Genetic Architecture Underpinning Yield Components and Seed Mineral–Nutrients in Sesame
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Phenotypic Characterization
2.3. Seed Nutrient Concentration Analysis
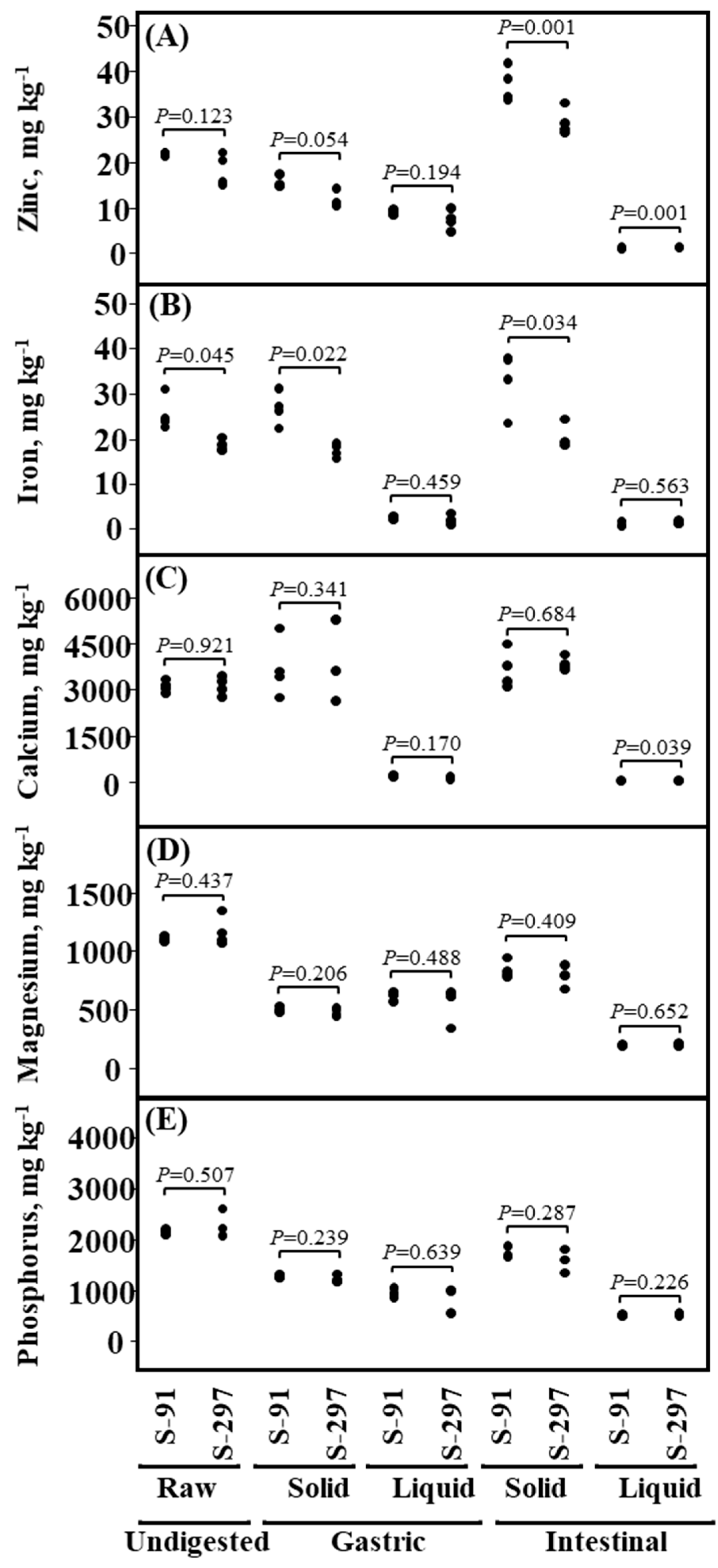
2.4. In-Vitro Digestion Analysis
2.5. QTL Analysis
2.6. Statistical Analyses of Phenotypic Data
3. Results
3.1. Wide Phenotypic Variation in Morphological and Seed Mineral-Nutrient Concentrations
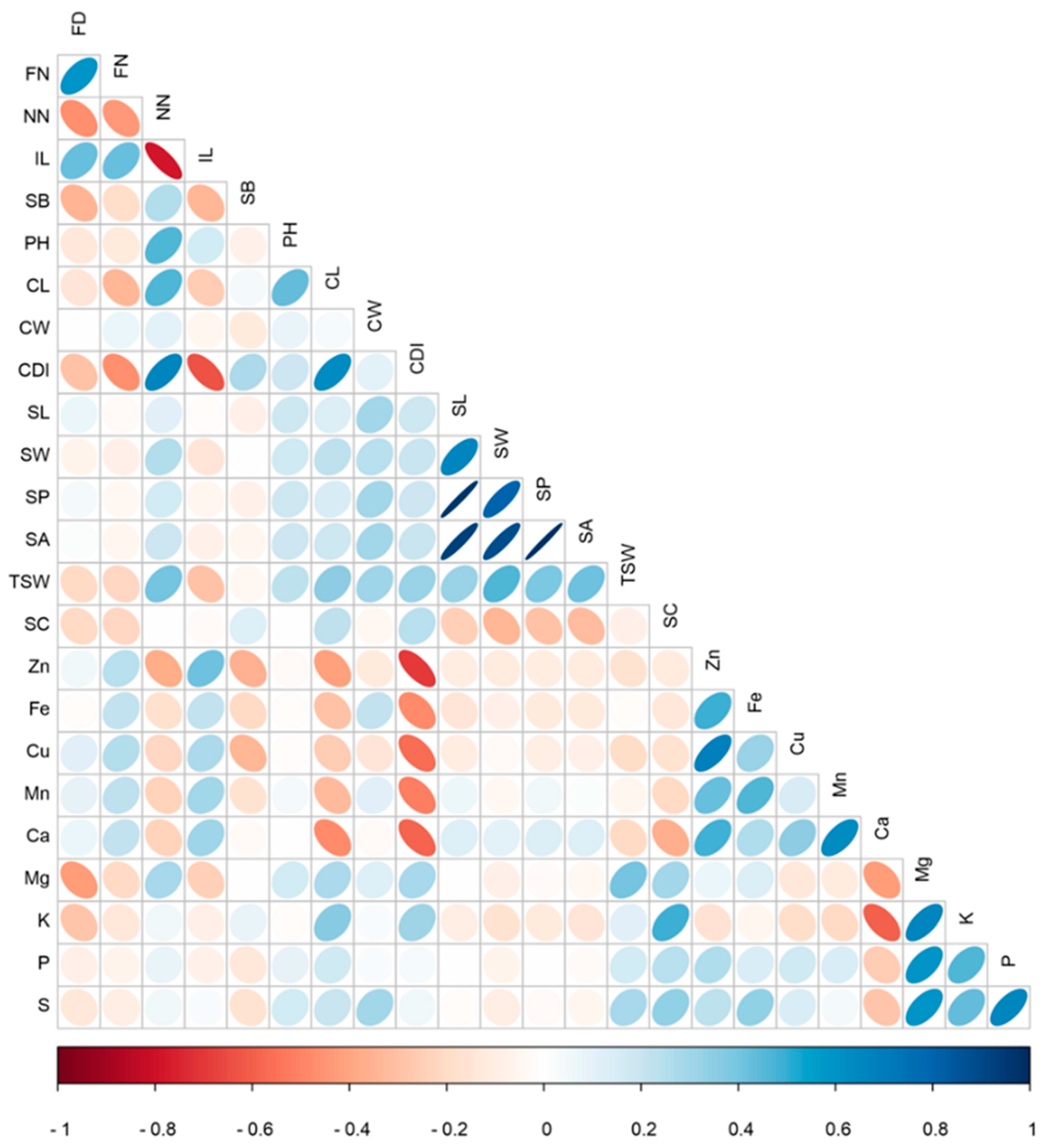
3.2. S-91 × S-297 Population Exhibited Range of Interrelationships between Plant Morphological and Seed Quality Traits
3.3. Constructing an High-Density Genetic Map and QTL Analysis
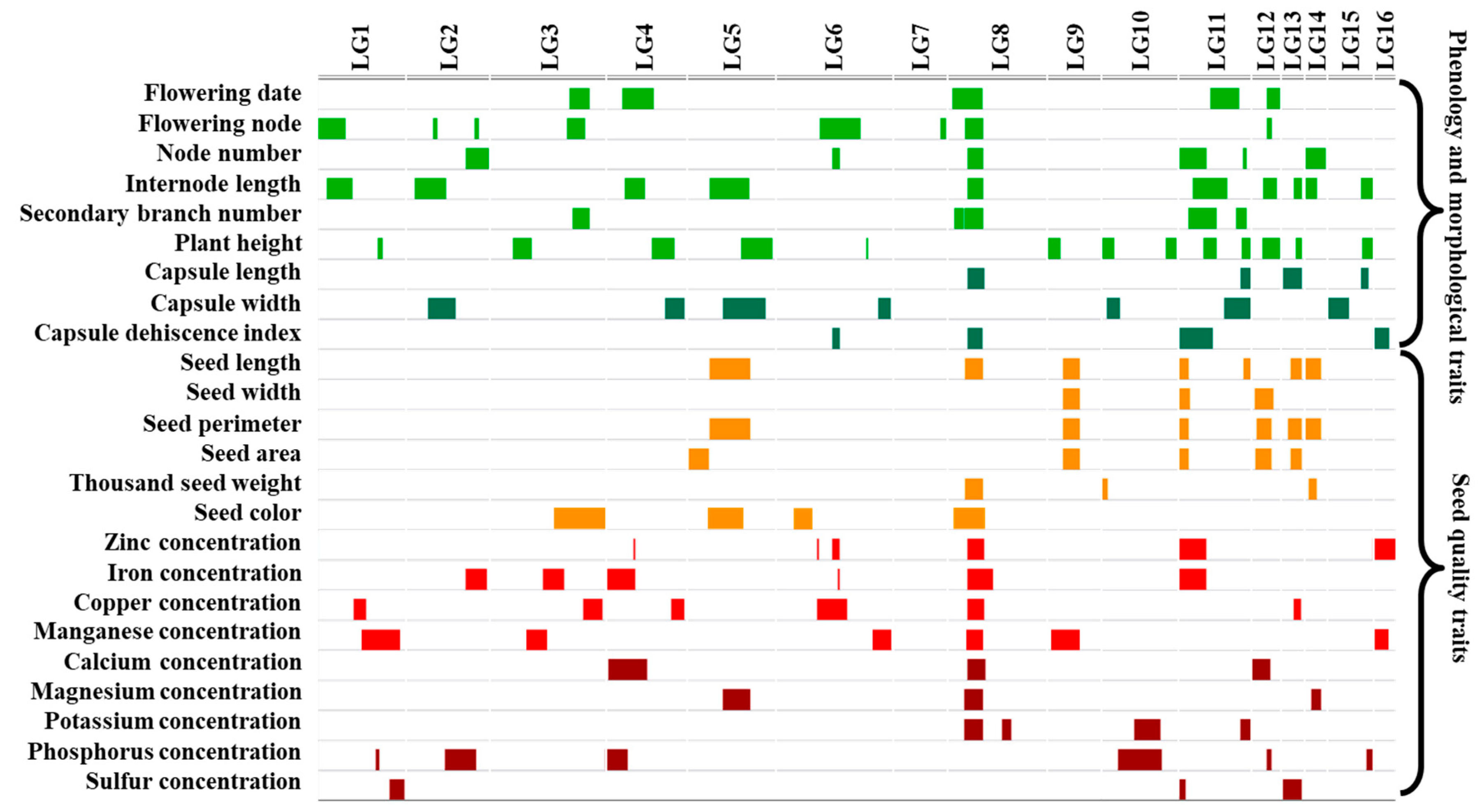
3.4. QTL for Phenology and Morphological Traits
3.5. QTL for Seed Quality Traits
3.6. Candidate Genes Associated with Seed Mineral-Nutrient Concentration
3.7. Bio-Accessibility of Seed Mineral–Nutrients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gharibzahedi, S.M.T.; Jafari, S.M. The importance of minerals in human nutrition: Bioavailability, food fortification, processing effects and nanoencapsulation. Trends Food Sci. Technol. 2017, 62, 119–132. [Google Scholar] [CrossRef]
- Müller, O.; Krawinkel, M. Malnutrition and health in developing countries. Can. Med. Assoc. J. 2005, 173, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Garg, M.; Sharma, N.; Sharma, S.; Kapoor, P.; Kumar, A.; Chunduri, V.; Arora, P. Biofortified crops generated by breeding, agronomy, and transgenic approaches are improving lives of millions of people around the world. Front. Nutr. 2018, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Platel, K.; Srinivasan, K. Bioavailability of micronutrients from plant foods: An update. Crit. Rev. Food Sci. Nutr. 2015, 56, 1608–1619. [Google Scholar] [CrossRef] [PubMed]
- White, P.J.; Broadley, M.R. Biofortification of crops with seven mineral elements often lacking in human diets-iron, zinc, copper, calcium, magnesium, selenium and iodine. New Phytol. 2009, 182, 49–84. [Google Scholar] [CrossRef]
- Lott, J.N.; Ockenden, I.; Raboy, V.; Batten, G.D. Phytic acid and phosphorus in crop seeds and fruits: A global estimate. Seed Sci. Res. 2000, 10, 11–33. [Google Scholar] [CrossRef]
- Gadri, Y.; Williams, L.E.; Peleg, Z. Tradeoffs between yield components promote crop stability in sesame. Plant Sci. 2020, 295, 110105. [Google Scholar] [CrossRef]
- Çağırgan, M.I. Selection and morphological characterization of induced determinate mutants in sesame. Field Crop. Res. 2006, 96, 19–24. [Google Scholar] [CrossRef]
- Mushtaq, A.; Hanif, M.A.; Ayub, M.A.; Bhatti, I.A.; Jilani, M.I. Sesame. In Medicinal Plants of South Asia; Hanif, M.A., Nawaz, H., Khan, M.M., Byrne, H.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 601–615. [Google Scholar]
- Uzun, B.; Arslan, Ç.; Furat, Ş. Variation in fatty acid compositions, oil content and oil yield in a germplasm collection of sesame (Sesamum indicum L.). J. Am. Oil Chem. Soc. 2008, 85, 1135–1142. [Google Scholar] [CrossRef]
- Pathak, N.; Rai, A.K.; Kumari, R.; Thapa, A.; Bhat, K.V. Sesame Crop: An Underexploited Oilseed Holds Tremendous Potential for Enhanced Food Value. Agric. Sci. 2014, 5, 519–529. [Google Scholar] [CrossRef]
- Johnson, L.A.; Suleiman, T.M.; Lusas, E.W. Sesame protein: A review and prospectus. J. Am. Oil Chem. Soc. 1979, 56, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Dimitrios, B. Sources of natural phenolic antioxidants. Trends Food Sci. Technol. 2006, 17, 505–512. [Google Scholar] [CrossRef]
- Bedigian, D. Sesame: The genus Sesamum; CRC Press, Taylor and Francis Group: Boca Raton, FL, USA, 2011. [Google Scholar]
- Tripathy, S.K.; Kar, J.; Sahu, D. Advances in sesame (Sesamum indicum L.) breeding. In Advances in Plant Breeding Strategies: Industrial and Food Crops; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2019; Volume 6, pp. 577–635. [Google Scholar]
- Pham, T.D.; Geleta, M.; Bui, T.M.; Bui, T.C.; Merker, A.; Carlsson, A.S. Comparative analysis of genetic diversity of sesame (Sesamum indicum L.) from Vietnam and Cambodia using agro-morphological and molecular markers. Hereditas 2011, 148, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Basak, M.; Uzun, B.; Yol, E. Genetic diversity and population structure of the Mediterranean sesame core collection with use of genome-wide SNPs developed by double digest RAD-Seq. PLoS ONE 2019, 14, e0223757. [Google Scholar] [CrossRef]
- Pandey, S.K.; Das, A.; Rai, P.; Dasgupta, T. Morphological and genetic diversity assessment of sesame (Sesamum indicum L.) accessions differing in origin. Physiol. Mol. Biol. Plants 2015, 21, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Whan, A.P.; Smith, A.B.; Cavanagh, C.R.; Ral, J.-P.F.; Shaw, L.M.; Howitt, C.A.; Bischof, L. GrainScan: A low cost, fast method for grain size and colour measurements. Plant Methods 2014, 10, 23. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carriere, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food–an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. Genomic plant DNA preparation from fresh tissue-CTAB method. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Wang, L.; Xia, Q.; Zhang, Y.; Zhu, X.; Zhu, X.; Li, D.; Ni, X.; Gao, Y.; Xiang, H.; Wei, X.; et al. Updated sesame genome assembly and fine mapping of plant height and seed coat color QTLs using a new high-density genetic map. BMC Genom. 2016, 17, 1–13. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2012, 14, 178–192. [Google Scholar] [CrossRef]
- Peleg, Z.; Cakmak, I.; Ozturk, L.; Yazici, A.; Jun, Y.; Budak, H.; Korol, A.B.; Fahima, T.; Saranga, Y. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat × wild emmer wheat RIL population. Theor. Appl. Genet. 2009, 119, 353–369. [Google Scholar] [CrossRef]
- Ragel, P.; Raddatz, N.; Leidi, E.O.; Quintero, F.J.; Pardo, J.M. Regulation of K+ nutrition in plants. Front. Plant Sci. 2019, 10, 281. [Google Scholar] [CrossRef] [PubMed]
- Khan, G.A.; Bouraine, S.; Wege, S.; Li, Y.; De Carbonnel, M.; Berthomieu, P.; Poirier, Y.; Rouached, H. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 2014, 65, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Connolly, E.L.; Campbell, N.H.; Grotz, N.; Prichard, C.L.; Guerinot, M.L. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003, 133, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Sherman, T.; Fromm, H. Cyclic nucleotide-gated channels in plants. FEBS Lett. 2007, 581, 2237–2246. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, S.A.; Lee, J.; Guerinot, M.L.; An, G. Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 2010, 29, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Tulchinsky, T.H. Micronutrient deficiency conditions: Global health issues. Public Heal. Rev. 2010, 32, 243–255. [Google Scholar] [CrossRef]
- Furat, S.; Uzun, B. The use of agro-morphological characters for the assessment of genetic diversity in sesame (Sesamum indicum L.). Plant Omics 2010, 3, 85–91. [Google Scholar]
- Arriel, N.H.C.; Di Mauro, A.O.; Arriel, E.F.; Unêda-Trevisoli, S.H.; Costa, M.M.; Bárbaro, I.M.; Muniz, F.R.S. Genetic divergence in sesame based on morphological and agronomic traits. Crop Breed. Appl. Biotechnol. 2007, 7, 253–261. [Google Scholar] [CrossRef]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef]
- Cai, G.; Yang, Q.-Y.; Chen, H.; Yang, Q.; Zhang, C.; Fan, C.; Zhou, Y. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci. Rep. 2016, 6, 21625. [Google Scholar] [CrossRef] [PubMed]
- Langham, D.R. Phenology of sesame. In Issues in New Crops and New Uses; Janick, J., Whipkey, A., Eds.; ASHS Press: Alexandria, VA, USA, 2007; pp. 144–182. [Google Scholar]
- Zhao, F.; Su, Y.; Dunham, S.; Rakszegi, M.; Bedo, Z.; McGrath, S.; Shewry, P. Variation in mineral micronutrient concentrations in grain of wheat lines of diverse origin. J. Cereal Sci. 2009, 49, 290–295. [Google Scholar] [CrossRef]
- Menkir, A. Genetic variation for grain mineral content in tropical-adapted maize inbred lines. Food Chem. 2008, 110, 454–464. [Google Scholar] [CrossRef] [PubMed]
- Pinson, S.R.M.; Tarpley, L.; Yan, W.; Yeater, K.; Lahner, B.; Yakubova, E.; Huang, X.-Y.; Zhang, M.; Guerinot, M.L.; Salt, D.E. Worldwide genetic diversity for mineral element concentrations in rice grain. Crop Sci. 2015, 55, 294–311. [Google Scholar] [CrossRef]
- Mamo, B.E.; Barber, B.L.; Steffenson, B.J. Genome-wide association mapping of zinc and iron concentration in barley landraces from Ethiopia and Eritrea. J. Cereal Sci. 2014, 60, 497–506. [Google Scholar] [CrossRef]
- Chatzav, M.; Peleg, Z.; Ozturk, L.; Yazici, A.; Fahima, T.; Cakmak, I.; Saranga, Y. Genetic diversity for grain nutrients in wild emmer wheat: Potential for wheat improvement. Ann. Bot. 2010, 105, 1211–1220. [Google Scholar] [CrossRef]
- Garcia-Oliveira, A.L.; Tan, L.; Fu, Y.; Sun, C. Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 2009, 51, 84–92. [Google Scholar] [CrossRef]
- Blair, M.W.; Astudillo, C.; Grusak, M.A.; Graham, R.; Beebe, S.E. Inheritance of seed iron and zinc concentrations in common bean (Phaseolus vulgaris L.). Mol. Breed. 2008, 23, 197–207. [Google Scholar] [CrossRef]
- Grotz, N.; Guerinot, M.L. Molecular aspects of Cu, Fe and Zn homeostasis in plants. Biochim. Biophys. Acta BBA-Mol. Cell Res. 2006, 1763, 595–608. [Google Scholar] [CrossRef]
- Ajeesh Krishna, T.P.; Maharajan, T.; Victor Roch, G.; Ignacimuthu, S.; Ceasar, S.A. Structure, function, regulation and phylogenetic relationship of ZIP family tansporters of plants. Front. Plant Sci. 2020, 11, 662. [Google Scholar] [CrossRef]
- Šimić, D.; Sudar, R.; Ledenčan, T.; Jambrović, A.; Zdunić, Z.; Brkić, I.; Kovačević, V. Genetic variation of bioavailable iron and zinc in grain of a maize population. J. Cereal Sci. 2009, 50, 392–397. [Google Scholar] [CrossRef]
- Shunmugam, A.; Bock, C.; Arganosa, G.C.; Georges, F.; Gray, G.R.; Warkentin, T.D. Accumulation of Phosphorus-containing compounds in developing seeds of low-phytate pea (Pisum sativum L.) mutants. Plants 2015, 4, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Abebe, Y.; Bogale, A.; Hambidge, K.M.; Stoecker, B.J.; Bailey, K.; Gibson, R.S. Phytate, zinc, iron and calcium content of selected raw and prepared foods consumed in rural Sidama, Southern Ethiopia, and implications for bioavailability. J. Food Compos. Anal. 2007, 20, 161–168. [Google Scholar] [CrossRef]
- Cichy, K.A.; Caldas, G.V.; Snapp, S.S.; Blair, M.W. QTL Analysis of Seed Iron, Zinc, and Phosphorus Levels in an Andean Bean Population. Crop Sci. 2009, 49, 1742–1750. [Google Scholar] [CrossRef]
- Stangoulis, J.C.; Huynh, B.-L.; Welch, R.M.; Choi, E.-Y.; Graham, R.D. Quantitative trait loci for phytate in rice grain and their relationship with grain micronutrient content. Euphytica 2007, 154, 289–294. [Google Scholar] [CrossRef]
- Zhang, H.; Miao, H.; Wei, L.; Li, C.; Duan, Y.; Xu, F.; Qu, W.; Zhao, R.; Ju, M.; Chang, S. Identification of a SiCL1 gene controlling leaf curling and capsule indehiscence in sesame via cross-population association mapping and genomic variants screening. BMC Plant Biol. 2018, 18, 296. [Google Scholar] [CrossRef] [PubMed]
- Gaymard, F.; Pilot, G.; Lacombe, B.; Bouchez, D.; Bruneau, D.; Boucherez, J.; Michaux-Ferrière, N.; Thibaud, J.-B.; Sentenac, H. Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 1998, 94, 647–655. [Google Scholar] [CrossRef]
- Zelman, A.K.; Dawe, A.; Gehring, C.; Berkowitz, G.A. Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels. Front. Plant Sci. 2012, 3, 95. [Google Scholar] [CrossRef]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta BBA-Biomembr. 2000, 1465, 190–198. [Google Scholar] [CrossRef]
- Rossander-Hultén, L.; Brune, M.; Sandström, B.; Lönnerdal, B.; Hallberg, L. Competitive inhibition of iron absorption by manganese and zinc in humans. Am. J. Clin. Nutr. 1991, 54, 152–156. [Google Scholar] [CrossRef]
- Hallberg, L.; Hulthén, L. Prediction of dietary iron absorption: An algorithm for calculating absorption and bioavailability of dietary iron. Am. J. Clin. Nutr. 2000, 71, 1147–1160. [Google Scholar] [CrossRef]
- Seshadri, S. Prevalence of micronutrient deficiency particularly of iron, zinc and folic acid in pregnant women in South East Asia. Br. J. Nutr. 2001, 85, S87. [Google Scholar] [CrossRef]
- Scheers, N. Regulatory Effects of Cu, Zn, and Ca on Fe absorption: The intricate play between nutrient transporters. Nutrients 2013, 5, 957–970. [Google Scholar] [CrossRef]
- Mulet-Cabero, A.-I.; Egger, L.; Portmann, R.; Ménard, O.; Marze, S.; Minekus, M.; Le Feunteun, S.; Sarkar, A.; Grundy, M.M.L.; Carrière, F.; et al. A standardised semi-dynamic in vitro digestion method suitable for food-an international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef]
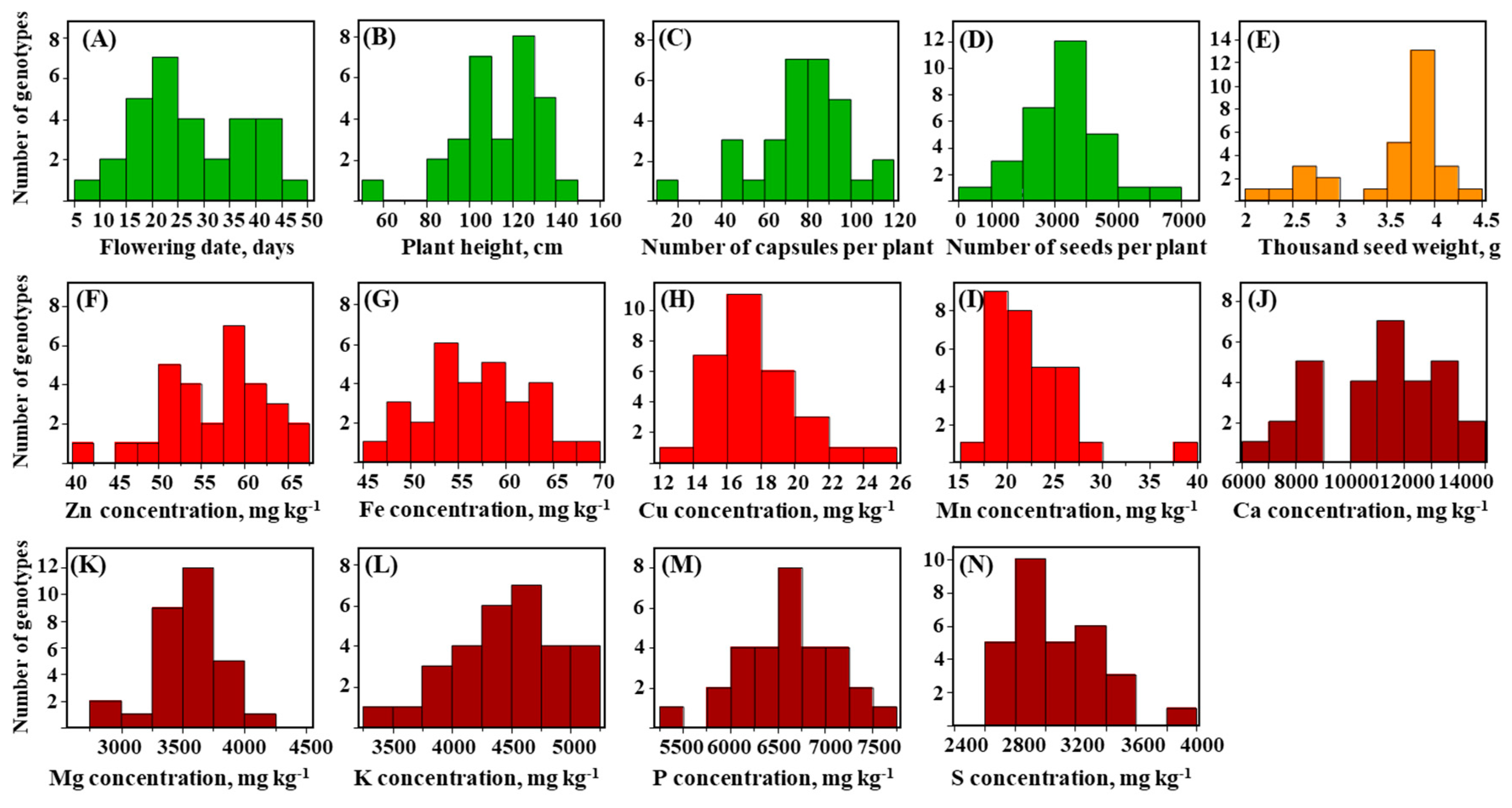
| Trait | Code | S-91 | S-297 | F2 Population | |
|---|---|---|---|---|---|
| Mean | Range | ||||
| Flowering date (days) | FD | 25 ± 0.14 | 18 ± 0.11 | 17.8 | 14–29 |
| Flowering Node | FN | 5 ± 0.7 | 3 ± 0.2 | 3.4 | 2–5 |
| Node Number | NN | 29 ± 1.9 | 36 ± 1.3 | 34.1 | 20–43 |
| Internode Length (cm) | IL | 4.9 ± 0.1 | 4.1 ± 0.2 | 4.1 | 2.7–6.5 |
| Number of Secondary Branches | SB | 1 ± 0.5 | 0 ± 0.2 | 0.7 | 0–5 |
| Plant Height (cm) | PH | 142 ± 11.7 | 147 ± 8.3 | 134.2 | 100–170 |
| Capsule Length (mm) | CL | 42.9 ± 1.3 | 28.8 ± 0.7 | 34.5 | 20.6–48.0 |
| Capsule Width (mm) | CW | 6.46 ± 0.22 | 5.71 ± 0.1 | 6.34 | 5.22–8.43 |
| Capsule Dehiscence Index | CDI | 1 | 5 | 4.2 | 1–5 |
| Seed Length (mm) | SL | 3.31 ± 0.02 | 3.41 ± 0.03 | 3.50 | 3.08–3.86 |
| Seed Width (mm) | SW | 2.1 ± 0.0 | 2.1 ± 0.0 | 2.1 | 1.9–2.3 |
| Seed Perimeter (mm) | SP | 10.6±0.1 | 10.9 ± 0.1 | 11.21 | 10.1–12.3 |
| Seed Area (mm2) | SA | 5.32 ± 0.11 | 5.56 ± 0.10 | 5.80 | 4.75–6.90 |
| Thousand Seed Weight (g) | TSW | 3.35 ± 0.14 | 2.97 ± 0.07 | 3.11 | 0.97–3.99 |
| Seed Color (RGB index) | SC | 153.1 ± 2.3 | 159.1 ± 2.5 | 155.3 | 128.1–198.3 |
| Zinc (mg/kg) | Zn | 87.5 ± 5.5 | 58.4 ± 1.7 | 71.1 | 45.5–108.6 |
| Iron (mg/kg) | Fe | 97.3 ± 11.22 | 76.4 ± 6.6 | 82.9 | 53.8–116.0 |
| Copper (mg/kg) | Cu | 23.2 ± 1.1 | 14.5 ± 1.0 | 17.2 | 11.5–23.5 |
| Manganese (mg/kg) | Mn | 19.3 ± 0.5 | 17.8 ± 1.6 | 16.0 | 11.8–22.2 |
| Calcium (mg/kg) | Ca | 12,591.2 ± 397.9 | 11,334.6 ± 947.9 | 10,693.6 | 4171.2–19,857.3 |
| Magnesium (mg/kg) | Mg | 4109.7 ± 122.2 | 4357.9 ± 90.8 | 4232.9 | 3433.0–4749.7 |
| Potassium (mg/kg) | K | 5577.2 ± 149.9 | 5519.8 ± 214.9 | 5434.4 | 4061.4–7201.0 |
| Phosphorus (mg/kg) | P | 8574.9 ± 239.4 | 9165.3 ± 81.1 | 8945.3 | 7777.9–9895.5 |
| Sulfur (mg/kg) | S | 3635.7 ± 64.1 | 3607.9 ± 183.2 | 3703.5 | 3082.6–4248.0 |
| Trait | #QTL | LOD a | PVE (%) b | Favorable Allele c | |
|---|---|---|---|---|---|
| S-91 | S-297 | ||||
| Plant Phenology and Morphology | |||||
| Flowering date | 6 | 2.1–4.92 | 6.3–14 | 0 | 6 |
| Flowering node | 8 | 2.56–7.57 | 7.6–20.7 | 2 | 6 |
| Node number | 6 | 2.59–19.46 | 7.7–45 | 1 | 5 |
| Internode length | 10 | 2–14.93 | 5.0–31.2 | 1 | 9 |
| Number of secondary branches | 5 | 3.03–4 | 8.9–11.6 | 0 | 5 |
| Plant height | 13 | 2–4.66 | 5.3–11.9 | 10 | 3 |
| Capsule Morphology | |||||
| Capsule length | 4 | 2.39–19.73 | 7.1–45.4 | 3 | 1 |
| Capsule width | 7 | 2.1–3.99 | 6.2–11.5 | 4 | 3 |
| Capsule dehiscence index | 4 | 2.66–95.1 | 6.3–76.8 | 4 | 0 |
| Seed Morphology | |||||
| Seed length | 7 | 2–6.4 | 5.9–17.8 | 2 | 5 |
| Seed width | 3 | 2.81–5.11 | 8.3–14.5 | 1 | 2 |
| Seed perimeter | 6 | 2.45–6.35 | 7.2–17.7 | 3 | 3 |
| Seed area | 5 | 2.3–5.77 | 6.8–16.2 | 2 | 3 |
| Seed Quality | |||||
| Thousand seed weight | 3 | 2.63–4.77 | 7.7–13.6 | 2 | 1 |
| Seed color | 4 | 2.14–12.09 | 6.3–31 | 0 | 4 |
| Seed Zn concentration | 6 | 2.43–19.92 | 7.2–45.7 | 5 | 1 |
| Seed Fe concentration | 6 | 2.05–7.94 | 6.1–21.6 | 5 | 1 |
| Seed Cu concentration | 6 | 2.13–11.8 | 6.3–30.4 | 6 | 0 |
| Seed Mn concentration | 6 | 2.4–8.17 | 7.1–22.2 | 4 | 1 |
| Seed Ca concentration | 3 | 2.05–7.84 | 6.1–21.4 | 2 | 1 |
| Seed Mg concentration | 3 | 2.28–4.16 | 6.8–12 | 1 | 2 |
| Seed K concentration | 4 | 2.55–3.7 | 7.5–10.7 | 2 | 2 |
| Seed P concentration | 6 | 2–2.65 | 5.9–7.8 | 5 | 1 |
| Seed S concentration | 3 | 2.45–4.34 | 7.2–12.5 | 2 | 1 |
| Total | 134 | 67 | 66 | ||
| LG | Interval | Trait | CG ID | Annotated Function |
|---|---|---|---|---|
| LG8 | 4550182–7125811 | FD, FN, NN, IL, SB, CL, CDI, SL, TSW, SC, Zn, Fe, Cu, Mn, Ca, Mg, K | LOC105167760 | Potassium channel SKOR-like |
| LOC105167785 | Potassium channel SKOR | |||
| LOC105167762 | Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase 2-like | |||
| LOC105167788 | Ethylene-responsive transcription factor 1B-like | |||
| LOC105167789 | Ethylene-responsive transcription factor 1B-like | |||
| LOC105167791 | Ethylene-responsive transcription factor 1B-like | |||
| LOC105167765 | Transcription repressor KAN1 | |||
| LOC105167815 | Isocitrate dehydrogenase [NADP] | |||
| LG11 | 310665–1216709 | NN, CDI, SL, SW, SP, SA, Zn, Fe, S | LOC110012885 | Ethylene-responsive transcription factor ERF023-like |
| LOC105173138 | Cyclic nucleotide-gated ion channel 1-like | |||
| LOC105173087 | Cyclic nucleotide-gated ion channel 1-like | |||
| LOC105173088 | Cyclic nucleotide-gated ion channel 1-like | |||
| LOC105173140 | MYB-like transcription factor ETC3 | |||
| LOC105173141 | Probable WRKY transcription factor 30 | |||
| LOC105173122 | Heavy metal-associated isoprenylated plant protein 39-like | |||
| LOC105173253 | Probable polygalacturonase | |||
| LOC105173155 | Ferric reduction oxidase 2 | |||
| LOC105173156 | Ferric reduction oxidase 2-like | |||
| LOC105173161 | Ascorbate transporter | |||
| LG11 | 5566003–5767772 | NN, IL, SB, PH, CDI, Fe, Zn | LOC105173373 | Protein PHOSPHATE STARVATION RESPONSE 1-like |
| LOC105173380 | Citrate synthase | |||
| LG11 | 14205927–14426041 | NN, SB, PH, CL, CW, SL, P | LOC105174482 | Transcription factor LHW |
| LOC105174515 | Transcription factor MYB1 | |||
| LG16 | 14816-3048510 | CDI, Zn, Mn | LOC105178592 | Transcription factor bHLH30-like |
| LOC105178450 | Transcription factor TCP10-like | |||
| LOC105178476 | Nicotianamine aminotransferase A | |||
| LOC105178598 | WRKY transcription factor 6 | |||
| LOC105178495 | Transcription factor MYB101 | |||
| LOC105178506 | MYB-like transcription factor ETC1 | |||
| LOC105178507 | Isocitrate lyase | |||
| LOC105178516 | Aconitate hydratase | |||
| LOC105178613 | Calcium-binding protein PBP1-like | |||
| LOC105178537 | Transcription factor MYB39-like | |||
| LOC105178559 | Ethylene-responsive transcription factor 4-like | |||
| LOC105178589 | Zinc transporter 8-like | |||
| LOC105178590 | Zinc transporter 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teboul, N.; Gadri, Y.; Berkovich, Z.; Reifen, R.; Peleg, Z. Genetic Architecture Underpinning Yield Components and Seed Mineral–Nutrients in Sesame. Genes 2020, 11, 1221. https://doi.org/10.3390/genes11101221
Teboul N, Gadri Y, Berkovich Z, Reifen R, Peleg Z. Genetic Architecture Underpinning Yield Components and Seed Mineral–Nutrients in Sesame. Genes. 2020; 11(10):1221. https://doi.org/10.3390/genes11101221
Chicago/Turabian StyleTeboul, Naama, Yaron Gadri, Zipi Berkovich, Ram Reifen, and Zvi Peleg. 2020. "Genetic Architecture Underpinning Yield Components and Seed Mineral–Nutrients in Sesame" Genes 11, no. 10: 1221. https://doi.org/10.3390/genes11101221
APA StyleTeboul, N., Gadri, Y., Berkovich, Z., Reifen, R., & Peleg, Z. (2020). Genetic Architecture Underpinning Yield Components and Seed Mineral–Nutrients in Sesame. Genes, 11(10), 1221. https://doi.org/10.3390/genes11101221