Abstract
The pacu (Piaractus mesopotamicus) is a Neotropical fish with remarkable productive performance for aquaculture. Knowledge of genetic resources in Neotropical fish is essential for their applications in breeding programs. The aim of this study was to characterize the genetic diversity of seven farmed populations of pacu which will constitute the basis for a broodstock foundation for coming breeding programs in Brazil. Analysis of one wild population (Paraná River) was used as a reference to compare genetic parameters in the farmed populations. The analyses were performed using 32 single-nucleotide polymorphisms (SNP) and 8 simple sequence repeat (SSR) markers. No significant differences in genetic diversity between populations estimated through the number of alleles and allelic richness, observed heterozygosity, expected heterozygosity, and minimum allele frequency were detected (p > 0.05). Low genetic diversity was observed in all farmed stocks and the wild population. Moreover, we detected low genetic structure when comparing farmed and wild populations for SNPs (FST = 0.07; K = 3) and SSRs (FST = 0.08; K = 2). Analysis of molecular variance (AMOVA) demonstrated that genetic variation was mostly within populations. Kinship analysis showed that most fish farms included related individuals at a proportion of at least 25%. Our results suggest that the basal broodstock for pacu breeding programs should be founded with individuals from different fish farms for higher genetic diversity and to avoid inbreeding risks.
1. Introduction
The pacu (Piaractus mesopotamicus) is a Characiform fish with a wide natural distribution throughout La Plata basin which covers an area over five South American countries: Brazil, Uruguay, Bolivia, Paraguay, and Argentina. Wild populations of pacu are threatened by overfishing since this species is considered to have high commercial value, with large-scale catches occurring by industrial and recreational fisheries [1]. According to the latest official statistics on industrial fisheries in Brazil, wild catches of pacu represent 5% (11 thousand tons) of the total inland capture (243.8 thousand tons) [2], classifying it as a critically endangered species in Sao Paulo State in Brazil (Decree 56,031). In relation to aquaculture production, pacu is one of the most cultivated fish species in South America. In Brazil, pacu farming already represents the second largest harvest from fish production (about 21 thousand tons) among native species [3]. According to its productive characteristics, it is an omnivorous fish considered a gainful species for aquaculture due to its rapid growth and low-cost feeding habits [4]. Due to its productive viability, its production in aquaculture has increased even in other parts of the world, such as China, Myanmar, Thailand, and Vietnam [5,6].
Despite breeding programs being fundamental for the sustainable development of aquaculture, less than 10% of aquaculture production in the world is based on genetically improved stocks, limited to only a few species [7], and there is no production of native species from Latin America resulting from genetic selection. For successful breeding strategies, suitable pre-breeding programs have been fundamental to providing advantageous and enduring genetic gains for fish production [8]. Therefore, evaluations of genetic variability and genetic relationships in the base population are essential for effective control over generations, which reduces problems related to loss of genetic potential for selective breeding in farmed stocks [9].
Mass selection without appropriate genetic supervision usually intensifies the processes of genetic drift, determining increased inbreeding and a reduction in genetic variability. Consequently, negative impacts such as reduced growth rates, low survival, morphological deformities, and susceptibility to disease have been reported [10,11,12]. Thus, the evaluation of genetic diversity and population structure by molecular markers to assist directed mating is important for successful management in selective breeding programs [13,14].
In pacu, genetic diversity studies have been limited to evaluating wild populations using the mitochondrial DNA control region [15] and microsatellite markers (or simple sequence repeats, SSRs) [16,17]. Both types of studies have characterized wild populations of pacu as a panmictic stock with high gene flow among rivers.
In this study, seven farmed stocks and one wild population of P. mesopotamicus were examined by using 32 single-nucleotide polymorphisms (SNPs) and 8 SSRs to assess the genetic diversity, structure, and kinship as a support to designing a suitable pre-breeding program for this species.
2. Materials and Methods
2.1. Ethic Statement
This study was conducted in strict accordance with the recommendations of the National Council for Control of Animal Experimentation (CONCEA) (Brazilian Ministry for Science, Technology and Innovation) and was approved by the Animal Use Ethics Committee (CEUA), protocol no. 22.255/15. The present study was performed under authorization no. 33435-1 issued through the Chico Mendes Institute for the Conservation of Biodiversity, Brazilian Ministry for the Environment (ICMBio). Fin clips were collected from each fish under benzocaine anesthesia and all efforts were made to minimize suffering. Fin samples were stored in 95% ethanol at −20 °C.
2.2. Experimental Population
Individuals of seven different broodstocks from fish farms in Brazil (São Paulo State) were collected (FF1–FF7) (number of individuals shown in Table 1). The commercial identity and localization of the fish farms were kept confidential. Animals were individually tagged with transponders (passive integrated transponder tags (pit-tags), model full-duplex FDX-B, 134.2 kHz) and kept alive for subsequent management to create the basal broodstock for the genetic breeding program. Fish farms were mostly set up between the 1980s and 1990s. There are no records of selective mating, except for at FF4. Additionally, 34 individuals collected in the Paraná River, Brazil (latitude −21.39, longitude −51.93) and previously used for SNP validation [18] were used as the reference wild population (Table 1). According to previous studies of genetic structure, wild populations of pacu present high levels of genetic similarity, representing a panmictic unit [15,17]. Therefore, we considered that this particular wild population (Paraná River) would represent the panmictic stock of the natural populations, being used as a reference for comparison of the genetic parameters.

Table 1.
Mean of diversity parameters per population in pacu.
2.3. SSR and SNP Analysis
DNA extraction was performed using the saline extraction protocol [19]. DNA integrity was evaluated on 1% agarose gel and its purity was assessed using a NanoDrop One spectrophotometer (Thermo Fisher, Madison, USA). The DNA concentration was quantified using the Qubit dsDNA BR Assay kit (Life Technologies, Oregon, USA) and measured in a Qubit 3.0 Fluorometer (Invitrogen, Kuala Lumpur, Malaysia).
SSR genotyping was performed using eight microsatellite markers (Pm1, Pm3, Pm5, Pm11, Pm4, Pm6, Pm9, and Pm13) in two Multiplex PCR reactions according to Posner [20] and previously standardized using fluorescent labeled primers by CONICET Service (Stan CONICET No. 2353, CCT Rosario, Argentina; http://vinculacion.conicet.gov.ar/buscador-de-oferta-tecnologica/?id_ot=2353&tipo=3). Markers were genotyped in a 3730XL DNA analyzer (Applied Biosystems) in Macrogen (Korea). Allele scoring was performed with the Peak Scanner program (Applied Biosystems) using the GeneScan 500 LIZ Size Standard.
SNP genotyping was carried out using 32 markers obtained from the liver transcriptome [18]. Analysis was performed using the MassARRAY platform (Sequenom, San Diego, CA, USA) in CeGen (Spanish Genotyping National Center, Santiago de Compostela node, Spain) as described in Mastrochirico-Filho et al. [18]. SNP data from the wild population (WILD) were previously reported by Mastrochirico-Filho et al. [18].
2.4. Statistical Analysis
The presence of null alleles (Fnull) and allelic dropout in microsatellite loci were tested using MICRO-CHECKER 2.2.3 [21]. The number of alleles per locus (Na), number of private alleles (Np), and observed (Hobs) and expected (Hexp) heterozygosity were estimated using CERVUS 3.0 [22]. The minimum allele frequency (MAF), exact tests for deviation from the Hardy–Weinberg equilibrium (HWE) (Markov Chains of 100,000 steps), and linkage disequilibrium (LD) (p < 0.05) were performed using GENEPOP 4.0.11 [23]. The allelic richness (Ar) and inbreeding coefficient (FIS) were estimated using FSTAT 2.9.3.2 software [24]. Bonferroni correction was applied for multiple tests [25].
The effective population size (Ne) was estimated by the linkage disequilibrium method in NeESTIMATOR V2.01 [26] and was used for a gross estimation of inbreeding in the analyzed fish farms (∆F = 1/2Ne) [27]. Recent bottleneck events were evaluated by M-ratio testing [28] using ARLEQUIN version 3.5.2.2 [29]. Bottleneck analysis was performed only for SSR data because of the biallelic nature of the SNPs.
To estimate genetic differentiation between the stocks, global and pairwise FST values were calculated using FSTAT version 2.9.3.2 [24]. The significance of these values was estimated with 10,000 permutations.
Levels of admixture among stocks were inferred by estimating the optimum number of population clusters (K) [30] using STRUCTURE version 2.3.4 [31] without prior information about the population. Primarily, we determined the distribution of ΔK, an ad hoc statistic based on the rate of change in the log probability of data between successive K values. The range of clusters (K) was predefined from 1 to 8. The analysis was performed in 80 replicated runs (i.e., 10 replicates for each K value) using 500,000 iterations after a burn-in period of 100,000 runs. The most likely K value to explain the population structure was the modal value of this ΔK [30]. The outputs of STRUCTURE analysis were visualized through the STRUCTURE HARVESTER program [32]. The results of independent STRUCTURE runs were summarized and corrected for the best K using CLUMPP software version 1.1.2 [33].
The partitioning of variation at different hierarchical levels was calculated by analysis of molecular variance (AMOVA) in ARLEQUIN version 3.5.2.2 [29] using 10,000 permutations. Stocks were grouped according to the clusters obtained by STRUCTURE software.
Kinship coefficients (rxy) [34] and potential for each locus to exclude a false parent were estimated using both SSR and SNP loci by COANCESTRY v. 1.0.1.8 [35]. Parentage exclusion probabilities of loci were determined when one individual taken at random from the population was excluded as a parent when no parent was known (PE2). This analysis was performed to evaluate with reliability the pairwise relatedness between farmed breeders from each fish farm without any parental information. Threshold values of the kinship coefficient were adopted as lower values (rxy < 0.125) corresponding to unrelated individuals; intermediate values 0.125 ≤ rxy ≤ 0.375 were considered half-siblings; and rxy ˃ 0.375 were considered full siblings [36].
3. Results
The parameters of genetic variability for pacu populations determined by 8 SSR and 32 SNP markers are shown in Tables S1 and S2, respectively. The mean values of the population parameters and overall locus p values of HWE and FIS values are shown in Table 1.
For SSR loci, a total of 44 alleles were detected in the analyzed populations. The average number of alleles per locus was 3.84 ± 1.16, with numbers ranging between 2 and 7. Allelic richness ranged from 2.000 to 6.140, with average population values ranging between 3.331 ± 0.868 (FF1) and 3.975 ± 1.189 (FF7). Private alleles were detected in individuals belonging to FF2, FF4, FF7, and WILD.
Monomorphic SNPs were found more frequently in FF2 (six monomorphic SNPs) and FF4 (four monomorphic SNPs). By contrast, FF1, WILD, FF6, and FF7 showed only polymorphic SNPs and the highest MAF average values (0.285 ± 0.133, 0.275 ± 0.132, 0.273 ± 0.127, and 0.270 ± 0.144, respectively). On the other hand, FF3 (0.269 ± 0.138) was the fish farms with the lowest MAF average values.
The average Hobs was 0.502 ± 0.219 for SSRs and 0.391 ± 0.173 for SNPs. When the average Hexp was estimated, 0.525 ± 0.170 was found for SSRs and 0.379 ± 0.133 for SNPs. The genetic diversity values observed in the fish farms (measured as Ar, Hobs, and Hexp for SSR and as MAF, Hobs, and Hexp for SNPs) were not significantly different from the WILD values (p > 0.75 for Ar, Hobs, and Hexp, Kruskall–Wallis tests for SSRs; p > 0.90 for MAF, Hobs, and Hexp, Kruskall–Wallis tests for SNPs), which showed Ar = 3.420 ± 1.0005, Hobs = 0.441 ± 0.213, and Hexp = 0.434 ± 0.214 for SSRs and MAF = 0.275 ± 0.132, Hobs = 0.385 ± 0.171, and Hexp = 0.370 ± 0.130 for SNPs (Table 1).
All SNP loci were in accordance with HWE after Bonferroni correction (p = 0.001), even when populations were subjected to global tests for all loci. By contrast, some SSR loci departed from HWE after Bonferroni correction (p = 0.006), such as locus Pm4 (FF2, FF3, and FF6) and locus Pm5 (FF4). Null alleles were detected in the locus Pm4 (FF2, FF3, FF4, and FF6) and locus Pm11 (FF2) (Table S1). Neither scoring errors nor allele drop-out were detected. Four fish farms (FF1, FF2, FF4, and FF6) departed from HWE (p < 0.05) when applying global tests for SSRs. However, only FF4 presented significant Hardy–Weinberg disequilibrium after Bonferroni correction (p < 0.006).
Overall population FIS values varied similarly between SSR (−0.113 and 0.212) and SNP loci (−0.082 to 0.056). However, no significant deviations from zero of FIS values were found in the fish farms when Bonferroni correction was applied for SNPs (p = 0.001) and SSRs (p = 0.006). Additionally, FIS values were not considered significantly different from zero over all loci for both markers (p > 0.05), presenting low 95% confidence interval values for SSRs (−0.022 to 0.135) and SNPs (−0.075 to 0.011) when bootstrapping over loci analyses were performed.
Except for in FF2 and FF6, SNP markers showed LD between pairs of loci in most fish farms: FF1 (7 pairs), FF7 (5), WILD (4), FF5 (2), FF3 (1), and FF4 (1) (p < 0.001). For SSR markers, LD was detected in FF4 (5 pairs), FF2 (2), FF1 (1), FF5 (1), and WILD (1) (p < 0.01). However, in both markers, the observed LD was not homogeneous among populations or loci (Table S3).
Reduced effective population size was detected in fish farms when compared to WILD for both SNP and SSR sets (except for FF6 in SSR analysis). Ne estimation showed values ranging from 2.3 in FF1 to 20.2 in WILD using SNP markers. In relation to SSRs, Ne values ranged from 7.0 in FF4 to 60.8 in FF6 (Table 2). The rate of inbreeding (∆F) was detected by considering Ne estimates and resulted in the lowest values in FF6 for SSRs (0.01) and in WILD for SNPs (0.02) and the highest values in FF4 for SSRs (0.07) and in FF1 for SNPs (0.22). Evidence of recent reductions in population size was found in all fish farms (M-ratio < 0.68) by considering SSR analysis (Table 2) characterizing bottleneck events.

Table 2.
Bottleneck tests by M-ratio test. Effective population size (Ne) parameter based on heterozygosity excess and inbreeding coefficient (∆F = 1/2Ne).
After the detection of reduced effective population sizes, kinship evaluation showed that most of the fish farms had related individuals (full sibling and half-sibling) in a proportion of at least 25%, as estimated from both marker sets (Table 3; Figure 1), with significant probability of mating between related breeders. The percentage of related individuals was notable in FF4 (61.0% and 51.4% for SNPs and SSRs, respectively) with a high proportion of full sibling individuals (25.0% and 36.0% for SNPs and SSRs, respectively). Although FF7 showed a considerable proportion of related individuals, this farm showed the highest rate of unrelated individuals (65.1% and 74.3% for SNPs and SSRs, respectively) (Figure 1). Confidence intervals (95%) of pairwise relatedness for each dyad are presented in Table S4. The PE2 for each locus for SSRs ranged from 0.04 (locus Pm3) to 0.31 (loci Pm2 and Pm7) with an exclusion probability of 0.81 for all loci. Considering SNPs, PE2 values ranged from 0.01 (loci 41_428, 437_455, 458_2209, and 83_761) to 0.12 (loci 1013_445, 213_629, 260_818, 391_875, 470_159, and 4_231) with an exclusion probability of 0.07 for all loci (Table S5).

Table 3.
Kinship analysis for the 8 SSR and 32 SNP loci in farmed populations of pacu (Piaractus mesopotamicus) (FF1–FF7), according to Wang’s rxy coefficient. Values are represented in percentage.
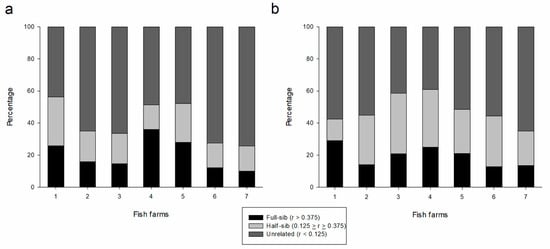
Figure 1.
Distribution of kinship values (rxy) by Wang’s estimator of fish farms (FF1–FF7) in SSR (a) and SNP (b) datasets. Threshold values for kinship analysis: unrelated (r < 0.125), half sibling (0.125 ≤ r ≤ 0.375), and full sibling (r > 0.375) individuals.
Global FST values suggested low genetic differentiation among populations when using SSRs (FST = 0.080, p < 0.006) or SNPs (FST = 0.067, p < 0.001). Pairwise FST values were also calculated for fish farms and significant differentiation was found between most pairs when considering SNP and SSR sets (p < 0.001 and p < 0.006, respectively) (Table 4). Low to high genetic differentiation was observed among the population pairs. Pairwise FST values for SSR loci were mostly significant (p < 0.006) and ranged between −0.002 and 0.204. The highest significant FST values (p < 0.006) were observed between WILD and FF4 (FST = 0.204) and FF2 and FF4 (FST = 0.143). Conversely, the lowest genetic differentiation was detected between FF3 and FF6 (FST = −0.002, p > 0.05). Regarding SNPs, the highest genetic differentiation (p < 0.001) was between FF2 and FF4 (FST = 0.146). Conversely, WILD and FF7 (FST = 0.032) registered the lowest genetic differentiation (p < 0.001).

Table 4.
Pairwise FST estimates from the 8 SSR loci (below diagonal) and from the 32 SNP loci (above diagonal) in the wild and farmed populations of pacu (Piaractus mesopotamicus).
To evaluate the level of admixture among samples, Bayesian model-based clustering analyses were performed based on the ΔK distribution by examining SSR and SNP loci. The selection of the estimated number of clusters in the dataset was based on the number of analyzed fish farms (K = 1 to 8). According to the analysis based on the Evanno method [30], the hypothesis of occurrence of K = 1 was discarded due to the higher −ln P(K) values found in all analyses (data not shown). The results showed that K values of 2 and 3 for SSRs and SNPs, respectively, were the most suitable to explain the population structure of pacu stocks (Figure 2). For the SNP dataset in K = 3, moderate clustering was found between fish farms, with three putative clusters composed of (1) FF1 and FF4; (2) FF2, FF3, and FF6; and (3) FF7 and WILD. In addition, FF5 seems to be represented as an admixture between the presented clusters (Figure 2a). However, considering the SSR analysis in K = 2, two genetic clusters were detected: (1) FF1, FF2, and WILD and (2) FF3, FF4, and FF5. The remaining fish farms (FF6 and FF7) were considered an admixture of both genetic groups (Figure 2b). Moreover, the analysis showed a structure for K = 5 in SSRs in which a clear cluster is composed of FF4. Hence, these analyses confirmed the estimated results of pairwise FST analyses.
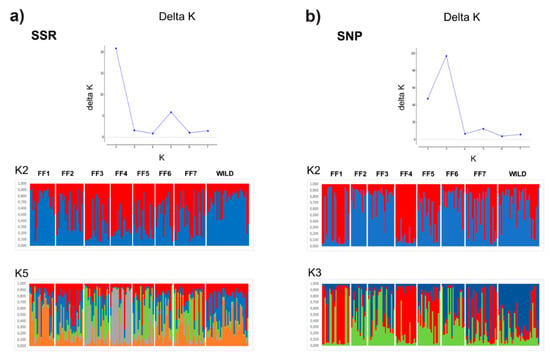
Figure 2.
ΔK statistics in STRUCTURE analysis as a function of the number of putative genetic clusters (K) for pacu populations. Coefficient of ancestry of pacu individuals collected from seven cultivated stocks (FF1–FF7) and one wild population (WILD), based on 8 SSR (a) and 32 SNP loci (b). Each vertical bar represents an individual. The fish farms are separated by vertical white lines. The color proportions of each bar correspond to individuals’ estimated membership fractions of each of the clusters.
Despite the genetic structure among populations being confirmed by STRUCTURE analysis, AMOVA analysis based on groups supported by the results of STRUCTURE showed that the higher percentage of genetic variation was assigned to differentiation within populations: 92.4% by SNPs (p < 0.001) and 91.1% by SSRs (p < 0.0001). The estimated variation between groups (FCT) was only 2.2% (p = 0.01) for SNPs and 3.9% (p = 0.02) for SSRs. In addition, the variation among populations and within groups (FSC) presented 5.3% of the genetic variation (p < 0.001) for SNPs and 5.0% (p < 0.0001) for SSRs.
4. Discussion
Currently, the methodology for breeding programs is well established in model species of worldwide aquaculture [37]. On the other hand, there are few studies applying genetic markers for the development of genetic selection programs in Neotropical fish. Several practices associated with Neotropical fish production, especially those related to the management of broodstock, may reduce the effective population size [38]. These practices are generally linked to the lack of registration and control of the broodstock, such as information on its origin, kinship, and mating record, and maintenance of the same stock over several generations, which results in increased susceptibility to inbreeding depression that compromises the foundation of hatchery stocks when starting breeding programs [39,40]. Therefore, our results demonstrate the importance of this study, which can be considered the baseline to create the basal broodstock for initial breeding programs of pacu, one of the most important native species of South American aquaculture.
Considering SSR loci, the analysis of genetic diversity parameters estimated on pacu broodstocks showed low values for the number of alleles (ranging around 4) and heterozygosity (ranging around 0.500). It was expected that cultivated stocks would present low genetic diversity values, as herein observed; these low values characterize populations with genetic drift events due to low effective population sizes and, consequently, recent bottleneck effects/founder events, similar to other studies in a related Neotropical species, Piaractus brachypomus [41]. However, the low levels of genetic diversity in farmed stocks were also shared by the WILD reference population. These values show lower results when compared to previous studies of genetic diversity in pacu natural populations based on SSRs [17] that reported a higher mean number of alleles (8.5 alleles) and higher heterozygosity values (Hexp ~ 0.600). Therefore, our hypothesis is that the WILD population may be threatened due to the high level of exploitation imposed by commercial and/or recreational fishing, since the region where the WILD population was collected is a famous fishing spot in Brazil [42]. Moreover, the Paraná River has been drastically impacted by dam barriers and pollution effects [17,42] which could negatively affect natural populations of pacu.
Although there is no comparative research of the genetic diversity of pacu populations using SNP markers, the values of heterozygosity (i.e., Hobs and Hexp) and MAF indicated low genetic variability in the farmed populations studied. This hypothesis was based on similar values found in WILD and sampled fish farms, considering that wild populations of pacu have no expressive heterozygosity values when using microsatellite loci. In addition, the diversity values herein observed were similar to those in other fish studies involving SNP analyses [43,44,45].
In general, farmed stocks have the tendency to show reduced genetic variability over generations due to artificial selection and a reduced number of breeders in the initial base population [11,46,47,48]. Therefore, it is important to evaluate how the genetic diversity can be maintained when considering the possibility of bottleneck events due to the low effective population size shown in fish farms. The significant bottlenecks detected for all stocks and their low Ne values (except for WILD) must receive special attention, since our results will serve to find a base population of pacu for upcoming breeding programs.
Kinship analysis has been an essential tool in genetic pre-breeding programs of fish to reduce inbreeding rates by directing the mating of unrelated individuals [49,50,51,52]. Except for FF7, which showed a high proportion of unrelated individuals for both markers (65.1% by SNPs and 74.3% by SSRs), all fish farms showed a substantial number of related individuals (half sibling or full sibling); this outcome results in a higher inbreeding risk, which can affect morphological and viability traits [10]. Thus, molecular identification of individuals is necessary to effectively monitor the genetic variability of the stocks and to assess how this variation can be maintained through selective mating [13,27]; this is especially the case in FF4, which presented a high proportion of pairwise kinship, the lowest value of effective population size, and a higher rate of inbreeding.
Impacts related to insufficient individuals used in hatchery productions and their interference with the genetic diversity of cultivated populations have been studied for important species cultivated worldwide, such as Atlantic salmon [53]. Similar studies in Neotropical species are fundamental and indispensable for ensuring the correct functioning of initial breeding programs, mainly due to the traditional practices related to the management of Neotropical broodstocks and the lack of genetic information of this species with high potential for production.
In the present study, our initial hypothesis was based on the fact that pacu farmed stocks did not have gene flow because the broodstocks are geographically isolated and producers frequently do not perform exchange of breeders among the fish farms. Therefore, we would expect higher values of FST estimates indicating genetic differentiation (driven by genetic drift and isolation), as detected in FF4, similar to in previous studies carried out in other related species such as P. brachypomus [41]. However, overall FST values suggested significant genetic differentiation of 0.067 by SNPs and 0.080 by SSRs (both values with p < 0.05), indicating low differentiation between the farmed stocks. This may be explained using two hypotheses: (i) stock foundation based on sharing of breeders among fish farms, resulting in genetic similarities between broodstocks, and/or (ii) stock foundation in the fish farms based on the capture of wild breeders, which are characterized as belonging to a panmictic unit due to the lack of genetic structure in natural populations [15,17], particularly because pacu have high gene flow capacity due to their migratory behavior in the wild.
The STRUCTURE analysis revealed differences between SNPs and SSRs, showing three and two clusters for the pacu stocks, respectively. In this study, the dataset of SNPs originated from the pacu liver transcriptome and these SNPs were annotated mostly in genes related to productive traits, including SNPs classified as non-synonymous mutations [18]. Therefore, both markers might differ in their ability to detect population structure; SNPs are mainly gene-associated while SSRs are expected to be neutral markers, which results in different mutation rates. Neutral markers, such as SSRs, are widely used to perform genetic variation analysis mainly in natural populations, while gene-associated markers could be more useful to analyze the variability of organisms in response to artificial selection [54,55]. This can explain the differentiation of FF1/FF4 in relation to the other fish farms as revealed by SNPs, particularly due to the breeding management practices carried out in these fish farms, such as mass selection to obtain individuals with better growth performance. To design suitable breeding programs in terms of genetic diversity in farmed pacu stocks, we also assume that it is better to use the information of the clusters generated by the SNPs. However, as SNPs are generally biallellic and with lower polymorphism compared to SSRs, additional SNPs markers need to be included in further analysis to achieve better conclusions about the genetic structure of pacu farmed stocks.
The results of this study are aimed to provide initial knowledge about the genetic profile of pacu stocks in different fish farms, considering the importance of pacu to South America aquaculture and the necessity to offer subsidies for the development of its production. The results provide information relevant to one of the most important cultivated Neotropical species. The genetic variability and differentiation of stocks and fish farm profiles considering the risks of inbreeding and the necessity of directed matings of the stocks should be known in order to take appropriate actions for the creation of the base population. In conclusion, the SNP and SSR sets showed their applicability in a pre-breeding program, particularly in delineating the formation of the best families in terms of genetic variability and genetic structure.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/668/s1, Table S1: Genetic variability parameters of 8 SSR loci in pacu populations, Table S2: Genetic variability parameters of the 32 SNP loci in pacu populations, Table S3: Linkage disequilibrium (LD) between the molecular markers, Table S4: Kinship values (rxy) by Wang’s estimator of fish farms (FF1–FF7) in SSR and SNP datasets with confidence intervals (95%) of pairwise relatedness for each dyad, Table S5: Parentage exclusion probabilities of loci for SSR and SNP datasets (PE2).
Author Contributions
Conceptualization, V.A.M.-F., F.d.P., G.V.V., F.F., P.M., F.P.-F., and D.T.H.; methodology, V.A.M.-F., F.d.P., and M.E.H.; formal analysis, V.A.M.-F., F.d.P., G.V.V., M.V., and D.T.H.; writing—original draft preparation, V.A.M.-F., F.d.P., G.V.V., M.V., and D.T.H.; writing—review and editing, V.A.M.-F., F.d.P., G.V.V., M.V., F.F., P.M., F.P.-F., and D.T.H.; project administration, D.T.H.; funding acquisition, V.A.M.-F., G.V.V., and D.T.H.
Funding
This research was funded by Pró-Reitoria de Pesquisa da UNESP (Prope 07/2015 – D.T.H.), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 446779/2014-8 and 305916/2015-7—D.T.H.; and 130262/2014-5—V.A.M.-F.), Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2014/03772-7—D.T.H. and 2014/12412-4—V.A.M.-F.), and the National Agency for the Promotion of Science and Technology of Argentina (ANPCyT) and the Government of Santa Fe province (grant number: PID 020-2013). F.d.P. is a student and G.V.V. is member of the researcher carrier from the National Council of Research of Argentina.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Resende, E.K. Migratory fishes of the Paraguay-Paraná basin excluding the Upper Paraná River. In Migratory Fishes of South America: Biology, Fisheries and Conservation States; Carolsfeld, J., Harvey, B., Ross, C., Baers, A., Eds.; World Bank: Victoria, BC, Canada, 2003; pp. 99–156. [Google Scholar]
- Ministério da Pesca e Aquicultura. Boletim Estatístico da Pesca e Aquicultura 2011; MPA: Brasília, Brazil, 2011; pp. 29–32. [Google Scholar]
- Instituto Brasileiro de Geografia e Estatística. Produção Pecuária Municipal 2016; IBGE: Rio de Janeiro, Brazil, 2017; Volume 44, pp. 1–51. [Google Scholar]
- Volkoff, H.; Sabioni, R.E.; Coutinho, L.L.; Cyrino, J.E.P. Appetite regulating factors in pacu (Piaractus mesopotamicus): Tissue distribution and effects of food quantity and quality on gene expression. Comp. Biochem. Physiol. Part A 2017, 203, 241–254. [Google Scholar] [CrossRef] [PubMed]
- Honglang, H. Freshwater fish seed resources in China. In Assessment of Freshwater Fish Seed Resources for Sustainable Aquaculture, FAO Fisheries Technical Paper No 501; Bondad-Reantaso, M.G., Ed.; FAO: Rome, Italy, 2007; pp. 185–199. [Google Scholar]
- FAO. The State of World Fisheries and Aquaculture 2010; FAO: Rome, Italy, 2010; pp. 3–87. [Google Scholar]
- Gjedrem, T.; Robinson, N.; Rye, M. The importance of selective breeding in aquaculture to meet future demands for animal protein: A review. Aquaculture 2012, 350–353, 117–129. [Google Scholar] [CrossRef]
- Lind, C.E.; Ponzoni, R.W.; Nguyen, N.H.; Khaw, H.L. Selective breeding in fish and conservation of genetic resources for aquaculture. Reprod. Domest. Anim. 2012, 47, 255–263. [Google Scholar] [CrossRef] [PubMed]
- Ponzoni, R.W. Genetic improvement effective dissemination: Keys to prosperous and sustainable aquaculture industries. In Development of Aquatic Animal Genetic Improvement and Dissemination Programs: Current Status and Action Plans; Ponzoni, R.W., Acosta, B.O., Ponniah, A.G., Eds.; WorldFish Center: Penang, Malásia, 2006; pp. 1–7. [Google Scholar]
- Kincaid, H.L. Inbreeding in fish populations used for aquaculture. Aquaculture 1983, 33, 215–227. [Google Scholar] [CrossRef]
- Li, L.; Lin, H.; Tang, W.; Liu, D.; Baolong, B.; Yang, J. Population genetic structure in wild and aquaculture populations of Hemibarbus maculates inferred from microsatellites markers. Aquac. Fish. 2017, 2, 78–83. [Google Scholar] [CrossRef]
- Xu, L.; Li, Q.; Xu, C.; Yu, H.; Kong, L. Genetic diversity and effective population size in successive mass selected generations of black shell strain Pacific oyster (Crassostrea gigas) based on microsatellites and mtDNA data. Aquaculture 2019, 500, 338–346. [Google Scholar] [CrossRef]
- Beaumont, A.R.; Hoare, K. Genetic considerations in the hatchery. In Biotechnology and Genetics in Fisheries and Aquaculture; Beaumont, A.R., Hoare, K., Eds.; Blackwell Science: Oxford, UK, 2003; pp. 73–90. [Google Scholar]
- Sui, J.; Luan, S.; Yang, G.; Chen, X.; Luo, K.; Gao, Q.; Wang, J.; Hu, H.; Kong, J. Genetic diversity and population structure of a giant freshwater prawn (Macrobrachium rosenbergii) breeding nucleus in China. Aquac. Res. 2018, 49, 2175–2183. [Google Scholar] [CrossRef]
- Iervolino, F.; Resende, E.K.; Hilsdorf, A.W.S. The lack of genetic differentiation of pacu (Piaractus mesopotamicus) populations in the Upper-Paraguay Basin revealed by the mitochondrial DNA D-loop region: Implications for fishery management. Fish. Res. 2010, 101, 27–31. [Google Scholar] [CrossRef]
- Calcagnotto, D.; Russello, M.; DeSalle, R. Isolation and characterization of microsatellite loci in Piaractus mesopotamicus and their applicability in other Serrasalminae fish. Mol. Ecol. Notes 2001, 1, 245–247. [Google Scholar] [CrossRef]
- Calcagnotto, D.; DeSalle, R. Population genetic structuring in pacu (Piaractus mesopotamicus) across the Paraná-Paraguay basin: Evidence from microsatellites. Neotrop. Ichthyol. 2009, 7, 607–616. [Google Scholar] [CrossRef]
- Mastrochirico-Filho, V.A.; Hata, M.E.; Sato, L.S.; Jorge, P.H.; Foresti, F.; Vera, M.; Martínez, P.; Porto-Foresti, F. SNP discovery from liver transcriptome in the fish Piaractus mesopotamicus. Conserv. Genet. Resour. 2016, 8, 109–114. [Google Scholar] [CrossRef]
- Villanova, G.V.; Vera, M.; Diaz, J.; Martinez, P.; Calcaterra, N.; Arranz, S. Isolation and characterization of 20 polymorphic microsatellite loci in the migratory freshwater fish Leporinus obtusidens (Characiformes: Anostomidae) using 454 shotgun pyrosequencing. J. Fish. Biol. 2015, 86, 1209–1217. [Google Scholar] [CrossRef] [PubMed]
- Posner, V. Desarrollo de Herramientas Biotecnológicas para el Cultivo de Pacú (Piaractus mesopotamicus). Bachelor’s Thesis, Universidad Nacional de Rosario, Rosario, Argentina, 2016. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Res. 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Heredity 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (version 1.2): A computer program to calculate F-statistics. J. Heredity 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.J.; Ovenden, J.R. NeEstimator V2: Re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Gjedrem, T.; Baranski, M. Selective Breeding in Aquaculture: An Introduction; Springer Science & Business Media: Amsterdam, The Netherlands, 2009; pp. 5–11. [Google Scholar]
- Garza, J.C.; Williamson, E.G. Detection of reduction in population size using data from microsatellite loci. Mol. Ecol. 2001, 10, 305–318. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. ARLEQUIN (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Jakobsson, M.; Rosenberg, N.A. CLUMPP: A cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 2007, 23, 1801–1806. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. An estimator for pairwise relatedness using molecular markers. Genetics 2002, 160, 1203–1215. [Google Scholar]
- Wang, J. COANCESTRY: A program for simulating, estimating and analyzing relatedness and inbreeding coefficients. Mol. Ecol. Resour. 2011, 11, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Martínez, P.; Fernández, J. Análisis de parentescos mediante el uso de marcadores moleculares. In Genética y Genómica en Acuicultura; Martínez, P., Figueras, A., Eds.; Publicaciones Científicas y Tecnológicas de la Fundación Observatorio Español de Acuicultura, CSIC: Madrid, Spain, 2009; pp. 241–308. [Google Scholar]
- Gjedrem, T.; Rye, M. Selection response in fish and shellfish: A review. Rev. Aquacult. 2018, 10, 168–179. [Google Scholar] [CrossRef]
- Alarcón, J.A.; Magoulas, A.; Georgakopoulos, T.; Zouros, E.; Alvarez, M.C. Genetic comparison of wild and cultivated European populations of the gilthead sea bream (Sparus aurata). Aquaculture 2004, 230, 65–80. [Google Scholar] [CrossRef]
- Duncan, N.J.; Sonesson, A.K.; Chavanne, H. Principles of finfish broodstock management in aquaculture: Control of reproduction and genetic improvement. In Advances in Aquaculture Hatchery Technology; Allan, G., Burnell, G., Eds.; Woodhead Publishing Limited: Cambridge, UK, 2013; pp. 23–75. [Google Scholar]
- Naish, K.A.; Seamons, T.R.; Dauer, M.B.; Hauser, L.; Quinn, T.P. Relationship between effective population size, inbreeding and adult fitness-related traits in a steelhead (Oncorhynchus mykiss) population released in the wild. Mol. Ecol. 2013, 22, 1295–1309. [Google Scholar] [CrossRef]
- Jorge, P.H.; Mastrochirico-Filho, V.A.; Hata, M.E.; Mendes, N.J.; Ariede, R.B.; Freitas, M.V.; Vera, M.; Porto-Foresti, F.; Hashimoto, D.T. Genetic characterization of the fish Piaractus brachypomus by microsatellites derived from transcriptome sequencing. Front. Genet. 2018, 9, 46. [Google Scholar] [CrossRef]
- Agostinho, A.A.; Pelicice, F.M.; Petry, A.C.; Gomes, L.C.; Júlio, H.F., Jr. Fish diversity in the upper Paraná River basin: Habitats, fisheries, management and conservation. Aquat. Ecosyst. Health 2007, 10, 174–186. [Google Scholar] [CrossRef]
- Vera, M.; Alvarez-Dios, J.A.; Fernandez, C.; Bouza, C.; Vilas, R.; Martinez, P. Development and validation of single nucleotide polymorphisms (SNPs) markers from two transcriptome 454-runs of turbot (Scophthalmus maximus) using high-throughput genotyping. Int. J. Mol. Sci. 2013, 14, 5694–5711. [Google Scholar] [CrossRef] [PubMed]
- Aykanat, T.; Johnston, S.E.; Orell, P.; Niemela, E.; Erkinaro, J.; Primmer, C.R. Low but significant genetic differentiation underlies biologically meaningful phenotypic divergence in a large Atlantic salmon population. Mol. Ecol. 2015, 24, 5158–5174. [Google Scholar] [CrossRef] [PubMed]
- Pocwierz-Kotus, A.; Bernas, R.; Kent, M.P.; Lien, S.; Leliuna, E.; Debowski, P.; Wenne, R. Restitution and genetic differentiation of salmon populations in the southern Baltic genotyped with the Atlantic salmon 7K SNP array. Genet. Sel. Evol. 2015, 47, 39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, L.; Shi, X.; Su, Y.; Meng, Z.; Lin, H. Loss of genetic diversity in the cultured stocks of the large yellow croaker, Larimichthys crocea, revealed by microsatellites. Int. J. Mol. Sci. 2012, 13, 5584–5597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhao, Z.; Xu, J.; Xu, P.; Bai, Q.; Yang, S.; Jiang, L.; Chen, B. Population genetic analysis of aquaculture salmonid populations in China using a 57K rainbow trout SNP array. PLoS ONE 2018, 13, e0202582. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, J.D.P.; Fazzi-Gomes, P.F.; Hamoy, I.G.; Santos, S.E.B.D.; Schneider, H.; Sampaio, I. Loss of genetic variability in the captive stocks of tambaqui, Colossoma macropomum (Cuvier, 1818), at breeding centres in Brazil, and their divergence from wild populations. Aquac. Res. 2018, 49, 1914–1925. [Google Scholar] [CrossRef]
- Sekino, M.; Sugaya, T.; Hara, M.; Taniguchi, N. Relatedness inferred from microsatellite genotypes as a tool for broodstock management of Japanese flounder Paralichthys olivaceus. Aquaculture 2004, 233, 163–172. [Google Scholar] [CrossRef]
- Porta, J.; Porta, J.M.; Martínez-Rodríguez, G.; Alvarez, M.C. Genetic structure and genetic relatedness of a hatchery stock of Senegal sole (Solea senegalensis) inferred by microsatellites. Aquaculture 2006, 251, 46–55. [Google Scholar] [CrossRef]
- Pino-Querido, A.; Hermida, M.; Vilariño, M.; Bouza, C.; Martínez, P. Statistical properties and performance of pairwise relatedness estimators using turbot (Scophthalmus maximus L.) family data. Aquac. Res. 2010, 41, 528–534. [Google Scholar] [CrossRef]
- Khadher, S.B.; Fontaine, P.; Milla, S.; Agnèse, J.F.; Teletchea, F. Genetic characterization and relatedness of wild and farmed Eurasian perch (Perca fluviatilis): Possible implications for aquaculture practices. Aquac. Rep. 2016, 3, 136–146. [Google Scholar] [CrossRef]
- Koljonen, M.L.; Tähtinen, J.; Säisä, M.; Koskiniemi, J. Maintenance of genetic diversity of Atlantic salmon (Salmo salar) by captive breeding programmes and the geographic distribution of microsatellite variation. Aquaculture 2002, 212, 69–92. [Google Scholar] [CrossRef]
- Seeb, J.E.; Carvalho, G.; Hauser, L.; Naish, K.; Roberts, S. Single-nucleotide polymorphism (SNP) discovery and applications of SNP genotyping in nonmodel organisms. Mol. Ecol. Res. 2011, 11 (Suppl. 1), 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Bai, J.; Li, Q. Comparative assessment of genomic SSR, EST–SSR and EST–SNP markers for evaluation of the genetic diversity of wild and cultured Pacific oyster, Crassostrea gigas Thunberg. Aquaculture 2014, 420–421, S85–S91. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).