Genetic Improvement of Cereals and Grain Legumes
Abstract
:Author Contributions
Funding
Conflicts of Interest
References
- Campbell, B.M.; Vermeulen, S.J.; Aggarwal, P.K.; Corner-Dolloff, C.; Girvetz, E.; Loboguerrero, A.M.; Ramirez-Villegas, J.; Rosenstock, T.; Sebastian, L.; Thornton, P.K. Reducing risks to food security from climate change. Glob. Food Secur. 2016, 11, 34–43. [Google Scholar] [CrossRef] [Green Version]
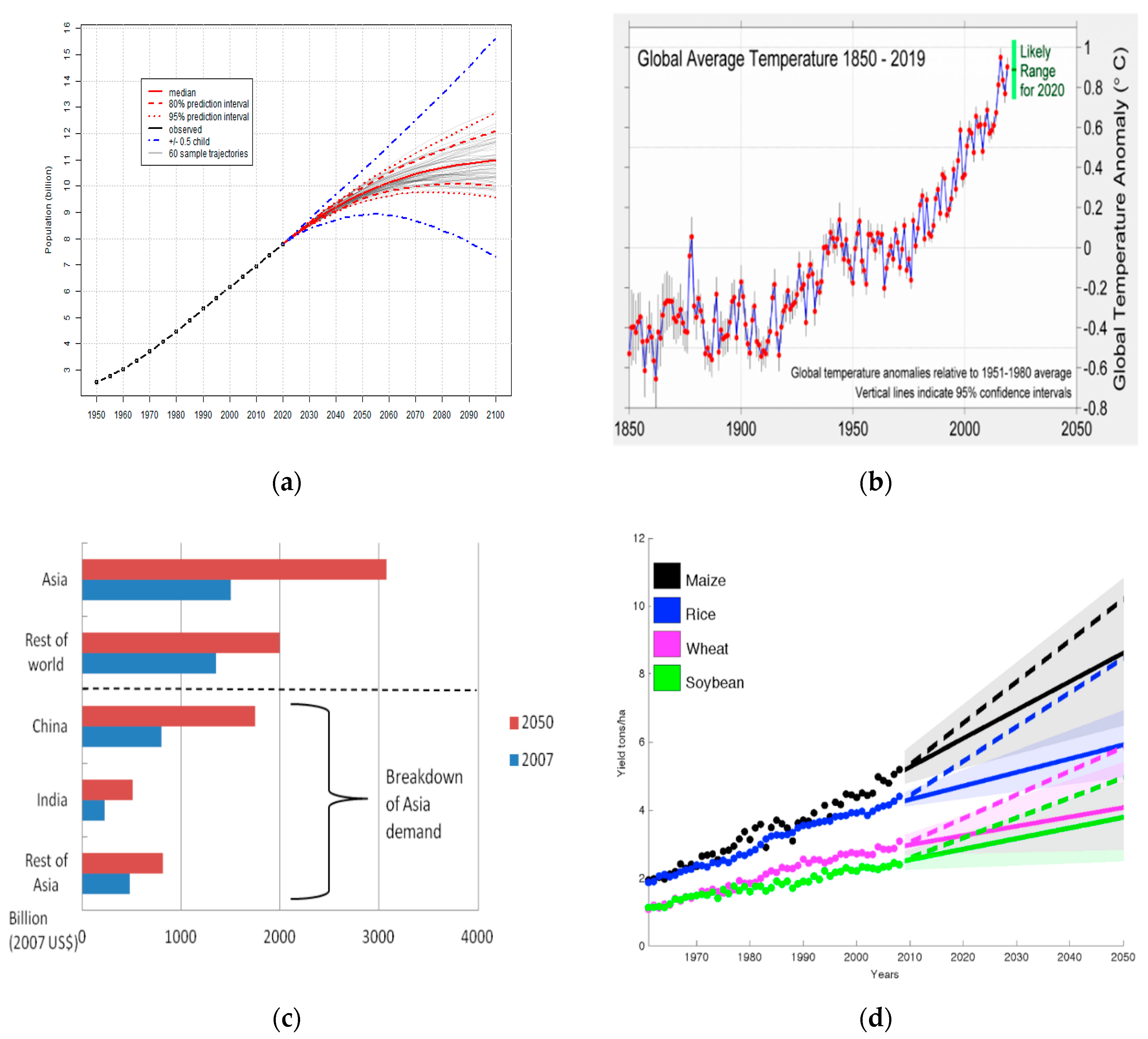
- Arora, N.K. Impact of Climate Change on Agriculture Production and Its Sustainable Solutions. Environ. Sustain. 2019, 2, 95–96. [Google Scholar] [CrossRef] [Green Version]
- Food and Agriculture Organization of the United Nations (FAO). High-level Expert Forum—How to Feed the World in 2050; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2009. [Google Scholar]
- Linehan, V.; Thorpe, S.; Andrews, N.; Beaini, F. Food demand to 2050: Opportunities for Australian agriculture–Algebraic description of agrifood model. In Proceedings of the Technical Annex to ABARES Outlook Conference Paper 12.4, Canberra, Australia, 6–7 March 2012. [Google Scholar]
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, e66428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challinor, A.J.; Watson, J.; Lobell, D.B.; Howden, S.; Smith, D.; Chhetri, N. A meta-analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 2014, 4, 287–291. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [Green Version]
- Dresselhaus, T.; Hückelhoven, R. Biotic and Abiotic Stress Responses in Crop Plants; Multidisciplinary Digital Publishing Institute: Basel, Switzerland, 2018. [Google Scholar]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Rasheed, A.; Hickey, L.T.; He, Z. Fast-forwarding genetic gain. Trends Plant Sci. 2018, 23, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Thudi, M.; Pandey, M.K.; Tardieu, F.; Ojiewo, C.; Vadez, V.; Whitbread, A.M.; Siddique, K.H.; Nguyen, H.T.; Carberry, P.S. Accelerating genetic gains in legumes for the development of prosperous smallholder agriculture: Integrating genomics, phenotyping, systems modelling and agronomy. J. Exp. Bot. 2018, 69, 3293–3312. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Li, R.; Xia, Y.; Bai, G.; Siddique, K.H.; Zhang, H.; Zheng, Y.; Yang, X.; Guo, P. Comparative Analysis of miRNA Expression Profiles between Heat-Tolerant and Heat-Sensitive Genotypes of Flowering Chinese Cabbage Under Heat Stress Using High-Throughput Sequencing. Genes 2020, 11, 264. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Hong, M.; Wan, H.; Luo, L.; Yu, Z.; Guo, R. Identification of Key Genes Involved in Embryo Development and Differential Oil Accumulation in Two Contrasting Maize Genotypes. Genes 2019, 10, 993. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wu, Q.; He, T.; Lan, J.; Ding, L.; Liu, T.; Wu, Q.; Pan, Y.; Chen, T. Transcriptomic and Metabolomic Changes Triggered by Fusarium solani in Common Bean (Phaseolus vulgaris L.). Genes 2020, 11, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heffner, E.L.; Lorenz, A.J.; Jannink, J.L.; Sorrells, M.E. Plant breeding with genomic selection: Gain per unit time and cost. Crop Sci. 2010, 50, 1681–1690. [Google Scholar] [CrossRef]
- Sorrells, M.E. Genomic selection in plants: Empirical results and implications for wheat breeding. In Advances in Wheat Genetics: From Genome to Field; Springer: Tokyo, Japan, 2015; pp. 401–409. [Google Scholar]
- Habyarimana, E.; Lopez-Cruz, M. Genomic Selection for Antioxidant Production in a Panel of Sorghum bicolor and S. bicolor× S. halepense Lines. Genes 2019, 10, 841. [Google Scholar] [CrossRef] [Green Version]
- Habyarimana, E.; Lopez-Cruz, M.; Baloch, F.S. Genomic Selection for Optimum Index with Dry Biomass Yield, Dry Mass Fraction of Fresh Material, and Plant Height in Biomass Sorghum. Genes 2020, 11, 61. [Google Scholar] [CrossRef] [Green Version]
- Kiranmayee, K.; Hash, C.T.; Sivasubramani, S.; Ramu, P.; Amindala, B.P.; Rathore, A.; Kishor, P.; Gupta, R.; Deshpande, S.P. Fine-Mapping of Sorghum Stay-Green QTL on Chromosome10 Revealed Genes Associated with Delayed Senescence. Genes 2020, 11, 1026. [Google Scholar] [CrossRef]
- Mace, E.; Singh, V.; Van Oosterom, E.; Hammer, G.; Hunt, C.; Jordan, D. QTL for nodal root angle in sorghum (Sorghum bicolor L. Moench) co-locate with QTL for traits associated with drought adaptation. Theor. Appl. Genet. 2012, 124, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, M.J.; Ismail, A.M.; McCouch, S.R.; Mackill, D.J. Marker assisted breeding. In Abiotic Stress Adaptation in Plants; Springer: Berlin/Heidelberg, Germany, 2009; pp. 451–469. [Google Scholar]
- Wani, S.H.; Choudhary, M.; Kumar, P.; Akram, N.A.; Surekha, C.; Ahmad, P.; Gosal, S.S. Marker-assisted breeding for abiotic stress tolerance in crop plants. In Biotechnologies of Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2018; Volume 3, pp. 1–23. [Google Scholar]
- Nadeem, M.A.; Gündoğdu, M.; Ercişli, S.; Karaköy, T.; Saracoğlu, O.; Habyarimana, E.; Lin, X.; Hatipoğlu, R.; Nawaz, M.A.; Sameeullah, M. Uncovering phenotypic diversity and DArTseq marker loci associated with antioxidant activity in common bean. Genes 2020, 11, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramalingam, J.; Savitha, P.; Alagarasna, G.; Renganathan, V.G.; Ramanathan, A.R.S. Improveent of stable restorer lines for blast resistance through functional marker in rice (Oryza sativa L.). Genes 2020, 11. in press. [Google Scholar]
- Macedo, R.; Sales, L.P.; Yoshida, F.; Silva-Abud, L.L.; Lobo, M. Potential worldwide distribution of Fusarium dry root rot in common beans based on the optimal environment for disease occurrence. PLoS ONE 2017, 12, e0187770. [Google Scholar] [CrossRef] [Green Version]
- D’Ippólito, S.; Martín, M.L.; Salcedo, M.F.; Atencio, H.M.; Casalongué, C.A.; Godoy, A.V.; Fiol, D.F. Transcriptome profiling of Fusarium solani f. sp. eumartii-infected potato tubers provides evidence of an inducible defense response. Physiol. Mol. Plant Pathol. 2010, 75, 3–12. [Google Scholar] [CrossRef]
- Hadwiger, L.A. Anatomy of a nonhost disease resistance response of pea to Fusarium solani: PR gene elicitation via DNase, chitosan and chromatin alterations. Front. Plant Sci. 2015, 6, 373. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhao, Q.; Cui, X.; Chen, R.; Li, X.; Qiu, B.; Ge, F. A transcriptome analysis uncovers Panax notoginseng resistance to Fusarium solani induced by methyl jasmonate. Genes Genom. 2019, 41, 1383–1396. [Google Scholar] [CrossRef]
- Voss-Fels, K.P.; Qian, L.; Gabur, I.; Obermeier, C.; Hickey, L.T.; Werner, C.R.; Kontowski, S.; Frisch, M.; Friedt, W.; Snowdon, R.J. Genetic insights into underground responses to Fusarium graminearum infection in wheat. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motameny, S.; Wolters, S.; Nürnberg, P.; Schumacher, B. Next generation sequencing of miRNAs–strategies, resources and methods. Genes 2010, 1, 70–84. [Google Scholar] [CrossRef] [Green Version]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muthusamy, M.; Kim, J.Y.; Yoon, E.K.; Kim, J.A.; Lee, S.I. BrEXLB1, a Brassica rapa Expansin-Like B1 Gene Is Associated with Root Development, Drought Stress Response, and Seed Germination. Genes 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marowa, P.; Ding, A.; Kong, Y. Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 2016, 35, 949–965. [Google Scholar] [CrossRef] [Green Version]
- Gullner, G.; Komives, T.; Király, L.; Schröder, P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front. Plant Sci. 2018, 9, 1836. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Wei, J.; Wu, Z.; Gao, J. Effects of Substrate-Binding Site Residues on the Biochemical Properties of a Tau Class Glutathione S-Transferase from Oryza sativa. Genes 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korir, N.K.; Han, J.; Shangguan, L.; Wang, C.; Kayesh, E.; Zhang, Y.; Fang, J. Plant variety and cultivar identification: Advances and prospects. Crit. Rev. Biotechnol. 2013, 33, 111–125. [Google Scholar] [CrossRef]
- Jördens, R. Progress of plant variety protection based on the International Convention for the Protection of New Varieties of Plants (UPOV Convention). World Pat. Inf. 2005, 27, 232–243. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, G.; Chen, B.; Du, H.; Zhang, F.; Zhang, H.; Wang, Q.; Geng, S. Candidate genes for first flower node identified in pepper using combined SLAF-seq and BSA. PLoS ONE 2018, 13, e0194071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, B.; Chen, Y.; Shaibu, A.S.; Zheng, H.; Sun, J. Molecular-Assisted Distinctness and Uniformity Testing Using SLAF-Sequencing Approach in Soybean. Genes 2020, 11, 175. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nawaz, M.A.; Chung, G. Genetic Improvement of Cereals and Grain Legumes. Genes 2020, 11, 1255. https://doi.org/10.3390/genes11111255
Nawaz MA, Chung G. Genetic Improvement of Cereals and Grain Legumes. Genes. 2020; 11(11):1255. https://doi.org/10.3390/genes11111255
Chicago/Turabian StyleNawaz, Muhammad Amjad, and Gyuhwa Chung. 2020. "Genetic Improvement of Cereals and Grain Legumes" Genes 11, no. 11: 1255. https://doi.org/10.3390/genes11111255
APA StyleNawaz, M. A., & Chung, G. (2020). Genetic Improvement of Cereals and Grain Legumes. Genes, 11(11), 1255. https://doi.org/10.3390/genes11111255





