone recycling have been uncovered (reviewed in [13]), it is not fully understood how H3K27me3 is reestablished following passage of the replication fork. PRC2 writes and reads H3K27me3, and colocalizes at replication forks, where it can aid DNA replication [39,40,41,42], suggesting a model of maintenance wherein PRC2 reestablishes H3K27me3 domains using recycled, H3K27me3-modified histones as a guide [43]. Recently, chromatin assembly factor 1 (CAF-1) was found to mediate the interaction between PRC2 and proliferating cell nuclear antigen (PCNA), a central component of DNA replication, in plants and animals [44,45]. In fungi, Msl1 is a component of CAF-1 in addition to its role as an accessory component of PRC2 [46]. PKL, a plant chromatin remodeling factor (CRF), promotes recycling of H3K27me3-modified histones following DNA replication [47]. Interactions among PRC2, CRFs, and DNA replication machinery have not been explored in fungi.
Contrasting evidence suggests that recycled H3K27me3 is not necessarily sufficient or required for long-term propagation of transcriptional repression. For example, in animals, lncRNAs have been suggested to target PRC2 to specific regions [17,48], something that has not been shown or even seriously explored in fungi. Specific DNA sequences or regions with certain physical characteristics, like polycomb response elements (PREs) in Drosophila [23,49,50] and hypomethylated CpG islands in mammals [51,52,53,54,55], recruit PRC2 and mediate transcriptional repression. Although similar mechanisms for H3K27me3 nucleation have not been identified in fungi, recent work in N. crassa showed that telomeric repeats can recruit H3K27me3 to naïve chromosome segments and spread H3K27me3 across dozens of kilobases [56]. Furthermore, depletion of H3K27me3 by mutation of PRC2 subunits can be faithfully reestablished following complementation of the causal mutation in fungi and animals [32,35,55]. From studies in mammals it is clear that H3K27me2 is wide-spread in chromatin (60–80% of all nucleosomes with H3), which suggested a “hit-and-run” activity (Figure 2) of PRC2 to generate H3K27me1/2; however, H3K27me3 activity may be more targeted. Together, this demonstrates that de novo PRC2 activity, likely conferred by sequence-specific binding, DNA structure, or an “epigenetic” assembly of RNA and protein factors, and maintenance activity work together to propagate H3K27me3-mediated repression in a manner dependent on the organism and the locus.
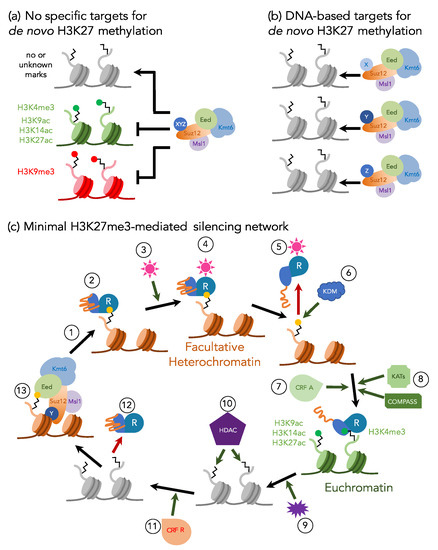
Figure 2.
Working models for de novo H3K27 methylation and a minimal H3K27me3-mediated silencing network. (a) De novo H3K27 methylation in fungi may occur in all regions not blocked by proteins or specific histone modifications (“default H3K27me1/2 model”) as suggested by others [9,17,34,64]. (b) A non-exclusive model predicts recognition of specific nucleic acid-based signals by chromatin-binding proteins (X, Y, and Z) that recruit the core PRC2 to specific genes or blocks of genes, presumably by using SUZ12 as an adaptor, thus yielding a diverse population of PRC2-adaptor complexes. (c) Based on the combination of data from fungi and plants (e.g., western blots, ChIP, histone binding assays, phenotyping, and effects of mutants that have been isolated), we propose that an adaptor protein (e.g., N. crassa EPR-1 [58], F. graminearum DIS2; unpublished results from our lab), which is similar to Arabidopsis SHL [60] and EBS [61], acts as a “Reader” (R) for both silent and active chromatin regions. PRC2 catalyzes H3K27me3 either de novo or by close association with the replication fork (1), and the Reader binds H3K27me3 (yellow dot) in facultative heterochromatin via its bromo-adjacent homology (BAH) domain (teal; 2). Exogenous or endogenous cues (pink star; 3) may result in binding to (4) and conformational changes (5) of the Reader. One prediction is that the Reader is temporarily removed from the H3 tail, while a histone demethylase (KDM; 6) removes H3K27me3; an alternative model suggests that the Reader remains bound to H3K27me3 until H3K4me3 binding is possible after action of the H3K4 methyltransferase, COMPASS (8) [10]. Conformational changes within the Reader’s carboxy-terminal motif (orange tail) makes the plant homeo domain (PHD) (blue) accessible, which allows Reader binding to H3K4me3, likely in combination with the activity of activating chromatin remodeling factors (CRF A; 7) and lysine acetyltransferases (KATs; 8) that may acetylate H3K9, H3K14, or H3K27, or a combination of them (green dot). Gene expression is altered based on unknown exogenous or endogenous cues (purple; 9). The Reader may directly or indirectly recruit histone deacetylase (HDAC; 10) complexes, which—in combination with repressive chromatin remodeling factors (CRF R; 11)—aid in Reader release from H3K4me3 (12). Chromatin-binding adaptors bound to core PRC2 re-methylate regions destined to be facultative heterochromatin (13).
4. Defining the Minimal H3K27me3 Network: Fungi as Excellent Genetic Models
Fungi are excellently suited for genetic analyses, and studies in at least three laboratories are under way to determine what other components are part of the H3K27 methylation network. Two recent papers describe new components of the fungal H3K27 methylation network. A forward genetics selection with N. crassa identified a novel fungal-specific protein, PRC2 ACCESSORY SUBUNIT (PAS) [57]. PAS promotes H3K27me2/3 and transcriptional repression in subtelomeric regions, similar to what had previously been observed for NPF/MSL1 [35,57]. While additional studies will be required to determine the functional conservation of PAS or PAS-like proteins in fungi, preliminary results suggest that this protein is weakly conserved and may have taxon-specific functions.
Studies in our lab have identified several mutants, called defective in silencing (dis), which exhibit phenotypes that are near-identical to those observed with kmt6 deletion strains. While our analyses are still incomplete and ongoing, one of these proteins, DIS2, resembles a protein also found recently in N. crassa, called EFFECTOR of POLYCOMB REPRESSION-1 (EPR-1) [58]. EPR-1 has a bromo-adjacent homology (BAH) domain followed by a plant homeo domain (PHD), both of which are able to bind modified or non-modified histone tails in other proteins [59]. In fact, the most similar two plant proteins, SHL [60] and EBS [61], have been proposed to bind H3K27me3 and H3K4me3 simultaneously (SHL) or sequentially (EBS), suggesting that this class of proteins may serve as a molecular switch from H3K4me3-enriched, active regions bound by the PHD, to H3K27me3-enriched, silent regions bound by the BAH domain [61]. The N. crassa EPR-1 protein may act in a similar fashion. All epr-1 mutants display gene expression changes that are similar to strains lacking PRC2 components, but surprisingly H3K27 methylation appeared unaffected [58]. By cytology, EPR-1 clusters in subtelomeric foci, and ChIP-seq showed that its genome-wide distribution is dependent on H3K27me3-enriched chromatin [58].
Our working model predicts additional factors that are expected to be necessary to complete a feedback loop that establishes and maintains H3K27 methylation until environmental or internal conditions require transcriptional activation (Figure 2). Apart from PRC2 and various associated proteins, a PHD–BAH domain containing “Reader” can alternatively bind to H3K27me3 and H3K4me3, and serves as a switch from a silent to active state. Altering the Readers’ binding activity may require a histone demethylase (KDM) to remove H3K27me2/3, as well as histone lysine acetyltransferases (KATs) and the H3K4 methyltransferase COMPASS complex [62,63], both of which would be required to convert long-term silencing into long-term activation. Recruitment of KATs and COMPASS may also require the activity of chromatin remodeling factors (CRFs) to generate more open chromatin structures or even evict nucleosomes with silencing H3K27me3 marks. Once active gene expression is no longer required, external or internal stimuli recruit HDACs, perhaps again in combination with CRFs, which generate the proper conditions for PRC2 recruitment.
ATP-dependent CRFs catalyze nucleosome sliding, disassembly, and exchange, which ultimately influences transcription via nucleosome positioning or DNA accessibility [65,66]. Interplay among CRFs and PRC2 is emerging as an integral part of H3K27me3-mediated repression in plants. One such relationship is the genetic interaction between PKL, a CHD subfamily II-type CRF [67], and CLF, an H3K27 methyltransferase in a plant PRC2 [68]. PKL is involved in transcriptional regulation of developmental genes [69,70] and associates with H3K27me3-enriched loci [47,71]. In root tissue, the relationship between PKL and CLF appears to be antagonistic, as PKL promotes cell proliferation in the meristem in opposition to CLF, which restricts cell proliferation [72]. Loss of PKL repressed developmental genes and showed increased H3K27me3 levels [72]. In aerial tissue, evidence suggests that PKL cooperates with CLF and PIE1, an SWR1-type CRF that incorporates the histone variant H2A.Z, to promote H3K27me3 and transcriptional repression [73]. Moreover, H2A.Z appears to colocalize with H3K27 methylation in plants and animals [5,73]. It was recently demonstrated that in N. crassa H2A.Z and the CRF, SWR-1, similarly promote H3K27 methylation, although H2A.Z and H3K27 methylation were not found to colocalize [74].
Recent evidence from animal studies also suggests that CRFs have various roles in controlling PRC2 activity (reviewed in [75,76]). The nucleosome remodeling and deacetylase (NuRD) complex mediates transcriptional repression through nucleosome invasion and histone deacetylation and is proposed to promote PRC2 recruitment [77,78]. NuRD consists of a CHD subfamily II-type CRF similar to PKL (e.g., Mi-2 in Drosophila) and NURF55, a homolog of Msl1, among other components [79,80]. In contrast, the name-giving SWI/SNF CRF complex promotes open chromatin in opposition to PRC2 activity [81,82,83,84]. Mutations in SWI/SNF components are associated with regional increases in H3K27me3 levels and transcriptional repression [75,76]. Taken together, evidence suggests that NuRD and SWI/SNF act in opposition to modify the chromatin environment and subsequently promote or antagonize PRC2. All CRFs mentioned above have highly conserved homologs in fungi [85], and although little is known about these proteins in filamentous fungi, similar interplay among CRFs and PRC2 is expected to have an important role in H3K27me3-mediated repression.
5. Functions of H3K27 Methylation in Fungi
5.1. Regulation of Transcription
Generally, transcriptional activity is lower or absent in chromatin enriched with H3K27me3 [6]. Transcriptional repression has thus been long thought of as the most important, if not only, function of H3K27me3 and PRC2 in fungi. The most compelling evidence for transcriptional regulation and the important functions of PRC2 within the fungi come from studies with Fusarium, where development and differentiation are severely disturbed by the expression or overexpression of ~2,500 genes following loss of PRC2 [32]. In N. crassa, 700–900 genes are enriched with H3K27me3, and ~150 genes are derepressed in PRC2 mutants [33,35], but many of these transcripts emanate from regions that are not usually enriched with H3K27me3, suggesting an indirect effect on transcription. In C. neoformans, ~75 transcripts increase in expression by more than threefold, and 71% of these are within 40 kb of telomeric regions [34]. In Z. tritici, ~1,300 genes are enriched for H3K27me3, but only 98 genes in H3K27me3 regions and 124 genes in total were upregulated in absence of H3K27me3 [37]. While direct quantitative comparisons between these datasets are difficult, it is clear that H3K27me3 is involved in transcriptional regulation in fungi. For example, we fully expect that transcriptional activation in the absence of H3K27me3 requires additional chromatin reorganization or the activity of context-specific transcription factors, or both.
How is H3K27me3-dependent transcriptional repression mediated? After discovery of the molecular activity of PRC2, a hierarchical model of catalysis and spreading of H3K27me3 by PRC2 and subsequent binding of PRC1 by virtue of its Pc subunit, which reads H3K27me3, was proposed [24]. It is now clear that the hierarchical “PRC2 first, then PRC1” model cannot reconcile all available data from all organisms that have been studied, and it has thus been rejected. Even in animals not all PRC1 variants contain chromo domain or “chromobox” (Cbx) proteins. PRC1 subunits RING1A/B form distinct subcomplexes (PRC1.1 to PRC1.6); only PRC1.2 and PRC1.4 contain a Cbx [86,87,88]. Lack of PRC2 has only mild effects on global PRC1-mediated ubiquitination levels of H2A lysine 119 (H2AK119ub1) [89], and some PRC1s are recruited independently of PRC2 and H3K27me3 [90]. For example, the histone demethylase, KDM2B, can mediate PRC1.1 recruitment via its CXXC motif, which can bind unmethylated CG-rich DNA [91,92,93], which in turn may recruit PRC2. H2AK119ub1 then modulates binding and catalytic activity of PRC2 through interaction with an accessory subunit, JARID2 [88,94,95], which suggests a feedback loop involving both canonical PRCs.
While many fungi have genes for the core PRC2 subunits, no fungal genome examined so far encodes known critical components of the canonical PRC1. There is no evidence for clear homologs of many PRC1 subunits, including KDM2B and JARID2, in fungal genomes. One catalytic activity of PRC1 is ubiquitination of H2A (H2AK118ub1 in flies, H2AK119ub1 in mammals, and H2AK121ub1 in plants), which is thought to result in transcriptional repression (reviewed in [13,17,48,96]). However, this model has not been investigated in any detail in fungi, simply because true Pc proteins are lacking from both fungi and plants [97,98]; thus, all facultative heterochromatin generated by “polycomb” PRC1 complexes must be generated by different mechanisms. In plants, the role of Pc may be at least partially fulfilled by LIKE-HETERCHROMATIN PROTEIN-1 (LHP1/TLF2), which functions in both PRC1 and PRC2 [98,99], but not in H3K9 methylation where canonical HP1 is required for normal function in both animals and fungi [100,101]. LHP1 promotes H3K27me3 spreading and maintenance via its function as H3K27me3 reader and its interaction with the plant Nurf55 homolog, MSI1 [98]. Another plant-specific protein, EMBRYONIC FLOWER-1 (EMF1), has been proposed to mediate H3K27me3-dependent silencing through chromatin compaction as a functional homolog for animal Psc proteins, and while EMF1 had been shown to interact with LHP1 in vitro, a recent study showed interactions with the BAH-domain containing H3K27me3 readers, SHL and EBS, in a BAH–EMF1 Complex to read H3K27me3 and induce transcriptional silencing [102]. In fungi, neither a chromo domain protein functionally similar to Pc or LHP1 nor a functional homolog of EMF1 have been found so far. Thus, it remains a complete mystery how exactly H3K27me3-mediated transcriptional repression is carried out in fungi; deciphering downstream effects of H3K27me3 on RNAP II activity will be an important goal for the next decade.
5.2. The Role of Facultative Heterochromatin During Development and Pathogenesis
Transcriptional regulation of important genes is considered the proximal cause for the requirement of PRC2-mediated H3K27 methylation for normal development in the plants and animals that have been studied. While the number of fungi, including plant pathogens, with experimental evidence for the presence of H3K27 methylation is rapidly increasing, the functional characterization of facultative heterochromatin is lagging behind. Loss of H3K27 methylation can slow growth and development in some fungi, most notably in the genus Fusarium. For example, deletion of genes encoding the core F. graminearum PRC2 subunits, kmt6/EZH2 [32], eed, or suz12 (Connolly and Freitag, unpublished results) resulted in severe defects in both linear growth and development. Colony morphology is severely disturbed, presumably because various pathways involved in signaling and asexual growth are expressed at improper times. Sexual development is completely abrogated in kmt6 mutants under normal conditions in this homothallic species [32]; in outcrosses with engineered strains kmt6 mutants are female sterile. Overexpression of numerous secondary metabolite clusters in kmt6 mutants may contribute to defects in development and pathogenicity [38]. In the rice pathogen, F. fujikuroi, attempts to delete kmt6 have been unsuccessful [103]. This suggests that PRC2-mediated gene silencing is essential for survival in this, and perhaps other, species. Quelling of kmt6 by introduction of constructs to generate siRNA from double-stranded RNA revealed severe defects in growth and sporulation [103]. Further work will delineate why PRC2 components are essential in some Fusarium species but not others.
In plant pathogens other than Fusarium, deletion of kmt6/EZH2 resulted in only minor defects in development. In the wheat pathogen Z. tritici, strains lacking H3K27me3 were indistinguishable from wild type in growth and stress responses (e.g., temperature, cell wall, osmotic, oxidative, genotoxic stress) except for a slight increase in melanization at elevated temperatures; pathogenicity on wheat was only mildly affected [37]. Deletion of kmt6 in the blast pathogen, Magnaporthe oryzae, abolished all H3K27me3, reduced asexual sporulation, and resulted in severe defects in pathogenicity on wheat and barley [104]. Epichloë festucae is a beneficial endosymbiont of perennial ryegrass (Lolium perenne). Deletion of ezhB/EZH2 abolished all H3K27me3 [105]. This fungus is well known for production of ergot alkaloids and indole diterpenes, which afford the host plant protection from insect and mammalian herbivory [106]. Genes underlying the production of ergot alkaloids and indole diterpenes are organized in the subtelomeric gene clusters, eas and ltm, respectively, and only expressed in planta [106,107]. In vitro suppression of eas and ltm genes was influenced by presence of heterochromatic marks, i.e., H3K9me3 or H3K27me3 [105]. H3K9me3 and H3K27me3 levels at eas and ltm were reduced in planta when compared to in vitro growth. Deletion of the H3K9-methyltransferase-encoding gene, clrD, or ezhB increased expression from both clusters in vitro, and double deletion resulted in additive effects [105]. However, in vitro metabolite production was not observed in any of the mutants [105]. Loss of EzhB on ryegrass resulted in increased expression of all tested genes in the eas cluster, including genes that were not derepressed in vitro, and increased production of the ergot alkaloid, ergovaline [105], suggesting a host-derived signal is required even in the absence of H3K27me3 repression. Strains lacking EzhB effectively colonized ryegrass yet negatively impacted plant development, while deletion of clrD abolished colonization of ryegrass [105], demonstrating that H3K9 and H3K27 methylation are somehow required for establishing and maintaining successful symbiosis. Dothistromin, a secondary metabolite produced from a developmentally-regulated pathway in the pine needle blight pathogen, Dothistroma septosporum, is regulated by H3K27me3 [108]. During development, dothistromin cluster genes were transcribed early and later repressed by H3K27me3. Although dothistromin is a key virulence factor in D. septosporum, the impact of kmt6 deletion on pathogenesis has not been determined [108].
Overt phenotypes are not observed in N. crassa or C. neoformans in the absence of H3K27me3 under normal laboratory growth conditions, but loss of accessory components, e.g., NPF/CAC-3/MSL1 in Neurospora and Msl1 or Ccc1 in Cryptococcus, resulted in minor growth reduction, presumably because these subunits are also found in several other complexes involved in chromatin regulation [34,35]. The important animal pathogens, Candida albicans and Aspergillus fumigatus, lack core PRC2 components and no H3K27 methylation has been detected [10]. So far, C. neoformans and Fusarium oxysporum are the only animal pathogens among the fungi in which H3K27me3 has been demonstrated [34,109], even though the role of facultative heterochromatin in pathogenesis has not been directly studied. In Cryptococcus, Msl1 is required for multiple virulence-associated phenotypes, including thermotolerance, cell wall stability, and melanin production, as well as full virulence in a murine infection model [46]. Loss of Hda1 causes defects in multiple virulence-associated phenotypes similar to MSL1 deletion, reduced survival in murine macrophages, and decreased virulence in a murine infection model [110]. These results emphasize the importance of determining the extent to which PRC2 influences pathogenesis. Taken together, all available evidence suggests that H3K27me3 has roles in growth and development under certain conditions in all fungi that have been studied, though the effects have been most obvious in the genus Fusarium. Furthermore, functional studies in pathogenic or symbiotic fungi have generally focused on specific and regional effects, so direct evidence for genome-wide chromatin changes during host infection remains scant.
5.3. H3K27me3 as a Driver for Genome Plasticity
Genome integrity is essential for the maintenance of cellular function and viability. Genome instability is a phenomenon that is frequently observed in malfunctioning cells associated with disease in many eukaryotes but has also been reported as advantageous for rapidly adapting organisms [111,112]. This suggests that genetic variability offers fitness advantages in certain environments that are selected for under specific conditions.
Pathogenic fungi are prominent examples of organisms with a high degree of genetic diversity [113]. Studies with human and plant pathogens showed extraordinary genome dynamics upon exposure to environmental stressors (e.g., fungicides) resulting in adaptation and increased fitness [114,115,116,117]. An important challenge in understanding genome dynamics is to elucidate mechanisms that confer or promote genetic variability.
Population genetic studies demonstrated that DNA sequences subject to rearrangements and instability are not randomly distributed within the genome, but rather specific regions show higher variability than others. Sequences located close to chromosomal ends, the subtelomeric regions, and sequences harboring active or mutated transposable elements (TEs) show a high degree of diversity, even between isolates of the same species [120,121,122]. These differences include a higher density of SNPs, presence/absence polymorphisms of genes and TEs, or structural rearrangements (Figure 3). Furthermore, various fungal plant pathogens contain conditionally dispensable, “accessory” chromosomes or sections of chromosomes, that, in contrast to the “core” genome, are present in only some isolates and sometimes of benefit to the species under certain environmental conditions [123,124,125]. Often, variation in these regions or chromosomes affects genes that are involved in rapid adaptation, e.g., pathogenicity factors or biosynthetic gene clusters that are required for the production of secondary metabolites [126,127,128,129].
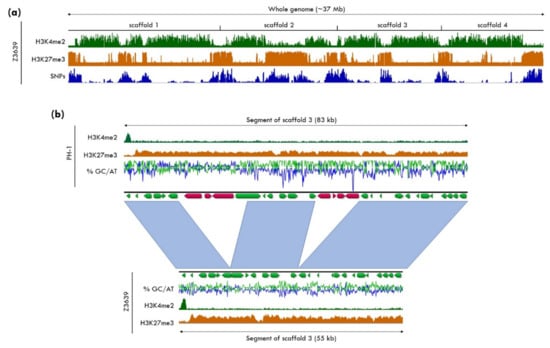
Figure 3.
Variability of H3K27me3 regions in different Fusarium graminearum isolates. (a) The genome of F. graminearum isolate Z3639 consists of four chromosomes (scaffolds 1–4) that are largely divided into blocks of euchromatin, marked by H3K4me2, and facultative heterochromatin, marked by H3K27me3. Genome comparison between the two newly completed F. graminearum genomes from two closely related isolates, Z3639, and the reference strain, PH-1 [118,119], revealed increased SNP density in H3K27me3-marked regions compared to euchromatic regions of the genome, as previously suggested [32,118]. (b) Sequence comparison within a large H3K27me3-enriched region between PH-1 and Z3639 shows that overall synteny between H3K27me3 regions is largely conserved between isolates but insertions and deletions of genes are frequently found in these regions. Note that scaffolds are arranged by size, so that “scaffold 3” refers to chromosome 4 and “scaffold 4” refers to chromosome 3 of previous genome assemblies. Complete genomes of Z3639 and PH-1 were sequenced and assembled in collaboration with the JGI, and complete ChIP-seq data sets from Z3639 will be part of a separate publication (Ridenour, Möller, and Freitag, in preparation).
Chromatin organization differs between highly variable regions and the more conserved core regions of genomes, and all evidence shows that H3K27 methylation is enriched in subtelomeric regions and on accessory chromosomes (Figure 1). There is mounting evidence linking both constitutive and facultative heterochromatin and genome organization within nuclei of filamentous fungi. Hi-C, a method to investigate three-dimensional genome organization, showed that in N. crassa, a mutant karyopherin (DIM-3) involved in the assembly of constitutive heterochromatin changed average chromosome structure [130], while a set-7/kmt6 deletion mutant resulted in the movement of subtelomeric regions to the interior of the nucleus and occasional disruption of centrochromatin, measured by the appearance of several CenH3 foci, rather than the usual single focus [131], in each nucleus [130,132]. Hi-C and RNA-seq were also used to study how the genome structure of E. festucae contributes to symbiosis with ryegrass [133]. Repetitive, AT-rich regions were considered to be in a more condensed chromatin state, and genes differentially expressed in planta were overrepresented near these regions. For example, the eas gene cluster (see Section 5.2) was found interspersed between AT-rich regions in a single topologically associated domain (TAD) in vitro [133], supporting the idea that facultative heterochromatin and genome organization have a role in regulating gene expression in a context-specific manner.
A series of studies documented that H3K27me3 is critical for conferring genome instability, which contributes to high genetic variability. In Z. tritici, H3K27me3-marked accessory chromosomes are highly unstable during both meiosis and mitosis; they are frequently lost from individuals but maintained in the population as a whole [36,117,134,135]. In the absence of H3K27me3 in kmt6 deletion mutants, the mitotic loss rate of accessory chromosomes decreased ~4-fold, suggesting a destabilizing effect caused by the presence of H3K27me3 [37]. Reintroduction of H3K27me3 in complementation strains restored loss rates back to the relatively high levels observed in the wild type (~7% of tested cells). Increased loss rates of an accessory chromosome, likely caused by increased H3K27me3 enrichment, were also reported in F. fujikuroi mutants that lacked the H3K36 methyltransferase, Ash1, supporting the hypothesis that the presence or imbalance of H3K27me3 decreases stability (the connection between methylation of H3K36 and H3K27 is discussed in Section 5.4) [136].
An imbalance of H3K27me3 as a consequence of loss of other histone marks that exclude H3K27me3 from specific chromosomal regions under standard lab conditions has been previously shown in N. crassa and C. neoformans. Genetic analyses revealed a genetic connection between constitutive and facultative heterochromatin in N. crassa [33]. Mutants defective in the H3K9 methylation system experience genotoxic stress, for example caused by methyl methanesulfonate (MMS) [137,138]. A selection scheme to isolate suppressors of this phenotype resulted in the identification of a deletion mutant of the PRC2 subunit, EED [33]; similar results were obtained by crossing existing single deletion strains of DIM-5/Kmt1 and SET-7/Kmt6 [139]. ChIP-seq analyses showed that H3K27me2/3 moved into regions previously occupied by H3K9me3 [33,139]. If only HP1 was lacking, constitutive heterochromatin was enriched with both H3K9me3 and H3K27me3 [139]. Mutants deficient in both H3K9 and H3K27 methylation rescued overt phenotypes, but accumulated γH2A and became more sensitive to camptothecin, a topoisomerase I inhibitor [33,139]. Combined, these results suggested that gaining H3K27me2/3 in regions that are usually enriched with H3K9me3 did not result in compensation for silencing of transposons or transcriptional dysfunction in regions that are usually occupied by H3K27me3, but rather that PRC2 modulates genotoxic stress genome-wide; almost all known transposons in N. crassa are inactivated by repeat-induced point mutation (RIP), which attracts H3K9me3, and, in turn, allows cytosine DNA methylation [140,141]. Similar results were obtained in C. neoformans. Mutants of ezh2/kmt6 and the novel ccc1 subunit were tested for conditional relocalization of H3K27me3 to constitutive heterochromatin; H3K27me3 moved into regions of constitutive heterochromatin in ccc1 mutants [34]. The authors proposed that binding of Ccc1 to H3K27me3 restricts relocalization, while in absence of Ccc1, EED binds to H3K9me3 resulting in H3K27me3 enrichment in H3K9me3 regions [34]. However, in N. crassa, the presence of H3K9me2/3 is not necessary for H3K27me3 relocalization, suggesting additional factors involved in this process. Recent reviews proposed models (see Figure 2) to explain these data [8,9].
Recent work in Z. tritici came to similar conclusions [37]. Loss of H3K9me3 in kmt1 deletion strains resulted in redistribution of H3K27me3 to regions that are normally covered by H3K9me3 [37]. Sequences that acquired H3K27me3 were shown to be involved in genome rearrangements in an evolution experiment that monitored genome stability in H3K27me3- and H3K9me3-deficient strains. However, in contrast to the situation in N. crassa, there was no increase in sensitivity to MMS in kmt1 deletion mutants, suggesting that even wild-type Z. tritici strains are exquisitely sensitive to MMS, at much lower concentrations than N. crassa, and that genotoxic effects are therefore difficult to compare. The chromosome loss in kmt1 deletion strains was twice as high as in the wild type, suggesting that relocalization of H3K27me3 resulted in genome destabilization, while removal of the redistributed H3K27me3 has a genome stabilizing effect because chromosome loss in the kmt1/kmt6 double deletion mutant was decreased to wild-type levels [37].
5.4. Interaction of H3K27me3 with Other Chromatin Marks
So far, relatively little is known about interactions of H3K27me3 with other chromatin marks and modifiers in fungi, but studies in flies [142] and mammals [143] demonstrated that H3K36 methylation can antagonize PRC2-mediated H3K27 methylation. In budding yeast, all H3K36 methylation is catalyzed by a single KMT, Set2, and while H3K36me3 is generally correlated with regions that are actively transcribed, the molecular effect of H3K36me3 is transcriptional repression [144]. Set2 associates with RNAP II and is involved in transcript quality control during both initiation and elongation [145]. In fission yeast, H3K36me3 reduces overall chromatin accessibility and promotes non-homologous end joining (NHEJ) in a cell cycle-specific manner [146]. A Neurospora set-2 mutant appeared to lack all H3K36 methylation by western blots, showed poor growth, poor conidiation, and was female sterile; an H3K36L substitution recapitulated these defects [147]. In F. graminearum, ChIP-seq showed that H3K36me3 was enriched at every single gene; euchromatic genes had H3K36me3 in 3’ regions of transcripts, while genes with H3K27me3 enrichment had H3K36me3 across the entire gene and the promoter [32].
Two recent studies in F. fujikuroi [136] and N. crassa [148] addressed the role of H3K36 methylation in transcriptional regulation, genome stability, and the relationship between H3K36me2/3 and facultative heterochromatin marked by H3K27me2/3. Many plants, fungi, and animals have additional methyltransferases involved in RNAP II-independent H3K36me2/3 [149]. In combination, N. crassa and F. fujikuroi Set2 and Ash1 are responsible for all H3K36me2/3. While the RNAP II-dependent Set2 marks euchromatic regions, Ash1-catalyzed H3K36me2/3 overlaps with poorly transcribed regions, specifically subtelomeric regions [148]. In the case of F. fujikuroi, Ash1-catalyzed H3K36me3 overlaps accessory chromosomes that are also characterized by the presence of H3K27me2/3 [136]. Loss of Ash1-, but not Set2-mediated H3K36me3, affected H3K27me2/3. In F. fujikuroi, deletion of Ash1 and subsequent regional loss of H3K36me2/3 increased H3K27me3 levels in several subtelomeric locations. Similarly, in N. crassa, loss of Ash1-mediated H3K36me2/3 enabled H3K27me2/3 to spread to neighboring sequences. Notably, if loss of H3K36me2/3 was accompanied by gene activation, H3K27me2/3 was also lost in some regions.
In F. fujikuroi, subtelomeric regions and an accessory chromosome, normally covered by H3K27me3 and Ash1-mediated H3K36me3, became unstable in ash1 deletion mutants as observed by chromosome breakage or even complete loss of the chromosome [136]. These findings indicate that RNAP II-independent H3K36 methylation is important to balance H3K27me2/3 levels, and that an increase and spread of H3K27me2/3 can have detrimental consequences for the cell.
Numerous studies in animals have addressed the interplay between 5-methylcytosine (5mC) and H3K27me2/3 distribution. H3K27me3 enrichment is reduced at normal target genes upon loss of 5mC, and relocalizes to loci with normally high 5mC levels [150], including imprinted genes [151], demethylated CpG islands (CGIs) [52], some TEs [152], and pericentric or centromeric regions [153], which can also involve PRC1 activity [95]. The major consequence that has been studied in animals is transcriptional reprogramming, which results in altered development or disease [154]. Similarly, H3K27me3 undergoes redistribution to demethylated transposons in Arabidopsis, mutants deficient for the CG-specific maintenance methyltransferase, MET1 [155]. One of the few accepted rules about recruitment of the two main PRC2 variants in animals is that they effectively bind unmethylated CGIs [17]. In N. crassa, the only fungus in which the relationship between 5mC and H3K27me3 has been investigated, loss of 5mC does not affect distribution of H3K27me2/3 [139].
So far, little is known about interactions of histone lysine acetylation (KAT) or deacetylation complexes, either HDACs or sirtuins, with PRC2. One of the two novel subunits of C. neoformans PRC2, Ccc1, is similar to proteins found in HDAC complexes and associates with the class II HDAC, Hda1 (Clr3); deletion of Hda1 results in reduced growth as observed for Ccc1 and de-repression of PRC2-silenced genes in subtelomeric regions [34,110]. While further work is clearly necessary, it is possible that PRC2/Ccc1 and HDACs co-operate in transcriptional regulation of developmental genes.
From this glimpse into connections between different chromatin marks, it is clear that our understanding of interactions between chromatin modifications and the function of H3K27 methylation is still rudimentary, and that it will take comparative studies in several organisms to uncover all existing connections.
5.5. Evolution of H3K27me3-Dependent Transcriptional Silencing from an Ancestral Immune System Function
Population genetic analyses as well as recent experimental work indicated that H3K27me3 is connected to sequence diversity and instability. An intriguing puzzle to solve in the near future is how H3K27me3 can confer instability on a molecular level, and why this mechanism evolved over evolutionary timescales. While functional studies on the molecular mechanism are still lacking, a recent study on early land plants suggested that H3K27me3 together with H3K9me3 might represent an ancient form of genome defense mechanism against transposons [156]. Regions enriched with H3K9me2/3 and DNA cytosine methylation are usually AT- and repeat-rich, and movement of H3K27me3 to these regions in absence of H3K9me3 hints to a compensatory genome defense mechanism that was previously proposed to act in animals [152]. Redundancy between PRC2 and the cytosine DNA methylation pathways ensures that transposons remain silent, yet the gain of H3K27 methylation in constitutive heterochromatin in N. crassa appeared deleterious rather than compensatory [8].
While H3K9me3 regions in fungi are AT-rich, this is not necessarily the case for H3K27me3 regions (Figure 4). Furthermore, the presence of H3K9me3 universally indicates the presence of an already silenced and inactivated transposon, whereas integration of an active transposon (and sometimes associated “hitch-hiking” genes) is predicted to recruit H3K9me3 only to AT-rich, repetitive regions [157,158,159] but not potentially harmful GC-rich genes. By comparing genomes and chromatin marks in different Z. tritici isolates, several H3K27me3-marked, isolate-specific regions were identified (Möller and Freitag, unpublished results). These were often flanked by H3K9me3-enriched regions and contained genes that are most similar to transposons or their “cargo” (Figure 4a). In contrast to the AT-rich H3K9me3 regions, the internal H3K27me3-covered regions harbored sequences that were slightly more GC-rich than the genome average. These sequences were transcriptionally repressed yet not completely silenced. Comparison of homologous loci in different Z. tritici isolates led to the hypothesis that H3K27me3 may function as an initial mechanism to mark “foreign” GC-rich DNA sequences to initiate transcriptional repression (Figure 4b). Over time, these H3K27me3 regions accumulate additional, mostly transition mutations that completely inactivate the now silenced genes, and eventually the GC content becomes so low that an H3K9me3 deposition mechanism similar to that envisioned in N. crassa [140] takes over, which also allows for wide-spread 5mC deposition [160,161] (Figure 4c). Isolate-specific, H3K27me3 regions should therefore be considered as meta-stable regions that, likely based on selection on the underlying genes, become permanently inactivated, neo-functionalized due to increased mutation rates, or conditionally repressed due to so far unknown dynamics of H3K27me3 silencing. This, or a similar system, may operate in other species as well, but detailed comparative studies in multiple strains of known lineages are lacking for other species. Detailed mechanistic studies on Z. tritici, F. graminearum, and N. crassa with several completely sequenced isolates is finally possible, and all three species are amenable to genetic, biochemical, and cytological studies.
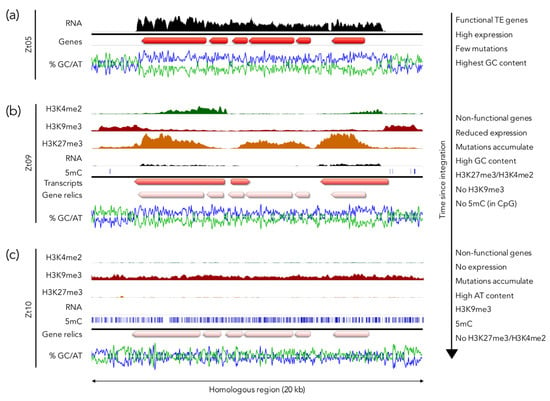
Figure 4.
H3K27me3 as part of an ancestral genome defense. Homologous regions on different chromosomes in three Z. tritici isolates illustrate a model for H3K27me3-mediated silencing and step-wise inactivation of GC-rich regions on a short evolutionary timescale. (a) A recent transposon insertion in a subtelomeric region in the isolate Zt05. The GC-rich gene cluster containing transposon-related genes is flanked by AT-rich repetitive sequences. The genes inside the cluster are highly expressed. ChIP-seq and 5mC data are currently not available for this isolate. (b) A homologous region in the isolate Zt09 is marked by H3K27me3 and H3K4me2 enrichment in the GC-rich region and H3K9me3 in the flanking AT-rich regions. Compared to Zt05, there are ~600 mutations, mainly transitions, in the GC-rich region. Some sequences are expressed but the expression level is relatively low, and the genes are likely non-functional due to the numerous mutations. Very few 5mC sites are present in H3K9me3 regions as this isolate is lacking the DNA methyltransferase Dim2 [162]. (c) The isolate Zt10 accumulated > 1000 mutations in a homologous region, decreasing the GC content to ~49% (~60% in Zt05). The entire region is covered by H3K9me3 and 5mC and there is no detectable transcriptional activity or H3K4me2. The isolate Zt10 contains a functional DNA methyltransferase Dim2 [162]. Data used to generate this figure are from [37,162,163], and Möller and Freitag, unpublished results.
6. Open Questions to be Resolved in the Near Future
How many different PRC2s exist in fungi? In animal cells there seem to be two core PRC2s (EZH1 or EZH2, plus EED and SUZ12) that result in the formation of a variety of subcomplexes [17]. In plants there are three EZH2 and three SUZ12 core subunits each that use a single EED homolog, yielding a minimum of three PRC2s [18]. These various complexes are expressed at different times and under varying conditions, having separate but partially overlapping functions. Biochemical studies have been carried out in N. crassa and C. neoformans but again only under standard laboratory growth conditions. Based on the number of putative DNA-binding “transcription factors” (from ~200 to ~700 in N. crassa and F. graminearum, respectively) and the increase in percentage of the genome enriched with H3K27me3 (from ~7 to ~32% in N. crassa and F. graminearum, respectively), we propose that there is an increased number of PRC2 adapter proteins that interact with SUZ12 to recruit PRC2 activity to its target regions in some fungi, e.g., F. graminearum. This large number of repressor complexes would be balanced by activator complexes that are coupled to HAT co-activator complexes.
How does H3K27me3 and facultative heterochromatin control fungal development? In plants and animals, H3K27me3 is involved in differentiation and development but no comprehensive time course studies across a complete life cycle have been carried out in fungi. This is an important knowledge gap that must be filled in the near future.
What happens to H3K27me3-enriched regions in pathogenic fungi in response to the host environment? We are largely naïve on this issue, as successful and routine in planta ChIP-seq is still extremely difficult. H3K27me3 covers more than 30% of the F. graminearum genome during growth in vitro, including regions enriched with pathogenesis-associated genes [32]. Many of the genes covered by H3K27me3 in vitro are derepressed in planta [32,164]; a similar situation has been observed in other pathogenic fungi (e.g., F. oxysporum and Verticillium dahliae [109,165]). This strongly suggests that fungal pathogens must redistribute H3K27me3 or relax transcriptional repression associated with H3K27me3 in response to the host environment to facilitate pathogenic development within the host. To enable future studies, we are designing a system that allows enrichment and purification of fungal nuclei from infected host plants, which will augment current efforts to determine how chromatin structure of plant pathogenic fungi changes in response to the host environment and how the host responds to this challenge.
How are chromatin domains organized in three-dimensional space within the nucleus? Existing Hi-C studies revealed that facultative heterochromatin has major impacts on nuclear organization, and we are currently only at the beginning of these studies. Hi-C investigations in double and triple mutants that are defective for several chromatin marks at the same time are underway.
How do we reconcile genome metastability, increase of point mutations, chromosome loss, and transcriptional effects at the evolutionary scale? Comparing existing evidence from all fungi demonstrates that transcriptional regulation is not the only role of PRC2. Using several strains of the same species and several species of the same genus is now possible in all four genera discussed here. Genome sequencing, resulting in complete genome assemblies, ChIP-seq, Hi-C, and RNA-seq, are now routine and no longer prohibitively expensive. We predict that numerous “lab evolution” studies will help to resolve the current uncertainties. To this end, we also have begun to investigate the mechanistic connections between spontaneous and induced DNA lesions, fixed mutations, replication, and H3K27me3 targeting. The surprising observation in Z. tritici suggesting that H3K27me3 generates a “metastable” genome makes this species an ideal model to uncouple transcriptional and genome stability effects in future studies.
Author Contributions
All authors were involved in conceptualization, writing, and editing of the review, and have read and agreed to the published version of the manuscript. Both MM and JBR provided original data for the writing of this manuscript; their contributions to this paper should be considered equal. All authors have read and agreed to the published version of the manuscript.
Funding
Research in the Freitag laboratory is funded by grants from the US National Science Foundation (NSF), grant numbers MCB 1515998 and MCB 1818006. JBR is supported by the USDA-AFRI Education and Workforce Development Program, grant number 2019-67012-29722. MM is funded by the Deutsche Forschungs-gemeinschaft (DFG), grant number MO 3755/1-1.
Acknowledgments
We gratefully acknowledge Allyson Erlendson for the original idea and design of our evolving working model, and all Freitag lab members for fruitful discussions. Strains Z3639 and PH-1 were sequenced in collaboration with the JGI (CSP504417); a separate publication is in preparation. We apologize to colleagues whose original work was not cited because of space limitations.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the analyses or interpretation of data, in the writing of the manuscript, or in the decision to publish results.
References
- Heitz, E. Das Heterochromatin der Moose I. Jahrbuecher Wiss. Bot. 1928, 69, 762–818. [Google Scholar]
- Allis, C.D. Epigenetics, 2nd ed.; Allis, C.D., Caparros, M.-L., Jenuwein, T., Reinberg, D., Lachner, M., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Freitag, M. The kinetochore interaction network (KIN) of ascomycetes. Mycologia 2016, 108, 485–505. [Google Scholar] [CrossRef][Green Version]
- Sullivan, B.; Karpen, G.H. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat. Struct. Mol. Boil. 2004, 11, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Grossniklaus, U.; Paro, R. Transcriptional Silencing by Polycomb-Group Proteins. Cold Spring Harb. Perspect. Boil. 2014, 6, a019331. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Phatale, P.A.; Bredeweg, E.L.; Connolly, L.R.; Pomraning, K.R.; Freitag, M. Epigenetics of Filamentous Fungi. In Encyclopedia of Molecular Cell Biology and Molecular Medicine; Wiley: Hoboken, NJ, USA, 2012; Volume 2, pp. 1063–1105. [Google Scholar]
- Lewis, Z.A. Polycomb Group Systems in Fungi: New Models for Understanding Polycomb Repressive Complex 2. Trends Genet. 2017, 33, 220–231. [Google Scholar] [CrossRef]
- Wiles, E.T.; Selker, E.U. H3K27 methylation: A promiscuous repressive chromatin mark. Curr. Opin. Genet. Dev. 2016, 43, 31–37. [Google Scholar] [CrossRef]
- Freitag, M. Histone Methylation by SET Domain Proteins in Fungi. Annu. Rev. Microbiol. 2017, 71, 413–439. [Google Scholar] [CrossRef]
- Żylicz, J.J.; Heard, E. Molecular Mechanisms of Facultative Heterochromatin Formation: An X-Chromosome Perspective. Annu. Rev. Biochem. 2020, 89. [Google Scholar] [CrossRef]
- Kuroda, M.I.; Kang, H.; De, S.; Kassis, J.A. Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming. Annu. Rev. Biochem. 2020, 89, 101146. [Google Scholar] [CrossRef]
- Hugues, A.; Jacobs, C.S.; Roudier, F. Mitotic Inheritance of PRC2-Mediated Silencing: Mechanistic Insights and Developmental Perspectives. Front. Plant. Sci. 2020, 11, 262. [Google Scholar] [CrossRef] [PubMed]
- Batista, R.A.; Köhler, C. Genomic imprinting in plants—revisiting existing models. Genome Res. 2020, 34, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Boil. 2019, 29, 660–671. [Google Scholar] [CrossRef]
- Schubert, D. Evolution of Polycomb-group function in the green lineage. F1000Research 2019, 8, 8. [Google Scholar] [CrossRef]
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18. [Google Scholar] [CrossRef]
- Pu, L.; Sung, Z.R. PcG and trxG in plants–friends or foes. Trends Genet. 2015, 31, 252–262. [Google Scholar] [CrossRef]
- Xiao, J.; Jin, R.; Yu, X.; Shen, M.; Wagner, J.D.; Pai, A.; Song, C.; Zhuang, M.; Klasfeld, S.; He, C.; et al. Cis and trans determinants of epigenetic silencing by Polycomb repressive complex 2 in Arabidopsis. Nat. Genet. 2017, 49, 1546–1552. [Google Scholar] [CrossRef]
- Smith, K.M.; Kothe, G.O.; Matsen, C.; Khlafallah, T.K.; Adhvaryu, K.K.; Hemphill, M.; Freitag, M.; Motamedi, M.; Selker, E.U. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics Chromatin 2008, 1, 5. [Google Scholar] [CrossRef]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef]
- Orlando, V.; Paro, R. Mapping polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 1993, 75, 1187–1198. [Google Scholar] [CrossRef]
- Schuettengruber, B.; Bourbon, H.-M.; Di Croce, L.; Cavalli, G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell 2017, 171, 34–57. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef]
- Cao, R.; Zhang, Y. SUZ12 Is Required for Both the Histone Methyltransferase Activity and the Silencing Function of the EED-EZH2 Complex. Mol. Cell 2004, 15, 57–67. [Google Scholar] [CrossRef]
- Erlendson, A.A.; Friedman, S.; Freitag, M. A Matter of Scale and Dimensions: Chromatin of Chromosome Landmarks in the Fungi. Fungal Kingd. 2017, 5, 571–597. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, X. Structural analysis of an active fungal PRC2. Nucleus 2016, 7, 284–291. [Google Scholar] [CrossRef]
- Jiao, L.; Liu, X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science 2015, 350, aac4383. [Google Scholar] [CrossRef]
- Kasinath, V.; Faini, M.; Poepsel, S.; Reif, D.; Feng, X.A.; Stjepanovic, G.; Aebersold, R.; Nogales, E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018, 359, 940–944. [Google Scholar] [CrossRef]
- Poepsel, S.; Kasinath, V.; Nogales, E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat. Struct. Mol. Boil. 2018, 25, 154–162. [Google Scholar] [CrossRef]
- Ciferri, C.; Lander, G.C.; Maiolica, A.; Herzog, F.; Aebersold, R.; Nogales, E. Molecular architecture of human polycomb repressive complex 2. ELife 2012, 1. [Google Scholar] [CrossRef]
- Connolly, L.R.; Smith, K.M.; Freitag, M. The Fusarium graminearum Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters. PLoS Genet. 2013, 9, e1003916. [Google Scholar] [CrossRef]
- Basenko, E.Y.; Sasaki, T.; Ji, L.; Prybol, C.J.; Burckhardt, R.M.; Schmitz, R.J.; Lewis, Z.A. Genome-wide redistribution of H3K27me3 is linked to genotoxic stress and defective growth. Proc. Natl. Acad. Sci. USA 2015, 112, E6339–E6348. [Google Scholar] [CrossRef] [PubMed]
- Dumesic, P.A.; Homer, C.M.; Moresco, J.J.; Pack, L.R.; Shanle, E.K.; Coyle, S.M.; Strahl, B.D.; Fujimori, D.G.; Yates, J.R.; Madhani, H.D. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell 2014, 160, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Jamieson, K.; Rountree, M.; Lewis, Z.A.; Stajich, J.E.; Selker, E.U. Regional control of histone H3 lysine 27 methylation in Neurospora. Proc. Natl. Acad. Sci. USA 2013, 110, 6027–6032. [Google Scholar] [CrossRef] [PubMed]
- Schotanus, K.; Soyer, J.L.; Connolly, L.R.; Grandaubert, J.; Happel, P.; Smith, K.M.; Freitag, M.; Stukenbrock, E.H. Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin 2015, 8, 41. [Google Scholar] [CrossRef]
- Möller, M.; Schotanus, K.; Soyer, J.L.; Haueisen, J.; Happ, K.; Stralucke, M.; Happel, P.; Smith, K.M.; Connolly, L.R.; Freitag, M.; et al. Destabilization of chromosome structure by histone H3 lysine 27 methylation. PLoS Genet. 2019, 15, e1008093. [Google Scholar] [CrossRef]
- Adpressa, D.A.; Connolly, L.R.; Konkel, Z.M.; Neuhaus, G.F.; Chang, X.L.; Pierce, B.R.; Smith, K.M.; Freitag, M.; Loesgen, S. A metabolomics-guided approach to discover Fusarium graminearum metabolites after removal of a repressive histone modification. Fungal Genet. Boil. 2019, 132, 103256. [Google Scholar] [CrossRef]
- Hansen, K.; Bracken, A.P.; Pasini, D.; Dietrich, N.; Gehani, S.S.; Monrad, A.; Rappsilber, J.; Lerdrup, M.; Helin, K. A model for transmission of the H3K27me3 epigenetic mark. Nature 2008, 10, 1291–1300. [Google Scholar] [CrossRef]
- Margueron, R.; Justin, N.; Ohno, K.; Sharpe, M.; Son, J.; Drury, W.J.; Voigt, P.; Martin, S.R.; Taylor, W.R.; De Marco, V.; et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 2009, 461, 762–767. [Google Scholar] [CrossRef]
- Petruk, S.; Sedkov, Y.; Johnston, D.M.; Hodgson, J.W.; Black, K.L.; Kovermann, S.K.; Beck, S.; Canaani, E.; Brock, H.W.; Mazo, A. TrxG and PcG Proteins but Not Methylated Histones Remain Associated with DNA through Replication. Cell 2012, 150, 922–933. [Google Scholar] [CrossRef]
- Del Olmo, I.; Lopez, J.A.; Vázquez, J.; Raynaud, C.; Piñeiro, M.; Jarillo, J.A. Arabidopsis DNA polymerase ϵ recruits components of Polycomb repressor complex to mediate epigenetic gene silencing. Nucleic Acids Res. 2016, 44, 5597–5614. [Google Scholar] [CrossRef]
- Reinberg, D.; Vales, L.D. Chromatin domains rich in inheritance. Science 2018, 361, 33–34. [Google Scholar] [CrossRef]
- Jiang, D.; Berger, F. DNA replication–coupled histone modification maintains Polycomb gene silencing in plants. Science 2017, 357, 1146–1149. [Google Scholar] [CrossRef]
- Cheng, L.; Zhang, X.; Wang, Y.; Gan, H.; Xu, X.; Lv, X.; Hua, X.; Que, J.; Ordog, T.; Zhang, Z. Chromatin Assembly Factor 1 (CAF-1) facilitates the establishment of facultative heterochromatin during pluripotency exit. Nucleic Acids Res. 2019, 47, 11114–11131. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.-H.; Maeng, S.; Strain, A.K.; Floyd, A.; Nielsen, K.; Heitman, J.; Bahn, Y.-S. Pleiotropic Roles of the Msi1-Like Protein Msl1 in Cryptococcus neoformans. Eukaryot. Cell 2012, 11, 1482–1495. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, H.; Bishop, B.; Ringenberg, W.; Muir, W.M.; Ogas, J. The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant. Physiol. 2012, 159, 418–432. [Google Scholar] [CrossRef]
- Holoch, D.; Margueron, R. Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem. Sci. 2017, 42, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.T.; Struhl, G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science 2017, 356, eaai8236. [Google Scholar] [CrossRef]
- Laprell, F.; Finkl, K.; Müller, J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science 2017, 356, 85–88. [Google Scholar] [CrossRef]
- Mendenhall, E.; Koche, R.P.; Truong, T.; Zhou, V.W.; Issac, B.; Chi, A.S.; Ku, M.; Bernstein, B.E. GC-Rich Sequence Elements Recruit PRC2 in Mammalian ES Cells. PLoS Genet. 2010, 6, e1001244. [Google Scholar] [CrossRef]
- Lynch, M.D.; Smith, A.J.H.; De Gobbi, M.; Flenley, M.; Hughes, J.R.; Vernimmen, U.; Ayyub, H.; A Sharpe, J.; A Sloane-Stanley, J.; Sutherland, L.; et al. An interspecies analysis reveals a key role for unmethylated CpG dinucleotides in vertebrate Polycomb complex recruitment. EMBO J. 2011, 31, 317–329. [Google Scholar] [CrossRef]
- Klose, R.J.; Cooper, S.; Farcas, A.M.; Blackledge, N.P.; Brockdorff, N. Chromatin Sampling—An Emerging Perspective on Targeting Polycomb Repressor Proteins. PLoS Genet. 2013, 9, e1003717. [Google Scholar] [CrossRef] [PubMed]
- Riising, E.M.; Comet, I.; Leblanc, B.; Wu, X.; Johansen, J.V.; Helin, K. Gene Silencing Triggers Polycomb Repressive Complex 2 Recruitment to CpG Islands Genome Wide. Mol. Cell 2014, 55, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Hojfeldt, J.; Laugesen, A.; Willumsen, B.M.; Damhofer, H.; Hedehus, L.; Tvardovskiy, A.; Mohammad, F.; Jensen, O.N.; Helin, K. Accurate H3K27 methylation can be established de novo by SUZ12-directed PRC2. Nat. Struct. Mol. Boil. 2018, 25, 225–232. [Google Scholar] [CrossRef]
- Jamieson, K.; McNaught, K.J.; Ormsby, T.; Leggett, N.; Honda, S.; Selker, E.U. Telomere repeats induce domains of H3K27 methylation in Neurospora. ELife 2018, 7. [Google Scholar] [CrossRef]
- McNaught, K.J.; Wiles, E.T.; Selker, E.U. Identification of a PRC2 Accessory Subunit Required for Subtelomeric H3K27 Methylation in Neurospora crassa. Mol. Cell. Boil. 2020, 40, 40. [Google Scholar] [CrossRef]
- Wiles, E.T.; McNaught, K.J.; Kaur, G.; Selker, J.M.L.; Ormsby, T.; Aravind, L.; Selker, E.U. Evolutionarily ancient BAH–PHD protein mediates Polycomb silencing. Proc. Natl Acad. Sci. USA 2020, 117, 11614–11623. [Google Scholar] [CrossRef]
- Yang, N.; Xu, R.-M. Structure and function of the BAH domain in chromatin biology. Crit. Rev. Biochem. Mol. Boil. 2012, 48, 211–221. [Google Scholar] [CrossRef]
- Qian, S.; Lv, X.; Scheid, R.N.; Lu, L.; Yang, Z.; Chen, W.; Liu, R.; Boersma, M.D.; Denu, J.M.; Zhong, X.; et al. Dual recognition of H3K4me3 and H3K27me3 by a plant histone reader SHL. Nat. Commun. 2018, 9, 2425. [Google Scholar] [CrossRef]
- Yang, Z.; Qian, S.; Scheid, R.N.; Lu, L.; Chen, X.; Liu, R.; Du, X.; Lv, X.; Boersma, M.D.; Scalf, M.; et al. EBS is a bivalent histone reader that regulates floral phase transition in Arabidopsis. Nat. Genet. 2018, 50, 1247–1253. [Google Scholar] [CrossRef]
- Govindaraghavan, M.; Anglin, S.L.; Osmani, A.H.; Osmani, S.A. The Set1/COMPASS Histone H3 Methyltransferase Helps Regulate Mitosis with the CDK1 and NIMA Mitotic Kinases in Aspergillus nidulans. Genetics 2014, 197, 1225–1236. [Google Scholar] [CrossRef]
- Miller, T.; Krogan, N.J.; Dover, J.; Erdjument-Bromage, H.; Tempst, P.; Johnston, M.; Greenblatt, J.F.; Shilatifard, A. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. USA 2001, 98, 12902–12907. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, Y.B.; Pirrotta, V. A new world of Polycombs: Unexpected partnerships and emerging functions. Nat. Rev. Genet. 2013, 14, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Hauk, G.; Berger, J.M. The role of ATP-dependent machines in regulating genome topology. Curr. Opin. Struct. Boil. 2016, 36, 85–96. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.Y.; Johnson, S.L.; Gamarra, N.; Narlikar, G.J. Mechanisms of ATP-Dependent Chromatin Remodeling Motors. Annu. Rev. Biophys. 2016, 45, 153–181. [Google Scholar] [CrossRef] [PubMed]
- Ho, K.K.; Zhang, H.; Golden, B.L.; Ogas, J. PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1829, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.; Puangsomlee, P.; Martin, M.; Long, D.; Meyerowitz, E.M.; Coupland, G. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 1997, 386, 44–51. [Google Scholar] [CrossRef] [PubMed]
- Eshed, Y.; Baum, S.F.; Bowman, J.L. Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 1999, 99, 199–209. [Google Scholar] [CrossRef]
- Ogas, J.; Kaufmann, S.; Henderson, J.; Somerville, C. PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 1999, 96, 13839–13844. [Google Scholar] [CrossRef]
- Zhang, H.; Rider, S.D.; Henderson, J.T.; Fountain, M.; Chuang, K.; Kandachar, V.; Simons, A.; Edenberg, H.J.; Romero-Severson, J.; Muir, W.M.; et al. The CHD3 Remodeler PICKLE promotes Trimethylation of Histone H3 Lysine 27. J. Boil. Chem. 2008, 283, 22637–22648. [Google Scholar] [CrossRef]
- Aichinger, E.; Villar, C.B.; Di Mambro, R.; Sabatini, S.; Köhler, C. The CHD3 Chromatin Remodeler PICKLE and Polycomb Group Proteins Antagonistically Regulate Meristem Activity in the Arabidopsis Root[C][W]. Plant. Cell 2011, 23, 1047–1060. [Google Scholar] [CrossRef]
- Carter, B.; Bishop, B.; Ho, K.K.; Huang, R.; Jia, W.; Zhang, H.; Pascuzzi, P.E.; Deal, R.B.; Ogas, J. The Chromatin Remodelers PKL and PIE1 Act in an Epigenetic Pathway That Determines H3K27me3 Homeostasis in Arabidopsis. Plant. Cell 2018, 30, 1337–1352. [Google Scholar] [CrossRef]
- Courtney, A.J.; Kamei, M.; Ferraro, A.R.; Gai, K.; He, Q.; Honda, S.; Lewis, Z.A. The histone variant H2A.Z is required to establish normal patterns of H3K27 methylation in Neurospora crassa. bioRxiv 2020. [Google Scholar] [CrossRef]
- Kadoch, C.; Copeland, R.A.; Keilhack, H. PRC2 and SWI/SNF Chromatin Remodeling Complexes in Health and Disease. Biochemistry 2016, 55, 1600–1614. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Brien, G.L.; Verrijzer, C.P. Dangerous liaisons: Interplay between SWI/SNF, NuRD, and Polycomb in chromatin regulation and cancer. Genome Res. 2019, 33, 936–959. [Google Scholar] [CrossRef]
- Egan, C.M.; Nyman, U.; Skotte, J.; Streubel, G.; Turner, S.; O’Connell, D.J.; Rraklli, V.; Dolan, M.J.; Chadderton, N.; Hansen, K.; et al. CHD5 is required for neurogenesis and has a dual role in facilitating gene expression and polycomb gene repression. Dev. Cell. 2013, 26, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Kuo, A.; Schalch, T.; Vogel, H.; Joshua-Tor, L.; McCombie, W.R.; Gozani, O.; Hammell, M.G.; Mills, A.A. Chd5 Requires PHD-Mediated Histone 3 Binding for Tumor Suppression. Cell Rep. 2013, 3, 92–102. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Torchy, M.; Hamiche, A.; Klaholz, B. Structure and function insights into the NuRD chromatin remodeling complex. Cell. Mol. Life Sci. 2015, 72, 2491–2507. [Google Scholar] [CrossRef] [PubMed]
- Kolla, V.; Naraparaju, K.; Zhuang, T.; Higashi, M.; Kolla, S.; Blobel, G.A.; Brodeur, G.M. The tumour suppressor CHD5 forms a NuRD-type chromatin remodelling complex. Biochem. J. 2015, 468, 345–352. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Lee, R.S.; Alver, B.H.; Haswell, J.R.; Wang, S.; Mieczkowski, J.; Drier, Y.; Gillespie, S.M.; Archer, T.C.; Wu, J.N.; et al. SMARCB1-mediated SWI/SNF complex function is essential for enhancer regulation. Nat. Genet. 2016, 49, 289–295. [Google Scholar] [CrossRef]
- Stanton, B.Z.; Hodges, H.C.; Calarco, J.P.; Braun, S.; Ku, W.L.; Kadoch, C.; Zhao, K.; Crabtree, G.R. SMARCA4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat. Genet. 2016, 49, 282–288. [Google Scholar] [CrossRef]
- Nakayama, R.T.; Pulice, J.; Valencia, A.; McBride, M.; McKenzie, Z.M.; Gillespie, M.A.; Ku, W.L.; Teng, M.; Cui, K.; Williams, R.; et al. SMARCB1 is required for widespread BAF complex–mediated activation of enhancers and bivalent promoters. Nat. Genet. 2017, 49, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Kadoch, C.; Williams, R.; Calarco, J.P.; Miller, E.L.; Weber, C.M.; Braun, S.; Pulice, J.; Chory, E.J.; Crabtree, G.R. Dynamics of BAF–Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat. Genet. 2016, 49, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Borkovich, K.A.; Alex, L.A.; Yarden, O.; Freitag, M.; Turner, G.E.; Read, N.; Seiler, S.; Bell-Pedersen, D.; Paietta, J.; Plesofsky, N.; et al. Lessons from the Genome Sequence of Neurospora crassa: Tracing the Path from Genomic Blueprint to Multicellular Organism. Microbiol. Mol. Boil. Rev. 2004, 68, 1–108. [Google Scholar] [CrossRef]
- Lagarou, A.; Mohd-Sarip, A.; Moshkin, Y.M.; Chalkley, G.E.; Bezstarosti, K.; Demmers, J.A.A.; Verrijzer, C.P. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genome Res. 2008, 22, 2799–2810. [Google Scholar] [CrossRef]
- Gao, Z.; Zhang, J.; Bonasio, R.; Strino, F.; Sawai, A.; Parisi, F.; Kluger, Y.; Reinberg, D. PCGF Homologs, CBX Proteins, and RYBP Define Functionally Distinct PRC1 Family Complexes. Mol. Cell 2012, 45, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Blackledge, N.P.; Farcas, A.M.; Kondo, T.; King, H.W.; McGouran, J.F.; Hanssen, L.L.; Ito, S.; Cooper, S.; Kondo, K.; Koseki, Y.; et al. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 2014, 157, 1445–1459. [Google Scholar] [CrossRef]
- Leeb, M.; Pasini, D.; Novatchkova, M.; Jaritz, M.; Helin, K.; Wutz, A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010, 24, 265–276. [Google Scholar] [CrossRef]
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678. [Google Scholar] [CrossRef]
- Farcas, A.M.; Blackledge, N.P.; Sudbery, I.; Long, H.; McGouran, J.F.; Rose, N.R.; Lee, S.; Sims, D.; Cerase, A.; Sheahan, T.W.; et al. KDM2B links the Polycomb Repressive Complex 1 (PRC1) to recognition of CpG islands. ELife 2012, 1. [Google Scholar] [CrossRef]
- He, J.; Shen, L.; Wan, M.; Taranova, O.; Wu, H.; Zhang, Y. Kdm2b maintains murine embryonic stem cell status by recruiting PRC1 complex to CpG islands of developmental genes. Nature 2013, 15, 373–384. [Google Scholar] [CrossRef]
- Wu, X.; Johansen, J.V.; Helin, K. Fbxl10/Kdm2b Recruits Polycomb Repressive Complex 1 to CpG Islands and Regulates H2A Ubiquitylation. Mol. Cell 2013, 49, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.T.C.; Müller, C.W.; Vermeulen, M.; Müller, J.; Müller, C.W. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Boil. 2014, 21, 569–571. [Google Scholar] [CrossRef] [PubMed]
- Cooper, S.; Dienstbier, M.; Hassan, R.; Schermelleh, L.; Sharif, J.; Blackledge, N.P.; De Marco, V.; Elderkin, S.; Koseki, H.; Klose, R.; et al. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep. 2014, 7, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Boil. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Shaver, S.; Mollano, J.A.C.; Cerny, R.L.; Cerutti, H. Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics 2010, 5, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Tong, A.; Yan, B.; Wang, X. Governing the Silencing State of Chromatin: The Roles of Polycomb Repressive Complex 1 in Arabidopsis. Plant. Cell Physiol. 2017, 58, 198–206. [Google Scholar] [CrossRef]
- Zhou, Y.; Tergemina, E.; Cui, H.; Förderer, A.; Hartwig, B.; James, G.V.; Schneeberger, K.; Turck, F. Ctf4-related protein recruits LHP1-PRC2 to maintain H3K27me3 levels in dividing cells in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 4833–4838. [Google Scholar] [CrossRef]
- Fischle, W.; Wang, Y.; Jacobs, S.A.; Kim, Y.; Allis, C.D.; Khorasanizadeh, S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003, 17, 1870–1881. [Google Scholar] [CrossRef]
- Honda, S.; Bicocca, V.; Gessaman, J.; Rountree, M.; Yokoyama, A.; Yu, E.Y.; Selker, J.M.L.; Selker, E.U. Dual chromatin recognition by the histone deacetylase complex HCHC is required for proper DNA methylation in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2016, 113, E6135–E6144. [Google Scholar] [CrossRef]
- Li, Z.; Fu, X.; Wang, Y.; Liu, R.; He, Y. Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat. Genet. 2018, 50, 1254–1261. [Google Scholar] [CrossRef]
- Studt, L.; Rösler, S.M.; Burkhardt, I.; Arndt, B.; Freitag, M.; Humpf, H.-U.; Dickschat, J.S.; Tudzynski, B. Knock-down of the methyltransferase Kmt6 relieves H3K27me3 and results in induction of cryptic and otherwise silent secondary metabolite gene clusters in Fusarium fujikuroi. Environ. Microbiol. 2016, 18, 4037–4054. [Google Scholar] [CrossRef]
- Pham, K.T.; Inoue, Y.; Vu, B.V.; Nguyen, H.H.; Nakayashiki, T.; Ikeda, K.; Nakayashiki, H. MoSET1 (Histone H3K4 Methyltransferase in Magnaporthe oryzae) Regulates Global Gene Expression during Infection-Related Morphogenesis. PLoS Genet. 2015, 11, e1005385. [Google Scholar] [CrossRef]
- Chujo, T.; Scott, B. Histone H3K9 and H3K27 methylation regulates fungal alkaloid biosynthesis in a fungal endophyte-plant symbiosis. Mol. Microbiol. 2014, 92, 413–434. [Google Scholar] [CrossRef]
- Tanaka, A.; Takemoto, D.; Chujo, T.; Scott, B. Fungal endophytes of grasses. Curr. Opin. Plant. Boil. 2012, 15, 462–468. [Google Scholar] [CrossRef]
- Schardl, C.L.; Young, C.A.; Hesse, U.; Amyotte, S.G.; Andreeva, K.; Calie, P.J.; Fleetwood, D.J.; Haws, D.C.; Moore, N.; Oeser, B.; et al. Plant-Symbiotic Fungi as Chemical Engineers: Multi-Genome Analysis of the Clavicipitaceae Reveals Dynamics of Alkaloid Loci. PLoS Genet. 2013, 9, e1003323. [Google Scholar] [CrossRef]
- Chettri, P.; Dupont, P.; Bradshaw, R. Chromatin-level regulation of the fragmented dothistromin gene cluster in the forest pathogen Dothistroma septosporum. Mol. Microbiol. 2018, 107, 508–522. [Google Scholar] [CrossRef]
- Fokkens, L.; Shahi, S.; Connolly, L.R.; Stam, R.; Schmidt, S.M.; Smith, K.M.; Freitag, M.; Rep, M. The multi-speed genome of Fusarium oxysporum reveals association of histone modifications with sequence divergence and footprints of past horizontal chromosome transfer events. bioRxiv 2018. [CrossRef]
- Brandao, F.; Esher, S.K.; Ost, K.S.; Pianalto, K.; Nichols, C.B.; Fernandes, L.; Bocca, A.L.; Poças-Fonseca, M.J.; Alspaugh, J.A. HDAC genes play distinct and redundant roles in Cryptococcus neoformans virulence. Sci. Rep. 2018, 8, 5209. [Google Scholar] [CrossRef]
- Bennett, R.J.; Forche, A.; Berman, J. Rapid Mechanisms for Generating Genome Diversity: Whole Ploidy Shifts, Aneuploidy, and Loss of Heterozygosity. Cold Spring Harb. Perspect. Med. 2014, 4, a019604. [Google Scholar] [CrossRef]
- Lee, J.J.-K.; Choi, Y.-L.; Kwon, M.; Park, P.J. Mechanisms and Consequences of Cancer Genome Instability: Lessons from Genome Sequencing Studies. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 283–312. [Google Scholar] [CrossRef]
- Möller, M.; Stukenbrock, E.H. Evolution and genome architecture in fungal plant pathogens. Nat. Rev. Genet. 2017, 15, 756–771. [Google Scholar] [CrossRef] [PubMed]
- Poláková, S.; Blume, C.; Zárate, J.Á.; Mentel, M.; Jørck-Ramberg, D.; Stenderup, J.; Piškur, J. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl. Acad. Sci. USA 2009, 106, 2688–2693. [Google Scholar] [CrossRef]
- Selmecki, A.; Forche, A.; Berman, J. Genomic Plasticity of the Human Fungal Pathogen Candida albicans. Eukaryot. Cell 2010, 9, 991–1008. [Google Scholar] [CrossRef] [PubMed]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R.D.; et al. Polyploidy can drive rapid adaptation in yeast. Nature 2015, 519, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Möller, M.; Habig, M.; Freitag, M.; Stukenbrock, E.H. Extraordinary Genome Instability and Widespread Chromosome Rearrangements during Vegetative Growth. Genetics 2018, 210, 517–529. [Google Scholar] [CrossRef] [PubMed]
- Cuomo, C.A.; Güldener, U.; Xu, J.-R.; Trail, F.; Turgeon, B.G.; Di Pietro, A.; Walton, J.D.; Ma, L.-J.; Baker, S.E.; Rep, M.; et al. The Fusarium graminearum Genome Reveals a Link between Localized Polymorphism and Pathogen Specialization. Science 2007, 317, 1400–1402. [Google Scholar] [CrossRef]
- King, R.; Urban, M.; Hammond-Kosack, M.C.U.; Hassani-Pak, K.; E Hammond-Kosack, K. The completed genome sequence of the pathogenic ascomycete fungus Fusarium graminearum. BMC Genom. 2015, 16, 544. [Google Scholar] [CrossRef]
- Badet, T.; Oggenfuss, U.; Abraham, L.; McDonald, B.A.; Croll, D. A 19-isolate reference-quality global pangenome for the fungal wheat pathogen Zymoseptoria tritici. BMC Boil. 2020, 18, 12–18. [Google Scholar] [CrossRef]
- Chiara, M.; Fanelli, F.; Mulè, G.; Logrieco, A.F.; Pesole, G.; Leslie, J.F.; Horner, D.S.; Toomajian, C. Genome Sequencing of Multiple Isolates Highlights Subtelomeric Genomic Diversity within Fusarium fujikuroi. Genome Boil. Evol. 2015, 7, 3062–3069. [Google Scholar] [CrossRef]
- Faino, L.; Seidl, M.F.; Shi-Kunne, X.; Pauper, M.; Berg, G.C.V.D.; Wittenberg, A.H.; Thomma, B.P.H.J. Transposons passively and actively contribute to evolution of the two-speed genome of a fungal pathogen. Genome Res. 2016, 26, 1091–1100. [Google Scholar] [CrossRef]
- Miao, V.; Covert, S.; VanEtten, H. A fungal gene for antibiotic resistance on a dispensable (“B”) chromosome. Science 1991, 254, 1773–1776. [Google Scholar] [CrossRef] [PubMed]
- Galazka, J.M.; Freitag, M. Variability of chromosome structure in pathogenic fungi--of ’ends and odds’. Curr. Opin. Microbiol. 2014, 20, 19–26. [Google Scholar] [CrossRef]
- Soyer, J.L.; Balesdent, M.-H.; Rouxel, T.; Dean, R.A. to B or not to B: A tale of unorthodox chromosomes. Curr. Opin. Microbiol. 2018, 46, 50–57. [Google Scholar] [CrossRef]
- Coleman, J.J.; Rounsley, S.D.; Rodriguez-Carres, M.; Kuo, A.; Wasmann, C.C.; Grimwood, J.; Schmutz, J.; Taga, M.; White, G.J.; Zhou, S.; et al. The Genome of Nectria haematococca: Contribution of Supernumerary Chromosomes to Gene Expansion. PLoS Genet. 2009, 5, e1000618. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.-J.; Van Der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.-J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.B.; Ben M’Barek, S.; Dhillon, B.; Wittenberg, A.H.J.; Crane, C.F.; Hane, J.K.; Foster, A.J.; Van Der Lee, T.A.J.; Grimwood, J.; Aerts, A.; et al. Finished Genome of the Fungal Wheat Pathogen Mycosphaerella graminicola Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis. PLoS Genet. 2011, 7, e1002070. [Google Scholar] [CrossRef]
- Balesdent, M.; Fudal, I.; Ollivier, B.; Bally, P.; Grandaubert, J.; Eber, F.; Chevre, A.-M.; Leflon, M.; Rouxel, T. The dispensable chromosome of Leptosphaeria maculans shelters an effector gene conferring avirulence towards Brassica rapa. New Phytol. 2013, 198, 887–898. [Google Scholar] [CrossRef]
- Galazka, J.M.; Klocko, A.; Uesaka, M.; Honda, S.; Selker, E.U.; Freitag, M. Neurospora chromosomes are organized by blocks of importin alpha-dependent heterochromatin that are largely independent of H3K9me3. Genome Res. 2016, 26, 1069–1080. [Google Scholar] [CrossRef]
- Smith, K.M.; Galazka, J.; Phatale, P.A.; Connolly, L.R.; Freitag, M. Centromeres of filamentous fungi. Chromosom. Res. 2012, 20, 635–656. [Google Scholar] [CrossRef]
- Klocko, A.; Ormsby, T.; Galazka, J.M.; Leggett, N.; Uesaka, M.; Honda, S.; Freitag, M.; Selker, E.U. Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa. Proc. Natl. Acad. Sci. USA 2016, 113, 15048–15053. [Google Scholar] [CrossRef]
- Winter, D.J.; Ganley, A.R.D.; Young, C.A.; Liachko, I.; Schardl, C.L.; Dupont, P.; Berry, D.; Ram, A.; Scott, B.; Cox, M.P. Repeat elements organise 3D genome structure and mediate transcription in the filamentous fungus Epichloë festucae. PLoS Genet. 2018, 14, e1007467. [Google Scholar] [CrossRef] [PubMed]
- Wittenberg, A.H.J.; Van Der Lee, T.A.J.; Ben M’Barek, S.; Ware, S.B.; Goodwin, S.; Kilian, A.; Visser, R.G.F.; Kema, G.H.J.; Schouten, H.J. Meiosis Drives Extraordinary Genome Plasticity in the Haploid Fungal Plant Pathogen Mycosphaerella graminicola. PLoS ONE 2009, 4, e5863. [Google Scholar] [CrossRef]
- Habig, M.; Kema, G.H.J.; Stukenbrock, E.H. Meiotic drive of female-inherited supernumerary chromosomes in a pathogenic fungus. ELife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Janevska, S.; Baumann, L.; Sieber, C.; Münsterkötter, M.; Ulrich, J.; Kämper, J.; Güldener, U.; Tudzynski, B. Elucidation of the Two H3K36me3 Histone Methyltransferases Set2 and Ash1 in Fusarium fujikuroi Unravels Their Different Chromosomal Targets and a Major Impact of Ash1 on Genome Stability. Genetics 2017, 208, 153–171. [Google Scholar] [CrossRef]
- Lewis, Z.A.; Adhvaryu, K.K.; Honda, S.; Shiver, A.L.; Knip, M.; Sack, R.; Selker, E.U. DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC. PLoS Genet. 2010, 6, e1001196. [Google Scholar] [CrossRef]
- Sasaki, T.; Lynch, K.L.; Mueller, C.V.; Friedman, S.; Freitag, M.; Lewis, Z.A. Heterochromatin Controls H2A Localization in Neurospora crassa. Eukaryot. Cell 2014, 13, 990–1000. [Google Scholar] [CrossRef]
- Jamieson, K.; Wiles, E.; McNaught, K.J.; Sidoli, S.; Leggett, N.; Shao, Y.; Garcia, B.A.; Selker, E.U. Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Res. 2015, 26, 97–107. [Google Scholar] [CrossRef]
- Rountree, M.; Selker, E.U. DNA methylation and the formation of heterochromatin in Neurospora crassa. Heredity 2010, 105, 38–44. [Google Scholar] [CrossRef]
- Selker, E.U. Premeiotic instability of repeated sequences in Neurospora crassa. Annu. Rev. Genet. 1990, 24, 579–613. [Google Scholar] [CrossRef]
- Klymenko, T.; Müller, J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004, 5, 373–377. [Google Scholar] [CrossRef]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 Methylation Antagonizes PRC2-mediated H3K27 Methylation. J. Boil. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef]
- Strahl, B.D.; Grant, P.A.; Briggs, S.D.; Sun, Z.-W.; Bone, J.R.; Caldwell, J.A.; Mollah, S.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; et al. Set2 Is a Nucleosomal Histone H3-Selective Methyltransferase That Mediates Transcriptional Repression. Mol. Cell. Boil. 2002, 22, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Veerappan, C.S.; Avramova, Z.; Moriyama, E.N. Evolution of SET-domain protein families in the unicellular and multicellular Ascomycota fungi. BMC Evol. Boil. 2008, 8, 190. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.-H.; Schulze, J.M.; Jackson, J.; Hentrich, T.; Seidel, C.; Jaspersen, S.L.; Kobor, M.S.; Shilatifard, A. Dot1 and Histone H3K79 Methylation in Natural Telomeric and HM Silencing. Mol. Cell 2011, 42, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Adhvaryu, K.K.; Morris, S.A.; Strahl, B.D.; Selker, E.U. Methylation of Histone H3 Lysine 36 Is Required for Normal Development in Neurospora crassa. Eukaryot. Cell 2005, 4, 1455–1464. [Google Scholar] [CrossRef]
- Bicocca, V.; Ormsby, T.; Adhvaryu, K.K.; Honda, S.; Selker, E.U. ASH1-catalyzed H3K36 methylation drives gene repression and marks H3K27me2/3-competent chromatin. ELife 2018, 7. [Google Scholar] [CrossRef]
- Wagner, E.J.; Carpenter, P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Boil. 2012, 13, 115–126. [Google Scholar] [CrossRef]
- Wu, H.; Coskun, V.; Tao, J.; Xie, W.; Ge, W.; Yoshikawa, K.; Li, E.; Zhang, Y.; Sun, Y.E. Dnmt3a-Dependent Nonpromoter DNA Methylation Facilitates Transcription of Neurogenic Genes. Science 2010, 329, 444–448. [Google Scholar] [CrossRef]
- Lindroth, A.M.; Park, Y.J.; McLean, C.M.; Dokshin, G.; Persson, J.; Herman, H.; Pasini, D.; Miró, X.; Donohoe, M.E.; Lee, J.T.; et al. Antagonism between DNA and H3K27 Methylation at the Imprinted Rasgrf1 Locus. PLoS Genet. 2008, 4, e1000145. [Google Scholar] [CrossRef]
- Walter, M.; Teissandier, A.; Pérez-Palacios, R.; Bourc’His, D. An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. ELife 2016, 5. [Google Scholar] [CrossRef]
- Saksouk, N.; Barth, T.K.; Ziegler-Birling, C.; Olova, N.; Nowak, A.; Rey, E.; Mateos-Langerak, J.; Urbach, S.; Reik, W.; Torres-Padilla, M.-E.; et al. Redundant Mechanisms to Form Silent Chromatin at Pericentromeric Regions Rely on BEND3 and DNA Methylation. Mol. Cell 2014, 56, 580–594. [Google Scholar] [CrossRef] [PubMed]
- Reddington, J.; Perricone, S.M.; Nestor, C.E.; Reichmann, J.; Youngson, N.A.; Suzuki, M.; Reinhardt, D.; Dunican, D.S.; Prendergast, J.G.D.; Mjoseng, H.K.; et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Boil. 2013, 14, R25. [Google Scholar] [CrossRef]
- Deleris, A.; Stroud, H.; Bernatavichute, Y.; Johnson, E.; Klein, G.; Schubert, D.; Jacobsen, S.E. Loss of the DNA Methyltransferase MET1 Induces H3K9 Hypermethylation at PcG Target Genes and Redistribution of H3K27 Trimethylation to Transposons in Arabidopsis thaliana. PLoS Genet. 2012, 8, e1003062. [Google Scholar] [CrossRef]
- Montgomery, S.A.; Tanizawa, Y.; Galik, B.; Wang, N.; Ito, T.; Mochizuki, T.; Akimcheva, S.; Bowman, J.L.; Cognat, V.; Maréchal-Drouard, L.; et al. Chromatin Organization in Early Land Plants Reveals an Ancestral Association between H3K27me3, Transposons, and Constitutive Heterochromatin. Curr. Boil. 2020, 30, 573–588.e7. [Google Scholar] [CrossRef]
- Tamaru, H.; Selker, E.U. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 2001, 414, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.A.; Honda, S.; Khlafallah, T.K.; Jeffress, J.K.; Freitag, M.; Mohn, F.; Schübeler, D.; Selker, E.U. Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome Res. 2008, 19, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Honda, S.; Selker, E.U. Direct Interaction between DNA Methyltransferase DIM-2 and HP1 Is Required for DNA Methylation in Neurospora crassa. Mol. Cell. Boil. 2008, 28, 6044–6055. [Google Scholar] [CrossRef]
- Honda, S.; Lewis, Z.A.; Shimada, K.; Fischle, W.; Sack, R.; Selker, E.U. Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat. Struct. Mol. Boil. 2012, 19, 471–477. [Google Scholar] [CrossRef]
- Freitag, M.; Hickey, P.C.; Khlafallah, T.K.; Read, N.; Selker, E.U. HP1 Is Essential for DNA Methylation in Neurospora. Mol. Cell 2004, 13, 427–434. [Google Scholar] [CrossRef]
- Möller, M.; Habig, M.; Lorrain, C.; Feurtey, A.; Haueisen, J.; Fagundes, W.C.; Alizadeh, A.; Freitag, M.; Stukenbrock, E.H. Recent loss of the Dim2 cytosine DNA methyltransferase impacts mutation rate and evolution in a fungal plant pathogen. bioRxiv 2020. [CrossRef]
- Haueisen, J.; Möller, M.; Eschenbrenner, C.J.; Grandaubert, J.; Seybold, H.; Adamiak, H.; Stukenbrock, E.H. Highly flexible infection programs in a specialized wheat pathogen. Ecol. Evol. 2018, 9, 275–294. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, C.; Wang, C.; Chen, C.; Xu, J.-R.; Liu, H. Characterization of the Two-Speed Subgenomes of Fusarium graminearum Reveals the Fast-Speed Subgenome Specialized for Adaption and Infection. Front. Plant. Sci. 2017, 8, 1968. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.E.; Kramer, M.; Seidl, M.F.; Thomma, B.P. Chromatin features define adaptive genomic regions in a fungal plant pathogen. bioRxiv 2020. [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).