Abstract
RabGTPase activating proteins (RabGAP) are responsible for directing the deactivation of vesicular trafficking master regulators associated to plant development, the RabGTPase proteins. Recently, RabGAPs were identified in Arabidopsis and rice, but studies were not yet reported in tomato. Herein, we identified 24 RabGAP-encoding genes in cultivated tomato (Solanum lycopersicum) and its wild relative genomes (Solanum pimpinellifolium and Solanum pennellii). We analyzed them based on their exon-intron structures, conserved protein motifs, putative subcellular localizations, phylogenetic and gene duplications analyses, interaction networks, and gene expression patterns in tomato. Phylogenetic relationship analysis also indicated that RabGAP family is classified into seven subclasses, of which subclasses I and II are plant-exclusive. Furthermore, segmental duplication events and positive evolutionary forces are associated with the maintenance of the number and function of their members. On the other hand, the protein–protein interaction networks on tomato suggested that members of subclasses I, II, and III could be associated to endocytic traffic routes. In addition, the qRT-PCR experiments in S. lycopersicum and Solanum chilense exposed to a salt stress treatment validated the differential expression patterns of 20 RabGAP genes in different tissues, development stages, and stress conditions obtained through extensive microarray-based analyses. This work suggests the critical role of RabGAP family in the context of intracellular vesicular trafficking in tomato, particularly under conditions of abiotic stress. It also contributes to the breeding programs associated with the development of crops tolerant to salt stress.
1. Introduction
Abiotic stress represents the main environmental challenge in the production of agronomically important crops. Salt, drought, or heat, among other stresses, alter the internal homeostasis of plants, damaging their tissues and organs, as well as reducing their yield [1]. To counteract these negative effects, mechanisms for synthesis and accumulation of compatible osmolytes [2], detoxification of reactive oxygen species [3], mobilization of ion and water transporters [4,5], or synthesis and mobilization of lipids are activated [6]. The performance of these mechanisms depends on the efficient vesicular traffic between the different subcellular compartments [7,8,9]. The small GTPases of the Rab family (RabGTPases) regulate the vesicular traffic, alternating between an “active” state GTP-bound and an “inactive” state GDP-bound, as a molecular switch. The activated state is dependent on guanine nucleotide exchange factors (GEF) proteins, which facilitate the GDP dissociation by GTP [10]. Then, RabGTPase proteins are recognized by effector proteins such as homotypic fusion and protein sorting (HOPS), transport protein particle (TRAPP), and class C core vacuole/endosome tethering (CORVET) complexes, among others, helping membrane fusions [11]. The “inactive” state of RabGTPase is promoted by association with RabGAP proteins. The RabGAP proteins hydrolyze GTP, allowing RabGTPases to be bio-available for a new cycle of vesicular trafficking. In plants, each RabGTPase directs a particular traffic route, but the knowledge of the routes in which many RabGAP proteins participate is still very limited [12].
Unlike yeast and animals, little is known about the RabGAP proteins in plants, the mechanisms that they modulate, their transcriptional regulation, or their role in the stress tolerance. From a genomic analysis in yeast, all RabGAPs should have a conserved TBC-like domain (Tre-2/Bub2/Cdc16) with a hydrolytic activity directed by an arginine and glutamine that act as a dual-finger mechanism on GTP-bound RabGTPases [13]. Other domains associated with RabGAP proteins are the Rab3-GTPase_cat and Rab3GAP2_N domains, which are specific for RabGTPase proteins of subfamily D (RabD) [14]. In plants, 25 and 24 RabGAP genes have been identified in rice and Arabidopsis thaliana, respectively [15], whereas in Vitis vinifera, only five RabGAPs have been found by EST analysis [16]. In general, the RabGAP family has been subdivided into nine subclasses (I to IX), of which the subclasses III, IV, V, VI, and VIII are present in plants, yeast, insects, and animals, whereas the subclasses I and II are plant-specific [15].
During environmental stress the vesicular trafficking mechanisms are up-regulated. For example, RabGTPase genes are regulated in a positive way, increasing the endocytosis, the vesicular traffic, and the vacuolar compartmentation of ions [9]. Among them, Rab5 of Mesembryanthemum crystallinum, a salt tolerant species, or OsRab7 of rice, increase their expression during in cold, dehydration or salt stress [17,18]. Other studies have demonstrated that the over-expression of RabGTPase genes associated to endocytosis mechanisms, like RabG3e and RabF1 in A. thaliana or PgRAB7 in tobacco, also increase the vacuolar compartmentalization of Na+ ions and the transport of proteins between organelles [9,19,20]. Likewise, the participation of proteins associated to molecular switching during abiotic stress has been evaluated. Among them, the overexpression of SchRabGDI1 of Solanum chilense is capable of increasing the endocytosis and salt tolerance of A. thaliana [21]. While the absence of AtVPS9, a GEF protein, compromises its salt stress tolerance [22]. However so far, the OsGAP1 protein from rice is the only RabGAP protein characterized in the context of abiotic stress [23]. Specifically, OsGAP1 interacts with RAB11, being both essential for vesicle trafficking of OsVHA-a1 (a vacuolar H+-ATPase) from trans-Golgi network (TGN) to the central vacuole and salt stress tolerance [24].
The cultivated tomato (Solanum lycopersicum) is one of the most important crops in the world. However, abiotic stress is the main limitation of their yield, development and agronomic properties [25]. On the other hand, wild tomato species such as Solanum pimpinellifolium, Solanum habrochaites, Solanum pennellii, and S. chilense, among others, are tolerant species to abiotic stress and thrive in extreme environments due to differential expression of a diverse set of genes that activate molecular and physiological mechanisms that allow them to effectively adapt [26,27]. The recent sequencing of the S. pennellii, S. lycopersicum, and S. pimpinellifolium genomes provides an excellent opportunity to identify and analyze the RabGAP gene family in the context of abiotic stress [28,29]. In this sense, we have identified each RabGAP gene from these tomato species, reporting amino-acid characteristics, phylogenetics, and the interaction analysis between RabGTPase–RabGAP proteins. Furthermore, through a comprehensive study of the transcriptional profiles in different tissues and stages of development, as well as during salt stress conditions, we provide a global overview of its putative biological and physiological functions. This research supplies an important framework for future research in tomatoes as well as for the development of genetic improvement strategies.
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of RabGAPs from Tomatoes
To determine the sequences corresponding to RabGAP proteins, a local database was established using the proteomes of S. lycopersicum (SlRabGAP), S. pennellii (SpeRabGAP), and S. pimpinellifolium (SpiRabGAP) obtained from the SolGenomics Network database (https://solgenomics.net) [30]. Here, the RabGAP proteins were determined by the specific TBC domain (Pfam PF00566) as a profile for Hidden Markov chains (HMM), with a cut-off value of 1e−10 in the HMMERv3.1 software [31]. Each protein was manually validated by comparative analyses with the Solgenomics, BLASTp (https://blast.ncbi.nlm.nih.gov), CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), and Pfam databases (http://pfam.xfam.org/) [32].
To assign a name to each RabGAP protein and determine the subclass to which it belongs, the complete length of the amino-acid sequence was aligned with its closest homolog of A. thaliana using the software ClustalO [15,33]. The phylogenetic tree was constructed with MEGA7 software using the neighbor-joining method with 5000 iterations bootstrap [34].
2.2. Sequence Amino-Acid Properties, Synteny and Gene Duplication Analysis
The isoelectric point (pI) and molecular weight (MW) were predicted with Expasy Compute pI/Mw tool (http://web.expasy.org/compute_pi/). The exon-intron structure of the genes was performed with the GSDS software (http://gsds.cbi.pku.edu.cn) [35]. In addition, the conserved motifs in the amino-acid sequence were identified using the web tool MEME (http://meme-suite.org) and analyzed with InterPro Scan 5 (https://www.ebi.ac.uk/interpro/) [36,37]. Values of hydropathy (GRAVY value) were calculated with the help of Expasy ProtParam tool (http://expasy.org/tools/protparam.html). The subcellular locations were determined using CELLO V2.5 (http://cello.life.nctu.edu.tw) [38], Wolf PSORT (https://wolfpsort.hgc.jp) [39], and MultiLoc2 tools (https://abi.inf.uni-tuebingen.de/Services/MultiLoc2) [40]. The predictions of protein-protein interactions were made with the STRING database (https://string-db.org) [41].
Synteny analysis was carried out using the chromosomal locations of homologous and paralogous RabGAP genes in S. lycopersicum and A. thaliana obtained from the PGDD database (http://chibba.agtec.uga.edu/duplication/) [42]. The graphic representation was made using the web-based service ClicO FS [43]. Along with this, the selection pressure of RabGAP genes was determined by the ratio of nonsynonymous to synonymous nucleotide substitutions (Ka/Ks) between paralogs in S. lycopersicum. The approximate date of the duplication events was estimated using T = (Ks/2λ) × 10−6 million years ago (mya), based on the clock-like rates (λ) in Solanaceae of 1.5 × 10–8 [44].
2.3. RabGAP Gene Expression Profiles in Databases
Tissue-specific expression profiles of the RabGAP genes of S. lycopersicum and S. pennellii were obtained from the Bio-Analytic Resource database from the University of Toronto (http://bar.utoronto.ca). The data were obtained from flowers, shoots, leaves, vegetative meristems, seedling shoots, seedling roots, mature fruit, and developing fruit [45]. To establish a relation between the expression profiles of the RabGAP genes and the capacity of tolerance to abiotic stress among S. lycopersicum and its wild relatives (S. pimpinellifolium, S. habrochaites, and S. pennellii), values normalized in respect to S. lycopersicum were obtained from the “Tomato Expression” database (http://malooflab.phytonetworks.org/apps/tomato-expression/) [45]. In addition, to study the expression profiles of RabGAP genes in response to abiotic stress, we used the PLEX database (http://www.plexdb.org). The data were extracted from L6 (GEO accession GSE16401), L12 and L13 experiments (GEO accession GSE22304). The L6 experiment corresponded to a salt stress assay considering samples of leaves from 6-week-old S. lycopersicum and S. pimpinellifolium treated with NaCl 200 mM for 5 h [46]. The L12 experiment correspond to drought stress considering samples of leaves from four-week-old S. lycopersicum and S. habrochaites, previously non-irrigated for 14 days. Finally, the L13 experiment corresponded to a heat stress assay considering samples of leaves from four-week-old S. lycopersicum and S. habrochaites treated at 40 °C [47]. The profiles are relative to the control without stress.
2.4. Plant Material and Gene Expression Analysis
Seeds of S. chilense (Dunal) and S. lycopersicum were germinated in a mixture of perlite, vermiculite and peat moss (1:1:1), grown under greenhouse conditions at 23–25 ºC, with photoperiod of 16/8 h light/dark, irrigated with 400 mL every five days and fertilized with a commercial Hoagland’s solution (1/4 strength) every 10 days. At the sixth week, a saline stress of NaCl 300 mM was applied to a group of plants. Leaves and roots were collected at 0, 3, 6, 12, 24, 48, and 72 h for the gene expression analyses. Then, total RNA from both tissues of S. chilense was extracted, treated, and quantified following the protocol of San Martín-Davison et al. (2017) [21]. To obtain the first strand of cDNA, 2 μg of RNA was retro-transcribed in 20 μL of reaction using the First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) and following the manufacturer’s instructions.
Relative levels of gene expression were measured by qRT-PCR, using the Stratagene Mx3000p thermocycler (Agilent Technologies, Santa Clara, CA, USA). Maxima SYBR Green/ROX qRT-PCR Master MIX (Thermo Scientific) was used for all reactions according to the protocol described by the manufacturer. For each sample (three biological replicates), qRT-PCR was performed in triplicate (technical replicates) using 10 μL of Master MIX, 0.5 μL of 250 nM of primers, 1 μL of cDNA and nuclease-free water in a final volume of 20 μL [48]. The amplification was followed by a melting curve with continuous fluorescence measurement at 55 to 95 °C. The data were manually analyzed, and the expression normalized with the Ubiquitin3 gene [49]. All primers are listed in Table S1. The transcript levels of each gene were evaluated using the 2−ΔΔCT method [50]. The qRT-PCR data were analyzed using ANOVA (one-way analysis of variance) tests (with a significance of p-value < 0.05). Data management and standardization were performed with R version 3.2.5 [51].
3. Results
3.1. 24 Proteins Organized in 7 Subclasses Constitute the RabGAP Family in Tomato
An analysis of HMM on the proteomes of S. lycopersicum, S. pimpinellifolium, and S. pennellii allowed us to identify 24 putative RabGAP proteins in each species. They were named according to the closest ortholog of A. thaliana [15]. Among them, 21 RabGAPs possessed the TBC domain, two the Rab3-GTPase_cat domain and one the Rab3GAP2_N domain [14,52]. At the amino-acid level, all RabGAP proteins had the conserved residues of arginine and glutamine, critical for catalytic activity. The exceptions were RabGAP23b and those with the Rab3-GTPase_cat and Rab3GAP2_N domains, see in Figure S1. In S. lycopersicum, the RabGAP proteins had a length between 352 (SlRabGAP18) and 942 amino-acids (SlRab3GAP2) and a theoretical isoelectric point ranging from 4.79 (SlRabGAP6) to 8.97 (SlRabGAP18). Similar characteristics were also observed in the RabGAP protein families of S. pennellii and S. pimpinellifolium. On the other hand, the GRAVY index, associated to the hydrophobicity level of a protein, was less than 0, revealing that all RabGAPs are hydrophilic. In this same sense, the prediction of subcellular localization using three different web servers indicated that the main locations for all RabGAP proteins were in the nucleus and cytoplasm, as shown as in Table 1.

Table 1.
List of RabGTPase activating proteins (RabGAP) genes found in domesticated and wild tomatoes and their physical and chemical characteristics. Data obtained from Sol Genomics Network database. Gene prefixes represent Solanum lycopersicum (Sl), Solanum pennellii (Spe), and Solanum pimpinellifolium (Spi). Subcellular location: ML2, MultiLoc2; WPS, Wolf PSORT. Nuc, Nuclear; Cyt, Cytoplasmic; Chl, Chloroplast; MP, Plasma Membrane; ER, Endoplasmic Reticulum; and Mit, Mitochondria.
In respect to the classification of RabGAP proteins, the phylogenetic analysis evidenced that they are grouped in seven subclasses (I, II, III, IV, V, VI-Rab3GAP, and VIII), according to their homologs in A. thaliana, shown in Figure 1 [15]. This analysis revealed that the subclass with the most members was subclass I and those with the fewest members were subclasses III and V. In addition, subclasses II, V, and VIII of S. lycopersicum had fewer members than those of A. thaliana, while subclasses III and IV had more.
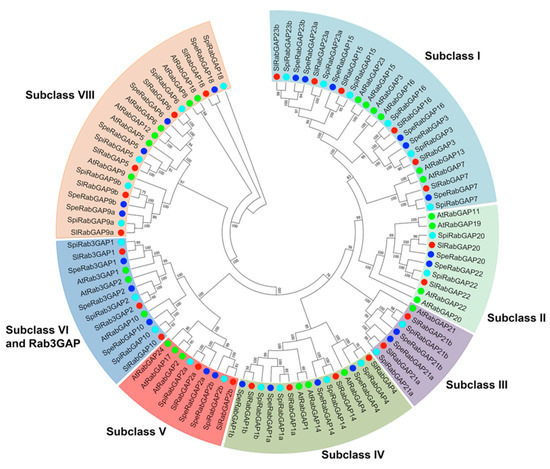
Figure 1.
Phylogenetic tree of tomato RabGAPs. Seven subclasses (I, II, III, IV, V, VI, and VIII) were identified. 26 RabGAP of A. thaliana [15] are represented in green circles. In red, 24 RabGAP of S. lycopersicum; in light blue, 24 RabGAP of S. pimpinellifolium; and in dark blue, 24 RabGAP of S. pennellii.
3.2. Events of Segmental Duplication Have Kept Constant the Number of Members of the RabGAP Family
To evidence the genomic organization and the role of duplication events in the evolutionary history of RabGAP family genes, synteny mapping, intron-exon structure identification and selection pressure analysis were performed, see in Figure 2. First, the genomic analysis revealed that the SlRabGAP genes were differentially distributed on 11 chromosomes of S. lycopersicum: One gene each on Sl-Chr1, -4, -5, -10, and -11; two genes each on Sl-Chr2 and Sl-Chr3; three genes each on Sl-Chr6, -9, and -12; and six genes on Sl-Chr7. Then, the comparative analysis between the exon-intron structures and their chromosome locations allowed us to identify seven events of segmental duplication (SlRabGAP20-SlRabGAP22, SlRabGAP21a-SlRabGAP21b, SlRabGAP1a-SlRabGAP1b, SlRabGAP9a-SlRabGAP9b, SlRabGAP5-SlRabGAP6, SlRabGAP2a-SlRabGAP2b, and SlRabGAP3-SlRabGAP16) and one triplication event between SlRabGAP23a, -23b, and -15. Along with the above, in all these cases the ratios of synonymous and non-synonymous nucleotide substitutions (Ka/Ks) were less than 1, indicating that the function of the genes has not diverged and has been maintained due to a stabilizing selection force. Finally, the estimated time in which these duplications occurred was 15 to 28 mya, see in Table 2.
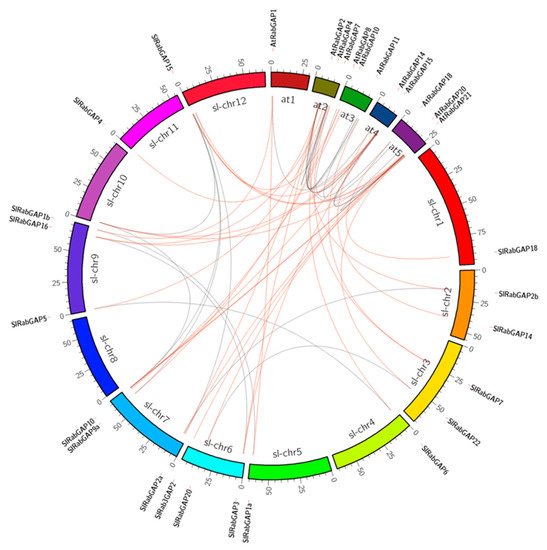
Figure 2.
Syntenic relationships among RabGAP. Paralogy and orthology relations between RabGAP from S. lycopersicum and Arabidopsis thaliana. Orthologous and paralogous genes are connected by red and gray lines, respectively.

Table 2.
Evolutionary parameters for duplicated RabGAP genes. Table shows Ka/Ks ratios, duplication and selection types and time of divergence.
In relation to the characterization of the RabGAP gene structure, the results showed that the number of introns is very diverse, see in Figure 3, Figure 3A. On average, the RabGAP genes had 13 introns, of which RabGAP2a, -20, and -22 had the lowest number (four introns), whereas Rab3GAP2 had the highest number (21 introns). On the other hand, the RabGAP proteins were characterized by having between four or five conserved motifs associated with the “catalytic core” from the TBC domain, as shown in Figure 3, Figure 3B. Exceptionally, the catalytic domain of subclass VI, specifically associated with Rab3, was present in three proteins—Rab3GAP1, Rab3GAP2, and RabGAP10—as seen as in Table 1.
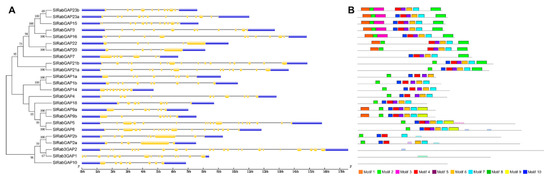
Figure 3.
Identification of exon-intron structure and conserved motifs in RabGAP from S. lycopersicum. (A). Exon-intron structure of RabGAP genes from S. lycopersicum. In yellow, exons; in blue, the 3′ and 5′ UTR regions; and the gray lines are introns. (B) Conserved motif of RabGAP proteins. Ten motifs were identified with MEME database. The gray lines are the complete amino-acid sequence represented proportionally. Each color represents a different motif. Together, the pink, red, purple, orange, and light blue motifs represent the TBC domain. The consensus sequences are detailed in Table S2.
3.3. In Silico Analysis of Protein-Protein Interactions Suggests the Association of RabGAP Proteins with Specific Vesicular Trafficking Pathway
In order to suggest specific vesicular trafficking pathways in which the RabGAP proteins could participate, we identified their possible affinities or physical interactions with the RabGTPase subfamilies of S. lycopersicum using the STRING database, see in Table S3. The results showed that in the exocytic pathway, SlRabGAP5, -6, -9a, and -9b could be interacting with RabGTPases of the subfamilies-A and E. In the anterograde pathway, RabGAP4 could be interacting with the subfamily-D, whereas in the retrograde pathway, SlRabGAP2a, -2b, -20, -21a, -21b, -23a, and -5 could be related with subfamily-H. In the endocytic pathway, only RabGAP18 could be interacting with subfamily-F, whereas SlRabGAP3, -7, -15, -16, -20, -21a, -21b, and -22 could be relating with subfamily-G (see in Figure 4).
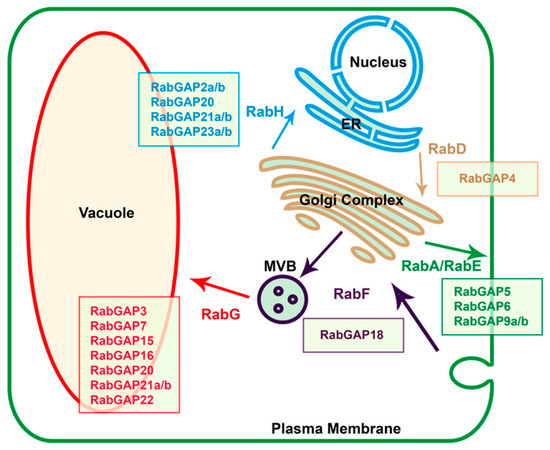
Figure 4.
Identification of proposed protein-protein interactions between RABs and RabGAPs. The vesicular trafficking routes directed by the RabGTPase subfamilies between the different organelles and membranes are represented by arrows. RabGAP proteins were associated to each RabGTPase using the protein-protein interaction functional networks provided by the STRING database [41]. ER: Endoplasmic Reticulum. MVB: Multi-Vesicular Body. All proposed protein-protein interaction networks are detailed in Table S4.
We also observed that some RabGAPs could form protein complexes. Homodimers are formed in the case of SlRabGAP1a, -1b, -3, -4, -9a, -9b, -14, -16, -21a, -21b, or -23a; heterodimers between SlRabGAP10, -18 or -23a; protein complexes between vacuolar protein sorting (VPS) and SlRabGAP3, -7, -15, -20, -21a, -21b, -22, -23a, or -23b. The complexes between proteins related to cell division checkpoint processes, such as SlRabGAP5, -6, -7, -9a, or -9b, and complexes with proteins that respond to hormones involved in stress, such as SlRabGAP5 or -6, were also identified (see Table S2).
3.4. Differential Expression of RabGAP Genes in Different Tissues and Development Stages of Cultivated and Wild Tomato Species
To explore patterns in the transcriptional regulation of the RabGAP genes, we analyzed the expression profiles in different organs and development stages of S. lycopersicum and S. pennellii (see Figure 5, File S1). We evidenced that the expression patterns of RabGAP genes in flowers, shoots, seedling shoots, seedling roots, and mature fruit of S. pennellii were inverse to that of S. lycopersicum. Furthermore, in almost all organs of S. lycopersicum the expression patterns were positive, while in S. pennellii, they were only positive in leaves and vegetative meristems. At the subclass level, no obvious differences were observed. However, we highlight that the most up-regulated RabGAP genes in S. lycopersicum were SlRabGAP23a in flowers, leaves, and vegetative meristems, SlRabGAP18 in stems and developing fruit, SlRabGAP2a in seedling shoots, and RabGAP15 in seedling roots. While in S. pennellii were SpRabGAP18 in seedling shoots, seedling roots, flowers and stems. SpRabGAP23a in leave, SpRabGAP3 in vegetative meristems, SpRabGAP15 in mature fruit, and SpRabGAP2a in developing fruit. It is also interesting to mention that RabGAP18, the only RabGAP possibly associated with RabGTPase of the F subfamily, was ubiquitously expressed in all organs and stages of development of both wild and cultivated tomato.
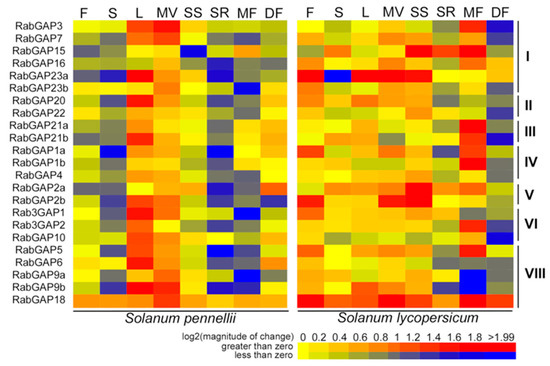
Figure 5.
Expression patterns of RabGAP genes in different tissues, organs and developmental stages of tomato. Induction of genes is represented in red and repression in blue. F: flowers, S: shoots, L: leave, MV: vegetative meristems, SS: seedling shoots, SR: seedling roots, MF: mature fruit, DF: developing fruit. All values are detailed in Table S5.
To evidence differential patterns of regulation between tomato and wild relatives in normal conditions, we analyzed the expression profiles of the RabGAP genes in leaves of S. pimpinellifolium, S. habrochaites, and S. pennellii, see in Figure 6. SpiRabGAP23a, ShaRabGAP3, and SpeRabGAP10 were the most up-regulated genes in each wild species, while SpiRab3GAP1, ShaRabGAP14, and SpeRabGAP16 were the most down-regulated. Studying an expression pattern associated with the degree of tolerance to abiotic stress of each wild species, we observed that RabGAP1a, -10, and -21a had positive correlations, whereas only RabGAP18 had a negative tendency. In addition, the analysis showed that both RabGAP2a and -22 represented about 35% of the number of transcripts of RabGAP family (see Table S6).
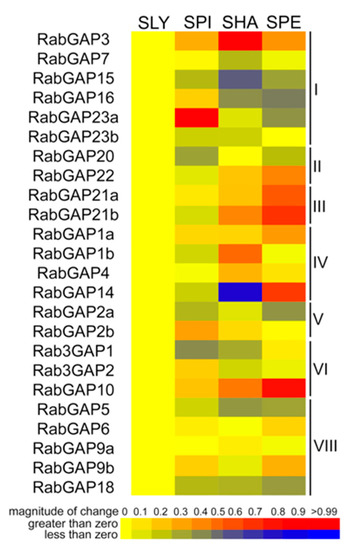
Figure 6.
Expression profiling of RabGAP genes in wild tomato species. Transcript levels of RabGAP genes from wild tomato species relative to S. lycopersicum are represented. Higher or lower gene expressions are shown in red and dark blue, respectively. Nomenclature: SLY: S. lycopersicum, SPI: S. pimpinellifolium, SHA: S. habrochaites and SPE: S. pennellii. All values are detailed in Table S6.
3.5. RabGAP Genes Associated with Endocytic and Pre-Vacuolar Trafficking Are Up-Regulated in Roots Subjected to Salt Stress
To obtain more information about the role of RabGAP genes under abiotic stress conditions, we analyzed their expression profiles in response to heat, drought and salt stress in S. lycopersicum and compared them with those results obtained in wild relatives. In this case, the only data available for the RabGAP3, -4, -18, -20, -21a, and -21b were found in different databases (see Figure 7). During heat stress, only RabGAP4 and -20 were as strongly induced in the tolerant species as RabGAP4, -21a, and -21b were in the S. lycopersicum. Interestingly, under conditions of drought stress, unlike heat stress, RabGAP3, -4, -18, -20, and -21b of tolerant species versus RabGAP21a and -21b of S. lycopersicum, were up-regulated. Finally, under salt stress, similar expression profiles were observed in the sensitive and tolerant species. In this case, RabGAP21a and -21b were the most up-regulated genes in both species.
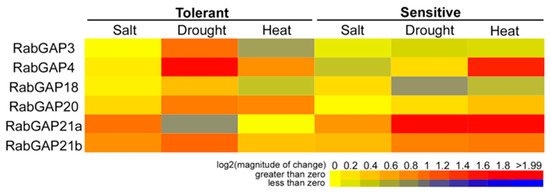
Figure 7.
Transcriptomic profiles of RabGAP genes from tomato species with different degrees of abiotic stress tolerance. Expression patterns of RabGAP genes in S. lycopersicum (sensitive species) and S. pimpinellifolium or S. habrochaites (tolerant species) in saline, drought and heat stress are represented. Higher or lower gene expressions versus the control, without stress, are shown in red and dark blue, respectively. Saline stress: S. lycopersicum versus S. pimpinellifolium (GEO accession GSE16401). RNA extraction from leaves of 6-week-old plants treated with NaCl 200 mM for 5 h [46]. Stress by drought: S. lycopersicum versus S. habrochaites (GEO accession GSE22304). Extraction of RNA from leaves of four-week-old plants, previously without irrigation for 14 days. Heat stress: S. lycopersicum versus S. habrochaites (GEO accession GSE22304). RNA extraction from leaves of four-week-old plants treated at 40 °C [47]. All values are detailed in Table S7.
Considering the low number of genes previously analyzed, and to learn more about the stress response and determine the possible biological effect of RabGAP genes of S. lycopersicum during salt stress, 20 RabGAP genes representing at least one gene of each subclass were selected for expression analysis by qRT-PCR (see Figure 8). Along with the above, we used S. chilense as the wild and halophyte species to contrast the analyses, as shown in Figure 9. The results showed that, in both leaves and roots of S. lycopersicum, four genes were up-regulated (SlRabGAP2b, -7, -9b, and -20), while 10 genes were induced in leaves, and three only in roots. At early times of salt stress, RabGAP2b and -22 in roots and RabGAP9b and -20 in leaves were the most up-regulated. Similarly, at late times, RabGAP3 and -22 in roots and SlRabGAP1a, -4, -10, and -18 in leaves had the highest expression levels. Interestingly, RabGAP3, -16, and -22 in roots and RabGAP1b in leaves managed to maintain a strong and stable induction throughout the experiment. In this context, it should be also noted that pairs of paralogous genes (RabGAP1a, -1b, -2a, -2b, -9a, -9b, -21a, and -21b) showed different expression patterns.
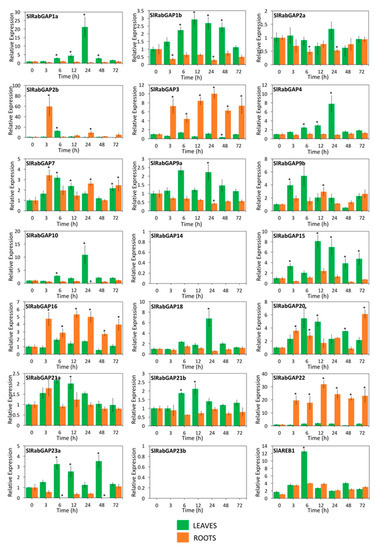
Figure 8.
Spatio-temporal expression patterns of RabGAP genes of S. lycopersicum exposed to salt stress. Relative levels of SlRabGAP gene transcripts in roots (orange bar) and leaves (green bar) of 10–12-week-old S. lycopersicum plants were determined at 0, 3, 6, 12, 24, 48, and 72 h after stress initiation with 300 mM NaCl. The relative levels of the salt-stress marker gene (SlAREB1) were measured. In SlRabGAP14 and -23b, transcripts were not detected. Columns and error bars represent the mean and standard deviation for three biological and three technical replicates. * p-value < 0.05.
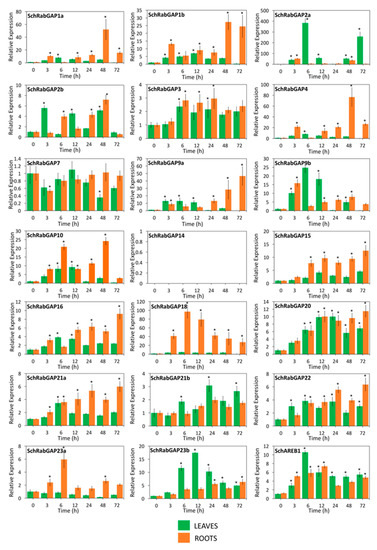
Figure 9.
Spatio-temporal expression patterns of RabGAP genes of S. chilense exposed to salt stress. Relative levels of RabGAP gene transcripts in roots (orange bar) and leaves (green bar) of 10–12 weeks old S. chilense plants were determined at 0, 3, 6, 12, 24, 48, and 72 h after stress initiation with 300 mM NaCl. The relative levels of the salt-stress marker gene (SchAREB1) were measured. In SlRabGAP14, transcripts were not detected. Columns and error bars represent the mean and standard deviation for three biological and three technical replicates. * p-value < 0.05.
The expression patterns of RabGAP genes in S. chilense revealed differences with those of S. lycopersicum, as seen as in Figure 9. In this species, 14 RabGAP genes significantly increased their relative expression in both leaves and roots during stress, whereas RabGAP15, -18, and -23a did it only in roots, and RabGAP21b only in leaves. We want to highlight the high and stable induction levels of RabGAP18 in roots of S. chilense throughout the salt stress, the inductions of RabGAP1a, -4, and -9a in roots at late times, and the large fluctuations in the expression of RabGAP2a in leaves. In addition, similar to S. lycopersicum, the expression levels of paralogous gene pairs also differed from the tissue in which they were expressed and their levels of induction.
4. Discussion
Within the Solanaceae family, the commercial tomato, S. lycopersicum, is the main cultivated vegetable in the world, but its sensitivity to abiotic stress negatively affects its productivity. On the other hand, wild species related to it such as S. pimpinellifolium, S. habrochaites, S. pennellii, and S. chilense are able to tolerate different degrees of abiotic stresses. A strategy of tolerance that S. chilense particularly uses could be associated with vesicular trafficking processes such as endocytosis, vacuolar compartmentalization of toxic ions, and damaged element recycling [21]. In this context, we have identified and characterized for the first time the family of RabGAP genes of different Solanaceaes through analyzing of the whole genome, mainly through transcriptional study. This allowed us to indicate the possible relationships between them and the components that direct the vesicular trafficking.
4.1. Overview of RabGAP Gene Family in the Tomato Genome
The RabGAP gene family encodes proteins capable of accelerating the hydrolysis of GTP to GDP bound to RabGTPase proteins, allowing them to direct a new round of vesicular trafficking [53]. In this work, 24 RabGAP genes were identified in each tomato species (S. lycopersicum, S. pennellii, and S. pimpinellifolium), a similar number of genes to that of other species such as rice or A. thaliana [15]. However, according to motifs and multiple alignment analyses, the results suggest that four proteins, including RabGAP23b, should not have RabGAP activity due to the absence of a conserved Arginine residue. In this regard, it has been evidenced that a point mutation on this residue in OsGAP1 (R450A) causes a severe loss of this activity against two RabGTPases—OsRab11 and OsRab8a—revealing its importance in catalytic activity [24].
Interestingly, the subclasses I and II of the RabGAP family are composed of six and two RabGAP genes, respectively. In addition, both families are potentially exclusive to plants and it is predicted that their evolution is an adaptation mechanism due to the loss of function related to the RasGAP family [15]. From the above, possible coevolution mechanisms between RabGTPases and RabGAPs, which have sculpted the endomembrane system, have been suggested [54]. On the other hand, the equivalent number of members of the RabGAP family between tomato and evolutionarily distant species, such as A. thaliana or rice, is an important question to raise. In this sense, it has been suggested that duplication and diversification events from five ancestral RabGAP genes have been the mechanisms that have kept the number of genes constant [15,28,55]. The exon-intron structure also provides relevant information to understand the evolution process of the RabGAP family. In this analysis, we found paralogous genes from subclasses I (RabGAP23b and -15), III (RabGAP21b and -21b), and VIII (RabGAP9a and -9b) located on chromosomes 7 and 12. The arrangements and size of exons and introns of these genes supports the hypothesis of an important segmental duplication event between both chromosomes, which has also been reported for members of the B-BOX, LEA, and GRAS families of S. lycopersicum [56,57,58]. Together, these data suggest that events of segmental duplications have played an important role in the gene evolution of the RabGAP gene family.
4.2. The New Subfamily VI and Rab3GAP Could Play an Important Role in Autophagy and Stress Tolerance
Regarding the characteristics of other identified genes, we found two new members of subclass VI with a Rab3GAP domain whose functional roles in plants have not yet been determined, see in Figure 1 and Table 1. However, homolog genes in animal cells participate in: (i) the deactivation of Rab3 [14], a homolog of RabD in plants, (ii) interaction with ATG8 in autophagy processes [59,60], (iii) or activation of a RabGTPase of subfamily C, acting as a guanine exchange factor in degradative pathways and macro-autophagy [61]. Intriguingly, a RabD of plants (RABD2A) localizes with proteins associated with salt-stress tolerance capacity, such as EHD1 or SYP41 [62,63]. In a similar way to expression patterns of Rab3GAP1, Rab3GAP2, and RabGAP10 of S. lycopersicum, the autophagy is a mechanism rapidly induced by salt stress [64]. These interesting characteristics suggest that the proteins of subclass VI can probably interact with RabD or ATG8 and play an important role during salt stress in tomato.
4.3. Endocytosis-Associated RabGAPs Could Be Mediating the Salt Stress Tolerance in Tomato
In general, the different RabGAP proteins of S. lycopersicum identified in this work were located in the nucleus or cytoplasm. Particularly, the predicted nuclear localization was consistent among RabGAP22 from S. lycopersicum and A. thaliana [65]. The patterns of expression of both genes are also similar. In both species, RabGAP22 is expressed in various tissues and stages of development, such as flowers, roots, or vegetative meristems. Along with this, RabGAP22 from A. thaliana can interact with peroxisome proteins such as AGT1 and be associated with photorespiration processes or adaptation mechanisms against abiotic stress [66]. According to this, the homolog of AtRabGAP22 in S. lycopersicum could have an important role in tomato roots when they are subjected to salt stress. On the other hand, the predicted subcellular location of RabGAP21a from S. lycopersicum, with respect to its rice homolog, OsGAP1 [23], is also coherent. In this case, the data suggest that SlRabGAP21a could be localized in the Trans-Golgi network and the pre-vacuolar compartments. Equally, it is interesting to observe that the expression patterns of OsGAP1 and RabGAP21a of S. lycopersicum are similar in leaves and roots when plants are subjected to salt stress [24]. These data suggest that RabGAP21 could also have an important role in a trafficking pathway that mobilizes transporters or proteins associated with the vacuole. Additionally, SlRabGAP21a could potentially interact with the TVP38 protein, which has been associated with the stability of the membranes [67]. The results for RabGAP18 are also consistent with the fact that the endocytosis mechanism is an inducible process against salt stress and regulated for RabGTPases of the subfamily-F [20,22]. To date, the RabGAP proteins that regulate this subfamily-F are unknown, however, the protein-protein interaction analysis suggested that RabGAP18 could be an exclusive regulator of this family. Interestingly, RabGAP18 is also strongly induced in the roots of S. chilense when they are exposed to salt stress in a similar manner to SchRabGDI1, whose overexpression in A. thaliana can increase its tolerance to salt stress [21]. In general, the various antecedents support the idea that RabGAP have a certain degree of affinity for RabGTPases, but the results equally show a high degree of promiscuity for some of them. In this sense, further analyses and experiments are needed to establish the vesicular trafficking routes in which the members of the RabGAP family of S. lycopersicum participate in order to finally understand their physiological functions during the abiotic stress.
5. Conclusions
In conclusion, we have identified 24 RabGAP genes in each S. lycopersicum, S. pennellii, and S. pimpinellifolium and classified them into seven subclasses. Two proteins with Rab3GAP domain were identified for the first time. All of them have the TBC domain characteristic of the RabGAP family. Evolutionarily segmental duplication and purifying selection have maintained constant the number and function of these genes in the S. lycopersicum genome. The results presented here show that the RabGAP proteins, such as RabGAP18 or RabGAP22, could be associating to different intracellular vesicular trafficking pathways and that the genes that encode them can change their expression depending on both endogenous and environmental stimuli. In this sense, the study of the regulation mechanisms of gene expressions, as well as the specific interactions with RabGTPase proteins could provide important information to understand their participation in tolerating abiotic stress in tomato species.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/10/9/638/s1, Figure S1: Multiple alignment of RabGAP sequences. RabGAP amino-acid sequences of S. lycopersicum and A. thaliana. In green, conserved amino-acids associated with catalytic activity, File S1: Expression patterns of RabGAP genes in different tissues and development stages of tomato (Tomato eFP Browser), Table S1: Primers used in this study, Table S2: Regular expression profile of the conserved motifs defined by MEME of SlRabGAP, Table S3: ID of RabGAP genes in A. thaliana used in phylogenetic analysis, Table S4: Protein-Protein interaction networks predicted with STRING database, Table S5: Relative expression of RabGAP genes in different tissues and development stages of S. lycopersicum and S. pennellii. Log2 (ratio), Table S6: Relative expression of RabGAP genes among different tomato species (normalized counts per million), Table S7: Relative expression of RabGAP genes from contrasting tomato species in different types of abiotic stresses.
Author Contributions
J.M.-E. and S.R.-L. conceived the project and designed the experiments; J.M.-E. and S.R.-L. performed the experiments; J.M.-E. and J.S.-C. analyzed the experiment data and wrote the manuscript. All authors read and approved the manuscript.
Funding
This work was funded by Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) Grants No. 1140636 and No.1170554. J.M.-E. was funded by CONICYT-CHILE doctoral fellowships Grant No. 21170702. J.S.-C. was the recipient of an Universidad de Talca doctoral fellowship.
Acknowledgments
We thank the Academic Writing Center in the Programa de Idiomas in the Universidad de Talca.
Conflicts of Interest
The authors declare no conflict of interest
References
- Cramer, G.R.; Urano, K.; Delrot, S.; Pezzotti, M.; Shinozaki, K. Effects of abiotic stress on plants: A systems biology perspective. BMC Plant Biol. 2011, 11, 163. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.H.; Murata, N. Glycinebetaine: An effective protectant against abiotic stress in plants. Trends Plant Sci. 2008, 13, 499–505. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Chan, Z. ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 2015, 6, 1925. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Sade, N.; Moshelion, M. Plant Aquaporins and Abiotic Stress. In PAMP Signals in Plant Innate Immunity; Springer Science and Business Media LLC: Berlin, Germany, 2017; pp. 185–206. [Google Scholar]
- Okazaki, Y.; Saito, K. Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 2014, 79, 584–596. [Google Scholar] [CrossRef] [PubMed]
- Einset, J.; Nielsen, E.; Connolly, E.L.; Bones, A.; Sparstad, T.; Winge, P.; Zhu, J.-K. Membrane-trafficking RabA4c involved in the effect of glycine betaine on recovery from chilling stress in Arabidopsis. Physiol. Plant. 2007, 130, 511–518. [Google Scholar] [CrossRef]
- Woollard, A.A.; Moore, I. The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 2008, 11, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Reddy, M.K.; Sopory, S.K.; Agarwal, P.K. Plant Rabs: Characterization, Functional Diversity, and Role in Stress Tolerance. Plant Mol. Biol. Rep. 2009, 27, 417–430. [Google Scholar] [CrossRef]
- Rehman, R.U.; Di Sansebastiano, G.P. Plant Rab GTPases in membrane trafficking and signaling. Plant Signaling: Understanding the Molecular Crosstalk; Hakeem, K., Rehman, R., Tahir, I., Eds.; Springer: New Delhi, India, 2014; pp. 51–73. [Google Scholar] [CrossRef]
- Vukašinović, N.; Žárský, V. Tethering complexes in the Arabidopsis endomembrane system. Front. Cell Dev. Biol. 2016, 4, 46. [Google Scholar] [CrossRef]
- Vernoud, V.; Horton, A.C.; Yang, Z.; Nielsen, E. Analysis of the Small GTPase Gene Superfamily of Arabidopsis1. Plant Physiol. 2003, 131, 1191–1208. [Google Scholar] [CrossRef]
- Pan, X.; Eathiraj, S.; Munson, M.; Lambright, D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 2006, 442, 303–306. [Google Scholar] [CrossRef] [PubMed]
- Aligianis, I.A.; Johnson, C.A.; Gissen, P.; Chen, D.; Hampshire, D.; Hoffmann, K.; Maina, E.N.; Morgan, N.V.; Tee, L.; Morton, J.; et al. Mutations of the catalytic subunit of RAB3GAP cause Warburg Micro syndrome. Nat. Genet. 2005, 37, 221–224. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.-Y.; Ramachandran, S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genom. 2006, 24, 235–251. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abbal, P.; Tesniere, C. Putative Vitis vinifera Rop- and Rab-GAP-, GEF-, and GDI-interacting proteins uncovered with novel methods for public genomic and EST database analysis. J. Exp. Bot. 2010, 61, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Schiene, K.; Dietz, K.-J. Characterization of a small GTP-binding protein of the rab 5 family in Mesembryanthemum crystallinum with increased level of expression during early salt stress. Plant Mol. Biol. 2000, 42, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Nahm, M.Y.; Kim, S.W.; Yun, D.; Lee, S.Y.; Cho, M.J.; Bahk, J.D. Molecular and Biochemical Analyses of OsRab7, a Rice Rab7 Homolog. Plant Cell Physiol. 2003, 44, 1341–1349. [Google Scholar] [CrossRef] [PubMed]
- Mazel, A.; Leshem, Y.; Tiwari, B.S.; Levine, A. Induction of Salt and Osmotic Stress Tolerance by Overexpression of an Intracellular Vesicle Trafficking Protein AtRab7 (AtRabG3e). Plant Physiol. 2004, 134, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Karim, S.; Zhang, H.; Aronsson, H. Arabidopsis RabF1 (ARA6) Is Involved in Salt Stress and Dark-Induced Senescence (DIS). Int. J. Mol. Sci. 2017, 18, 309. [Google Scholar] [CrossRef]
- San Martín-Davison, A.; Pérez-Díaz, R.; Soto, F.; Madrid-Espinoza, J.; González-Villanueva, E.; Pizarro, L.; Norambuena, L.; Tapia, J.; Tajima, H.; Blumwald, E.; et al. Involvement of SchRabGDI1 from Solanum chilense in endocytic trafficking and tolerance to salt stress. Plant Sci. 2017, 263, 1–11. [Google Scholar] [CrossRef]
- Baral, A.; Irani, N.G.; Fujimoto, M.; Nakano, A.; Mayor, S.; Mathew, M. Salt-Induced Remodeling of Spatially Restricted Clathrin-Independent Endocytic Pathways in Arabidopsis Root. Plant Cell 2015, 27, 1297–1315. [Google Scholar] [CrossRef]
- Heo, J.B.; Rho, H.S.; Kim, S.W.; Hwang, S.M.; Kwon, H.J.; Nahm, M.Y.; Bang, W.Y.; Bahk, J.D. OsGAP1 Functions as a Positive Regulator of OsRab11-mediated TGN to PM or Vacuole Trafficking. Plant Cell Physiol. 2005, 46, 2005–2018. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.S.; Im, C.H.; Kim, D.W.; Bahk, J.D. OsRab11 and OsGAP1 are essential for the vesicle trafficking of the vacuolar H+-ATPase OsVHA-a1 under high salinity conditions. Plant Sci. 2013, 198, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Solankey, S.S.; Singh, R.K.; Baranwal, D.K.; Singh, D.K. Genetic Expression of Tomato for Heat and Drought Stress Tolerance: An Overview. Int. J. Veg. Sci. 2014, 21, 496–515. [Google Scholar] [CrossRef]
- Loukehaich, R.; Wang, T.; Ouyang, B.; Ziaf, K.; Li, H.; Zhang, J.; Lu, Y.; Ye, Z. SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 2012, 63, 5593–5606. [Google Scholar] [CrossRef] [PubMed]
- Loyola, J.; Verdugo, I.; González, E.; Casaretto, J.A.; Ruiz-Lara, S. Plastidic isoprenoid biosynthesis in tomato: Physiological and molecular analysis in genotypes resistant and sensitive to drought stress. Plant Biol. 2012, 14, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Bolger, A.; Scossa, F.; Bolger, M.E.; Lanz, C.; Maumus, F.; Tohge, T.; Quesneville, H.; Alseekh, S.; Sørensen, I.; Lichtenstein, G.; et al. The genome of the stress-tolerant wild tomato species Solanum pennellii. Nat. Genet. 2014, 46, 1034–1038. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Eddy, S.R. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The protein families database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene features visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2016, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.; Binns, D.; Chang, H.-Y.; Fraser, M.; Li, W.; McAnulla, C.; McWilliam, H.; Maslen, J.; Mitchell, A.; Nuka, G.; et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef]
- Yu, C.-S.; Chen, Y.-C.; Lu, C.-H.; Hwang, J.-K. Prediction of protein subcellular localization. Proteins Struct. Funct. Bioinform. 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Blum, T.; Briesemeister, S.; Kohlbacher, O. MultiLoc2: Integrating phylogeny and Gene Ontology terms improves subcellular protein localization prediction. BMC Bioinform. 2009, 10, 274. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef]
- Cheong, W.-H.; Tan, Y.-C.; Yap, S.-J.; Ng, K.-P. ClicO FS: An interactive web-based service of Circos. Bioinformatics 2015, 31, 3685–3687. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Sun, W.; Yuan, Y.; Zhang, N.; Hayward, A.; Liu, Y.; Wang, Y. Phylogenetic analyses provide the first insights into the evolution of OVATE family proteins in land plants. Ann. Bot. 2014, 113, 1219–1233. [Google Scholar] [CrossRef] [PubMed]
- Koenig, D.; Jiménez-Gómez, J.M.; Kimura, S.; Fulop, D.; Chitwood, D.H.; Headland, L.R.; Kumar, R.; Covington, M.F.; Devisetty, U.K.; Tat, A.V.; et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc. Natl. Acad. Sci. USA 2013, 110, E2655–E2662. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Xu, X.; Zhu, H.; Liu, A.; Liu, L.; Li, J.; Hua, X. Comparative Transcriptomic Profiling of a Salt-Tolerant Wild Tomato Species and a Salt-Sensitive Tomato Cultivar. Plant Cell Physiol. 2010, 51, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Mishra, U.; Rai, A.; Kumar, R.; Singh, M.; Pandey, H.P. Gene expression analysis of Solanum lycopersicum and Solanum habrochaites under drought conditions. Genom. Data 2016, 9, 40–41. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pérez-Díaz, R.; Madrid-Espinoza, J.; Salinas-Cornejo, J.; González-Villanueva, E.; Ruiz-Lara, S. Differential Roles for VviGST1, VviGST3, and VviGST4 in Proanthocyanidin and Anthocyanin Transport in Vitis vinífera. Front. Plant Sci. 2016, 7, 1135. [Google Scholar] [CrossRef] [PubMed]
- Yáñez, M.; Cáceres, S.; Orellana, S.; Bastías, A.; Verdugo, I.; Ruiz-Lara, S.; Casaretto, J.A. An abiotic stress-responsive bZIP transcription factor from wild and cultivated tomatoes regulates stress-related genes. Plant Cell Rep. 2009, 28, 1497–1507. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Fukuda, M. TBC proteins: GAPs for mammalian small GTPase Rab? Biosci. Res. 2001, 31, 159–168. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Chen, J.-S.; Hsu, Y.-H.; Tsai, C.-H. A putative Rab-GTPase activation protein from Nicotiana benthamiana is important for Bamboo mosaic virus intercellular movement. Virology 2013, 447, 292–299. [Google Scholar] [CrossRef]
- Gabernet-Castello, C.; O’Reilly, A.J.; Dacks, J.B.; Field, M.C. Evolution of Tre-2/Bub2/Cdc16 (TBC) Rab GTPase-activating proteins. Mol. Biol. Cell 2013, 24, 1574–1583. [Google Scholar] [CrossRef] [PubMed]
- De Grassi, A.; Lanave, C.; Saccone, C. Genome duplication and gene-family evolution: The case of three OXPHOS gene families. Gene 2008, 421, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chu, Z.; Wang, X.; Li, Y.; Yu, H.; Li, J.; Lu, Y.; Li, H.; Ouyang, B. Genomic Organization, Phylogenetic and Expression Analysis of the B-BOX Gene Family in Tomato. Front. Plant Sci. 2016, 7, 473. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Zhao, T.; Xu, X.; Li, J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 2017, 5, 3955. [Google Scholar] [CrossRef]
- Spang, N.; Feldmann, A.; Huesmann, H.; Bekbulat, F.; Schmitt, V.; Hiebel, C.; Koziollek-Drechsler, I.; Clement, A.M.; Moosmann, B.; Jung, J.; et al. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy 2014, 10, 2297–2309. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Yu, J.-Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef]
- Feldmann, A.; Bekbulat, F.; Huesmann, H.; Ulbrich, S.; Tatzelt, J.; Behl, C.; Kern, A. The RAB GTPase RAB18 modulates macroautophagy and proteostasis. Biochem. Biophys. Res. Commun. 2017, 486, 738–743. [Google Scholar] [CrossRef]
- Bar, M.; Leibman, M.; Schuster, S.; Pitzhadza, H.; Avni, A. EHD1 Functions in Endosomal Recycling and Confers Salt Tolerance. PLoS ONE 2013, 8, e54533. [Google Scholar] [CrossRef]
- Yang, X.; Liao, C.; Tang, J.; Bassham, D.C. Overexpression of trans -Golgi network t- SNARE s rescues vacuolar trafficking and TGN morphology defects in a putative tethering factor mutant. Plant J. 2019. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, P.; Zhu, R.; Fu, J.; Su, J.; Zheng, J.; Wang, Z.; Wang, D.; Gong, Q. Autophagy Is Rapidly Induced by Salt Stress and Is Required for Salt Tolerance in Arabidopsis. Front. Plant Sci. 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Roos, J.; Bejai, S.; Oide, S.; Dixelius, C. RabGAP22 Is Required for Defense to the Vascular Pathogen Verticillium longisporum and Contributes to Stomata Immunity. PLoS ONE 2014, 9, e88187. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, J. Using Co-Expression Analysis and Stress-Based Screens to Uncover Arabidopsis Peroxisomal Proteins Involved in Drought Response. PLoS ONE 2015, 10, e0137762. [Google Scholar] [CrossRef] [PubMed]
- Keller, R.; Schneider, D. Homologs of the yeast Tvp38 vesicle-associated protein are conserved in chloroplasts and cyanobacteria. Front. Plant Sci. 2013, 4, 467. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).