Loss of a Fragile Chromosome Region leads to the Screwy Phenotype in Paramecium tetraurelia
Abstract
1. Introduction
2. Materials and Methods
2.1. Paramecium Strains, Cultivation, and Genetic Analysis
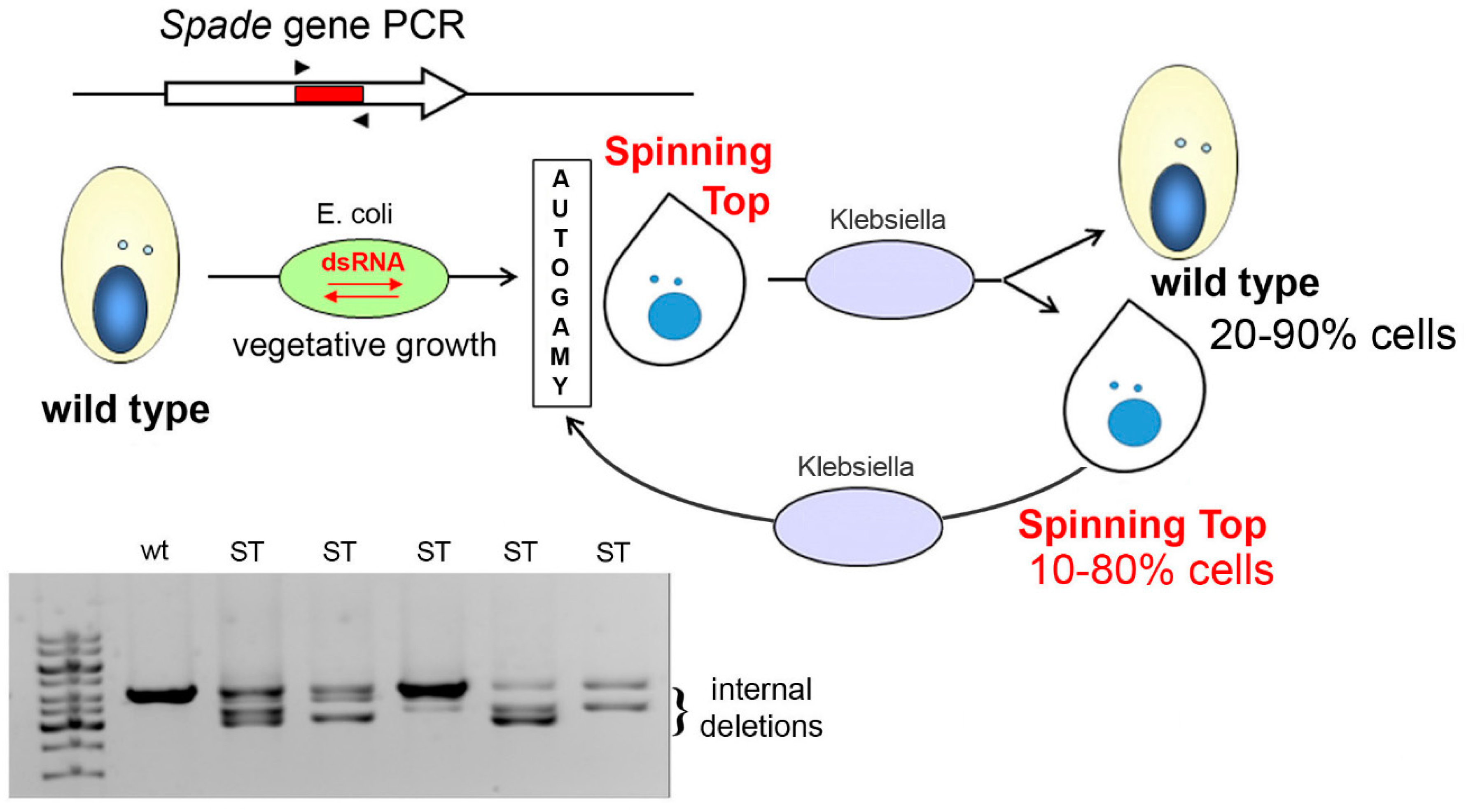
2.2. Spinning Top Mutant Macronuclear Genome Sequencing and Analysis
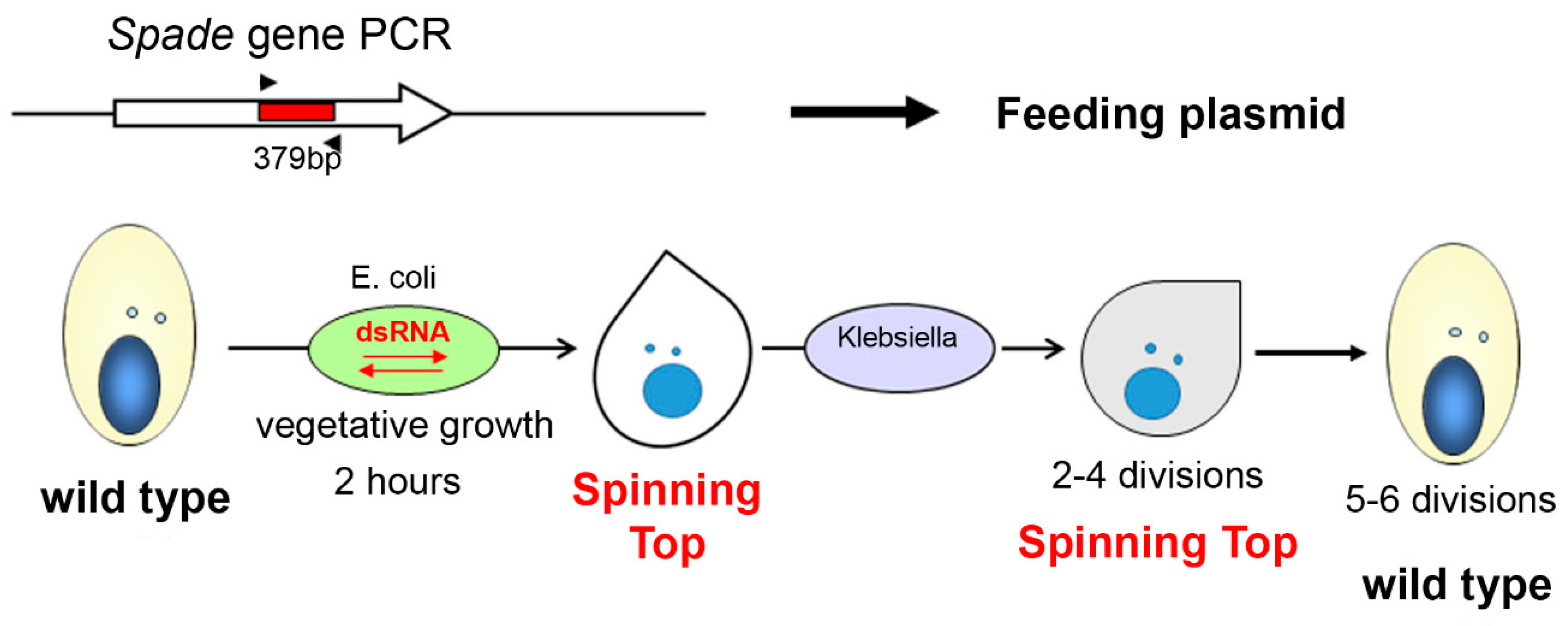
2.3. RNAi-Mediated Gene Silencing by dsRNA Feeding
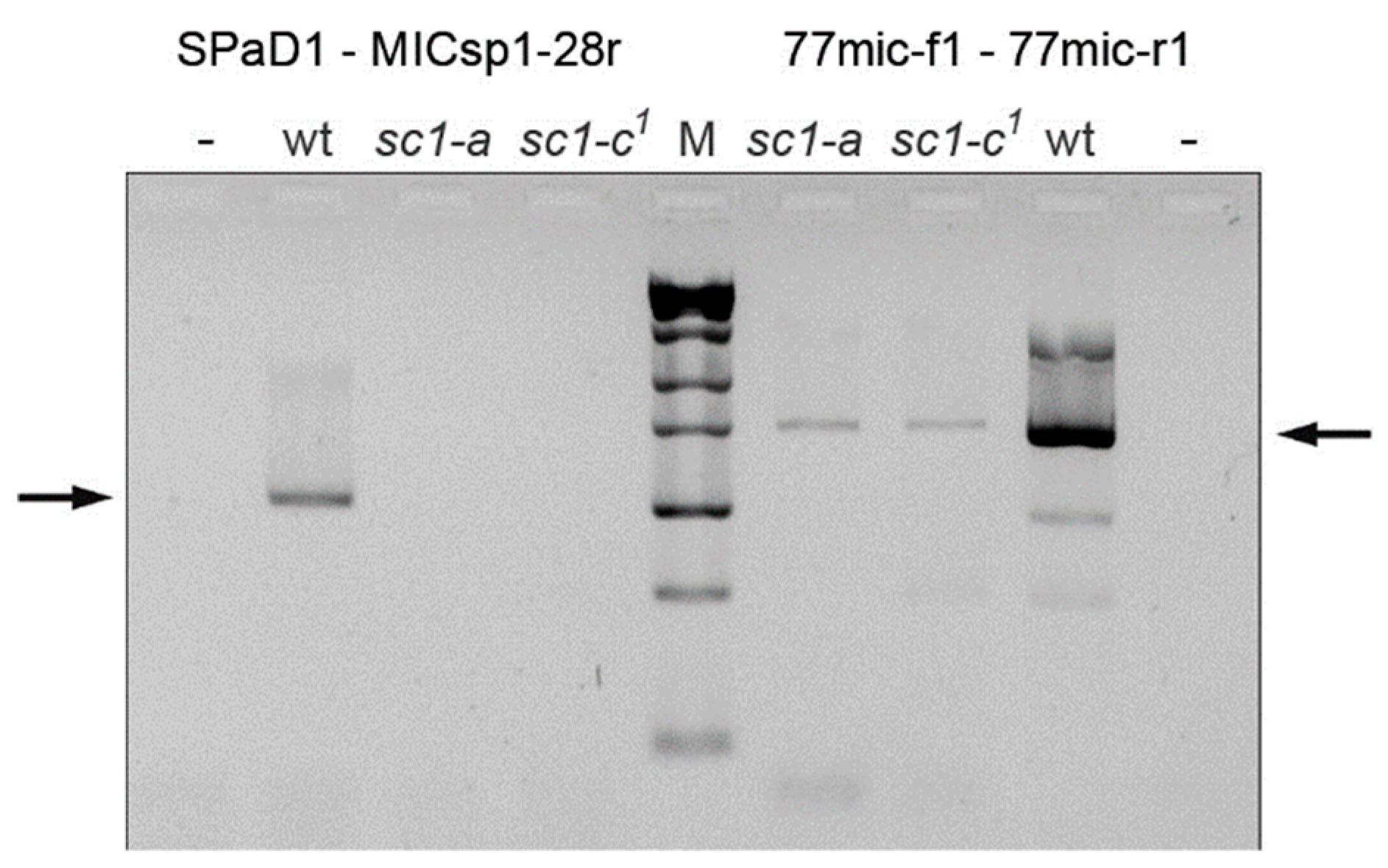
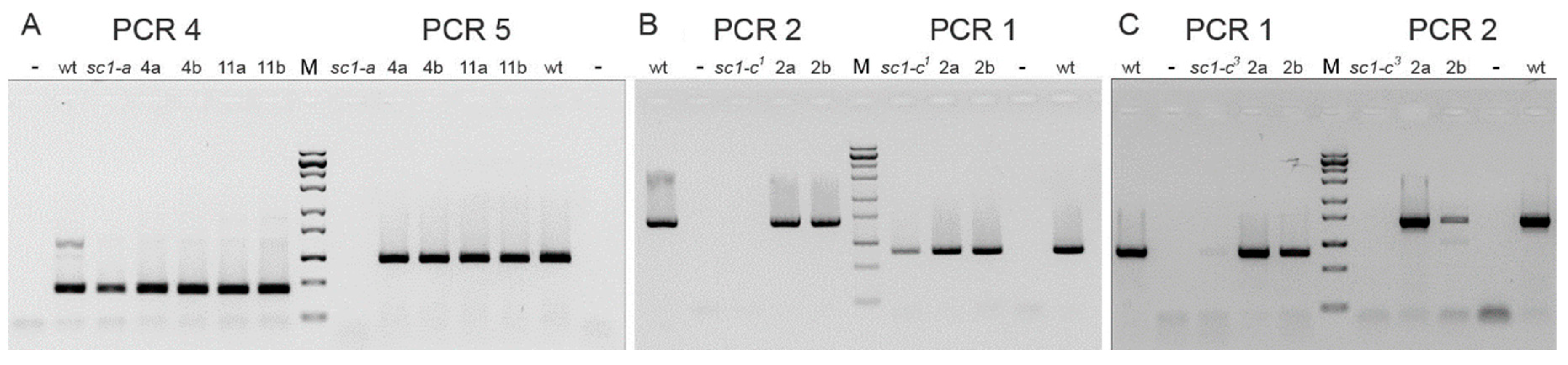
2.4. Localization of the Chromosome Breakage Point in the Mutant Strains
3. Results
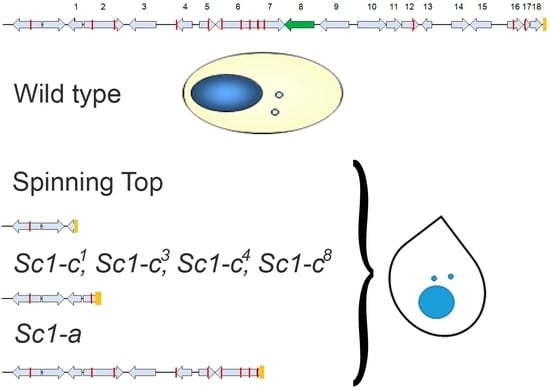
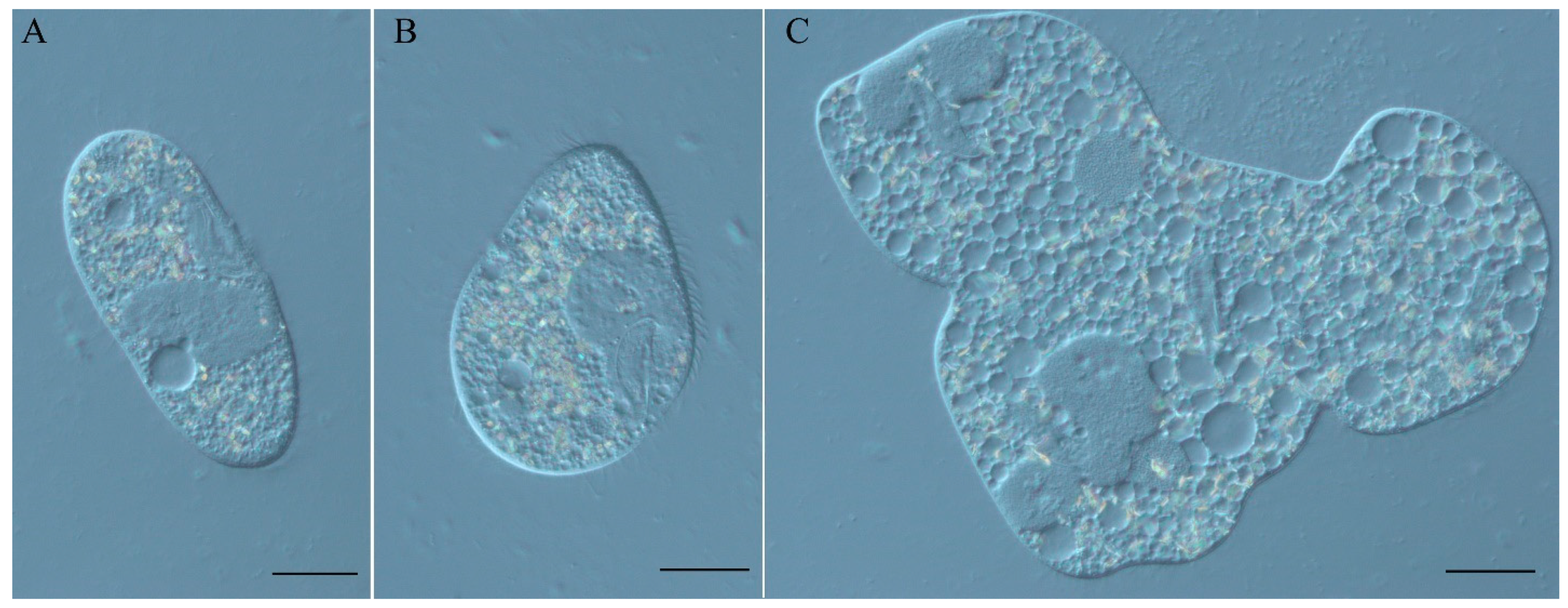
3.1. Characterization of the Spinning Top Mutation
3.2. Spade Gene Identification
3.3. Analysis of Other Screwy Mutants
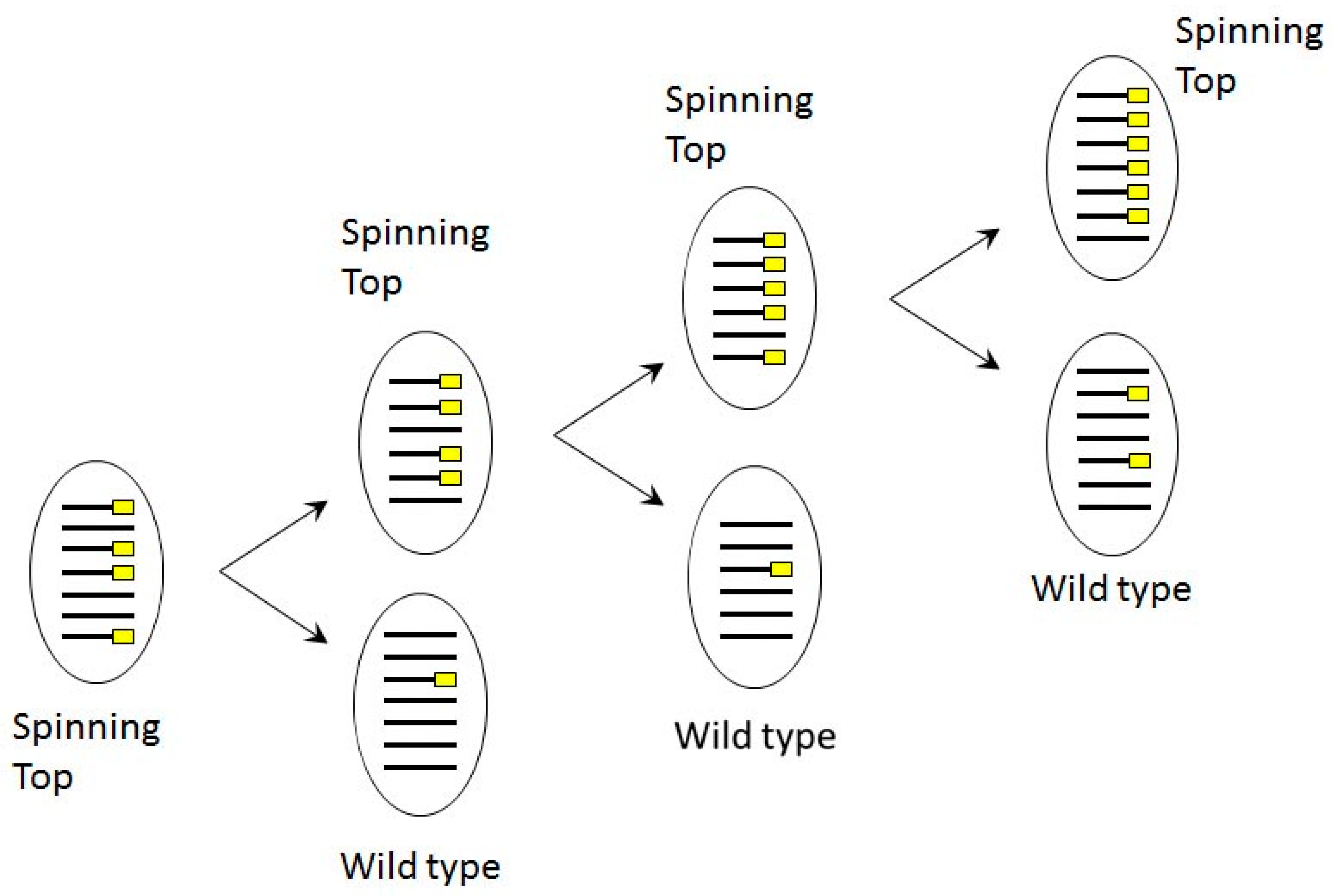
3.4. Induction and Inheritance of MAC Deletions at the sc1 Locus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Preer, J.R. Epigenetic mechanisms affecting macronuclear development in Paramecium and Tetrahymena. J. Eukaryot. Microbiol. 2000, 47, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Bétermier, M.; Duharcourt, S. Programmed rearrangement in ciliates: Paramecium. Microbiol. Spectr. 2014, 2. [Google Scholar] [CrossRef]
- Chalker, D.L.; Meyer, E.; Mochizuki, K. Epigenetics of Ciliates. In Epigenetics, 2nd ed.; Allis, C.D., Caparros, M.L., Jenuwein, T., Reinberg, D., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015; pp. 313–335. [Google Scholar]
- Bhullar, S.; Denby Wilkes, C.; Arnaiz, O.; Nowacki, M.; Sperling, L.; Meyer, E. A mating-type mutagenesis screen identifies a zinc-finger protein required for specific DNA excision events in Paramecium. Nucleic Acids Res. 2018, 46, 9550–9562. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, T.M. Paramecium aurelia. In Handbook of Genetics; King, R.C., Ed.; Plenum: New York, NY, USA, 1974; Volume 2, pp. 469–594. [Google Scholar]
- Pollack, S. Mutations affecting the trichocysts in Paramecium aurelia. I. Morphology and description of the mutants. J. Protozool. 1974, 21, 352–362. [Google Scholar] [CrossRef] [PubMed]
- Beisson, J.; Rossignol, M. The first case of linkage in Paramecium aurelia. Genet. Res. 1969, 13, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Whittle, J.R.; Chen, L. Cortical morphogenesis in Paramecium aurelia: Mutants affecting cell shape. Genet. Res. 1972, 19, 271–279. [Google Scholar] [CrossRef] [PubMed][Green Version]
- ParameciumDB. Available online: https://paramecium.i2bc.paris-saclay.fr (accessed on 04 July 2019).
- Beisson, J.; Bétermier, M.; Bre, M.H.; Cohen, J.; Duharcourt, S.; Duret, L.; Kung, C.; Malinsky, S.; Meyer, E.; Preer, J.R.; et al. Maintaining clonal Paramecium tetraurelia cell lines of controlled age through daily reisolation. Cold Spring Harb. Protoc. 2010, pdb–prot5361. [Google Scholar] [CrossRef]
- Galvani, A.; Sperling, L. RNA interference by feeding in Paramecium. Trends Genet. 2002, 18, 11–12. [Google Scholar] [CrossRef]
- Beisson, J.; Bétermier, M.; Bre, M.H.; Cohen, J.; Duharcourt, S.; Duret, L.; Kung, C.; Malinsky, S.; Meyer, E.; Preer, J.R.; et al. Silencing specific Paramecium tetraurelia genes by feeding double-stranded RNA. Cold Spring Harb. Protoc. 2010, pdb–prot5363. [Google Scholar] [CrossRef]
- Marker, S.; Carradec, Q.; Tanty, V.; Arnaiz, O.; Meyer, E. A forward genetic screen reveals essential and non-essential RNAi factors in Paramecium tetraurelia. Nucleic Acids Res. 2014, 42, 7268–7280. [Google Scholar] [CrossRef]
- Arnaiz, O.; Van Dijk, E.; Bétermier, M.; Lhuillier-Akakpo, M.; de Vanssay, A.; Duharcourt, S.; Sallet, E.; Gouzy, J.; Sperling, L. Improved methods and resources for paramecium genomics: Transcription units, gene annotation and gene expression. BMC Genom. 2017, 18, 483. [Google Scholar] [CrossRef]
- Creutz, C.E.; Tomsig, J.L.; Snyder, S.L.; Gautier, M.C.; Skouri, F.; Beisson, J.; Cohen, J. The copines, a novel class of C2 domain-containing, calcium-dependent, phospholipid-binding proteins conserved from Paramecium to humans. J. Biol. Chem. 1998, 273, 1393–1402. [Google Scholar] [CrossRef]
- Moniz, L.; Dutt, P.; Haider, N.; Stambolic, V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011, 6, 18. [Google Scholar] [CrossRef]
- Aury, J.M.; Jaillon, O.; Duret, L.; Noel, B.; Jubin, C.; Porcel, B.M.; Ségurens, B.; Daubin, V.; Anthouard, V.; Aiach, N.; et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006, 9, 171–178. [Google Scholar] [CrossRef]
- McGrath, C.; Gout, J.F.; Johri, P.; Doak, T.G.; Lynch, M. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 2014, 24, 1665–1675. [Google Scholar] [CrossRef]
- Garnier, O.; Serrano, V.; Duharcourt, S.; Meyer, E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol. Cell Biol. 2004, 24, 7370–7379. [Google Scholar] [CrossRef]
- Harbury, P.B.; Zhang, T.; Kim, P.S.; Alber, T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science 1993, 262, 1401–1407. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate filaments: Structure and assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef]
- Rose, A.; Schraegle, S.J.; Stahlberg, E.A.; Meier, I. Coiled-coil protein composition of 22 proteomes–differences and common themes in subcellular infrastructure and traffic control. BMC Evol. Biol. 2005, 5, 66. [Google Scholar] [CrossRef]
- Jewett, T.J.; Sibley, L.D. Aldolase forms a bridge between cell surface adhesins and the actin cytoskeleton in apicomplexan parasites. Mol. Cell 2003, 11, 885–894. [Google Scholar] [CrossRef]
- Huberts, D.H.; van der Klei, I.J. Moonlighting proteins: An intriguing mode of multitasking. Biochim. Biophys. Acta 2010, 1803, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Nowak, J.K.; Gromadka, R.; Juszczuk, M.; Jerka-Dziadosz, M.; Maliszewska, K.; Mucchielli, M.H.; Gout, J.F.; Arnaiz, O.; Agier, N.; Tang, T.; et al. Functional study of genes essential for autogamy and nuclear reorganization in Paramecium. Eukaryot. Cell 2011, 10, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Sonneborn, T.M. Gene action in development. Proc. R. Soc. Lond. B 1970, 176, 347–366. [Google Scholar] [CrossRef] [PubMed]
- Beisson, J. Preformed cell structure and cell heredity. Prion 2008, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Duharcourt, S.; Lepère, G.; Meyer, E. Developmental genome rearrangements in ciliates: A natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009, 25, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Rudman, B.; Preer, L.B.; Polisky, B.; Preer, J.R. Mutants affecting processing of DNA in macronuclear development in Paramecium. Genetics 1991, 129, 47–56. Available online: https://www.ncbi.nlm.nih.gov/pubmed/1936964 (accessed on 04 July 2019). [PubMed]
- Amar, L.; Dubrana, K. Epigenetic control of chromosome breakage at the 5’ end of Paramecium tetraurelia gene A. Eukaryot. Cell 2004, 3, 1136–1146. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.E.; Goldstein, E.S.; Kilpatrick, S.T. Lewin’s GENES XII, 12th ed.; Jones & Bartlett Learning: Burlington, MA, USA, 2017; pp. 1–838. [Google Scholar]
- Durkin, S.G.; Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007, 41, 169–192. [Google Scholar] [CrossRef]
- Zlotorynski, E.; Rahat, A.; Skaug, J.; Ben-Porat, N.; Ozeri, E.; Hershberg, R.; Levi, A.; Scherer, S.W.; Margalit, H.; Kerem, B. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003, 23, 7143–7151. [Google Scholar] [CrossRef]
- Guérin, F.; Arnaiz, O.; Boggetto, N.; Denby Wilkes, C.; Meyer, E.; Sperling, L.; Duharcourt, S. Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genom. 2017, 18, 327. [Google Scholar] [CrossRef]


| Mutant Strain Index */Strain of Origin | Original Mutant Name | Mutant Cell Line Index | Genotype | Obtained by | Origin of Mutation | Concomitant Phenotype Features | Independent Mutations in the Multiple-Marker Stocks |
|---|---|---|---|---|---|---|---|
| Mutants studied in this work | |||||||
| scr1-1/51 | sc | d4-136 | sc1-a | D. Cronkite [5] | nitrosoguanidine | not documented | none |
| scr1-1ci big/51 | sc | d4-75 | sc1-c1 | T. Sonneborn [5] | UV light | cigar-shaped functional trichocysts | big |
| 1-3ci/d4-2 | m1 | - | sc1-c3 | J. Beisson & M. Rossignol [7] | nitrosoguanidine | cigar-shaped functional trichocysts; slow growth | none |
| 1-4ci/d4-2 | m2 | - | sc1-c4 | J. Beisson & M. Rossignol [7] | nitrosoguanidine | cigar-shaped functional trichocysts; slow growth | ts401 |
| 1-8ci/d4-2 | 1-8ci | - | sc1-c8 | M. Rossignol | UV light | cigar-shaped functional trichocysts; slow growth | none |
| Spinning Top/51 | - | - | Spinning Top | S. Bhullar [4] | spontaneous mutation | not documented | none |
| Other mutants documented but not available anymore | |||||||
| -/51 | mut65 | d4-85 | sc1-b | C. Kung [8] | nitrosoguanidine | not documented | none |
| -/51 | mut80 | sc1-d | In genetic class [8] | nitrosoguanidine | not documented | none | |
| -/51 | 1961-38-8 | d4-137 | sc1-c2 | R. Kimball [5] | X rays | cigar-shaped functional trichocysts; cells large, spin less fast | none |
| -/51 | 1961-45-601 | d4-137 | sc1-c5 | R. Kimball [5] | X rays | cigar-shaped functional trichocysts | none |
| -/51 | - | d4-99 | sc1-c6 | D. Cronkite [5] | nitrosoguanidine | cigar-shaped functional trichocysts | none |
| -/51 | - | d4-186 | sc1-c7 | S. Pollack [5] | X rays | cigar-shaped functional trichocysts | none |
| -/51 | mut64 | - | sc64 ** | A. Whittle [8] | nitrosoguanidine | normal trichocysts | none |
| -/51 | mut66 | - | sc66 ** | A. Whittle [8] | nitrosoguanidine | normal trichocysts; rotating clockwise when swimming | none |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nekrasova, I.; Nikitashina, V.; Bhullar, S.; Arnaiz, O.; Singh, D.P.; Meyer, E.; Potekhin, A. Loss of a Fragile Chromosome Region leads to the Screwy Phenotype in Paramecium tetraurelia. Genes 2019, 10, 513. https://doi.org/10.3390/genes10070513
Nekrasova I, Nikitashina V, Bhullar S, Arnaiz O, Singh DP, Meyer E, Potekhin A. Loss of a Fragile Chromosome Region leads to the Screwy Phenotype in Paramecium tetraurelia. Genes. 2019; 10(7):513. https://doi.org/10.3390/genes10070513
Chicago/Turabian StyleNekrasova, Irina, Vera Nikitashina, Simran Bhullar, Olivier Arnaiz, Deepankar P. Singh, Eric Meyer, and Alexey Potekhin. 2019. "Loss of a Fragile Chromosome Region leads to the Screwy Phenotype in Paramecium tetraurelia" Genes 10, no. 7: 513. https://doi.org/10.3390/genes10070513
APA StyleNekrasova, I., Nikitashina, V., Bhullar, S., Arnaiz, O., Singh, D. P., Meyer, E., & Potekhin, A. (2019). Loss of a Fragile Chromosome Region leads to the Screwy Phenotype in Paramecium tetraurelia. Genes, 10(7), 513. https://doi.org/10.3390/genes10070513