Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Strains
2.2. Whole Mount In Situ Hybridization and Immunohistochemistry
2.3. Pharmaceutical Treatments and mRNA Overexpression
2.4. Histological Analysis and TUNEL Assay
2.5. BrdU Incorporation Assay
2.6. Cell Culture
2.7. RNA Interference and Immunofluorescence
2.8. Knockdown of ANKRD45 with the CRISPR/Cas9 System
2.9. Time-Lapse Assay
2.10. CCK8 Assay
2.11. Cell Cycle and Annexin V Apoptosis Assay
3. Results
3.1. Expression of Ankrd45 Is Enriched in Ciliated Organs
3.2. Ankrd45 Is Dispensable for Zebrafish Development
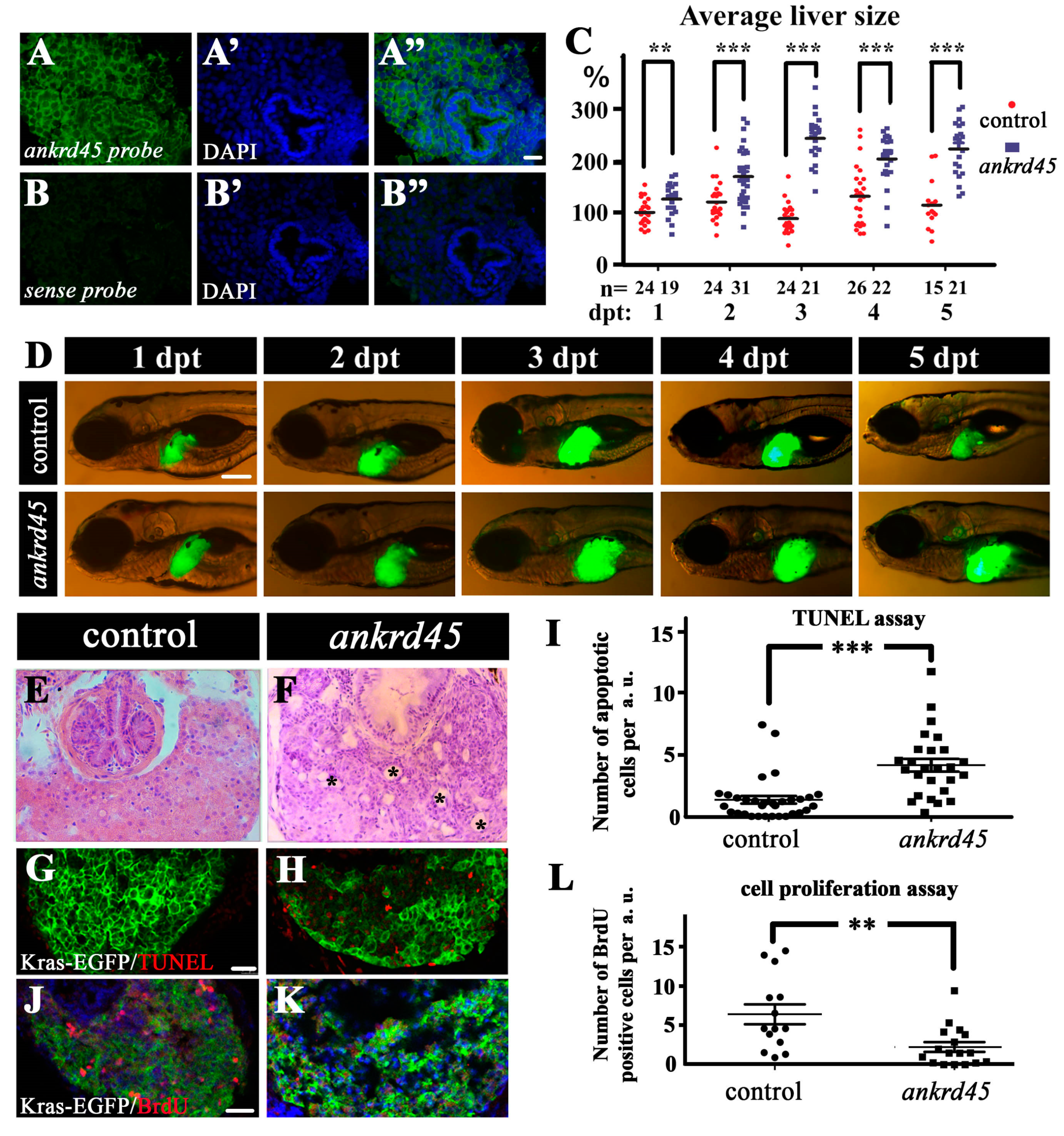
3.3. Liver Specific Expression of KrasG12V Induced Larger Livers in Ankrd45 Mutants
3.4. Enlarged Cystic Liver Is Due to Cell Degeneration in KrasG12V-Induced Ankrd45 Mutant
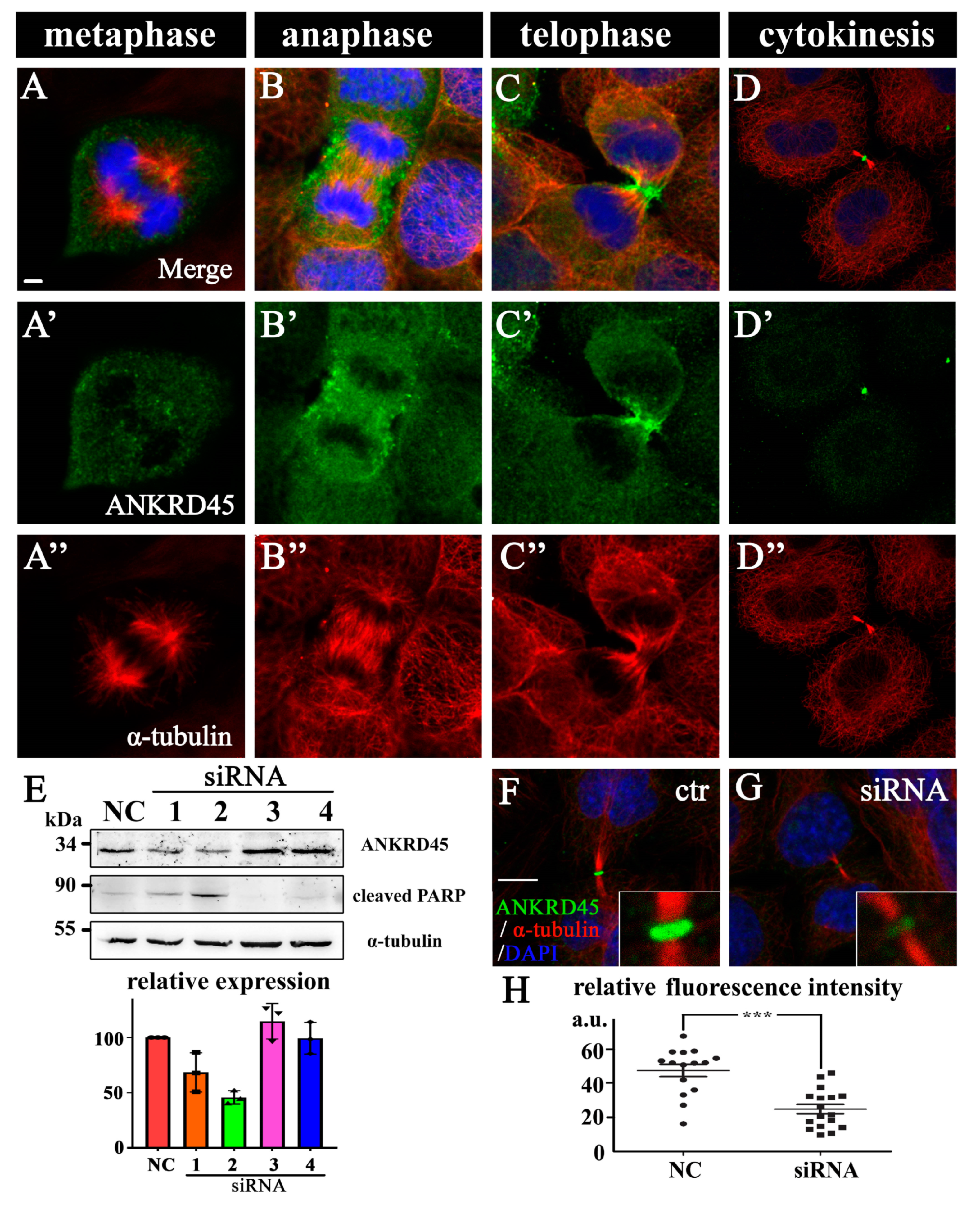
3.5. Dynamic Subcellular Localization of ANKRD45 During Mitosis
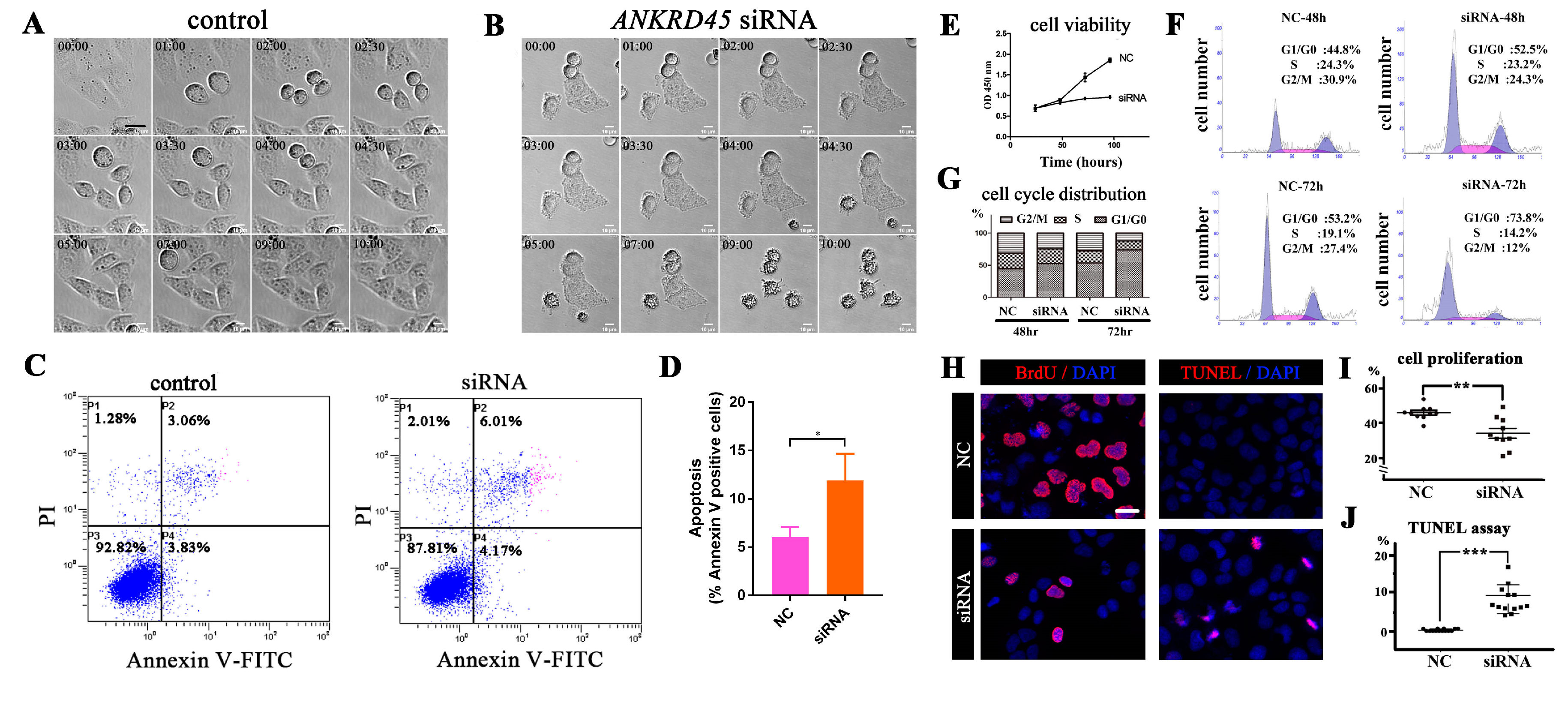
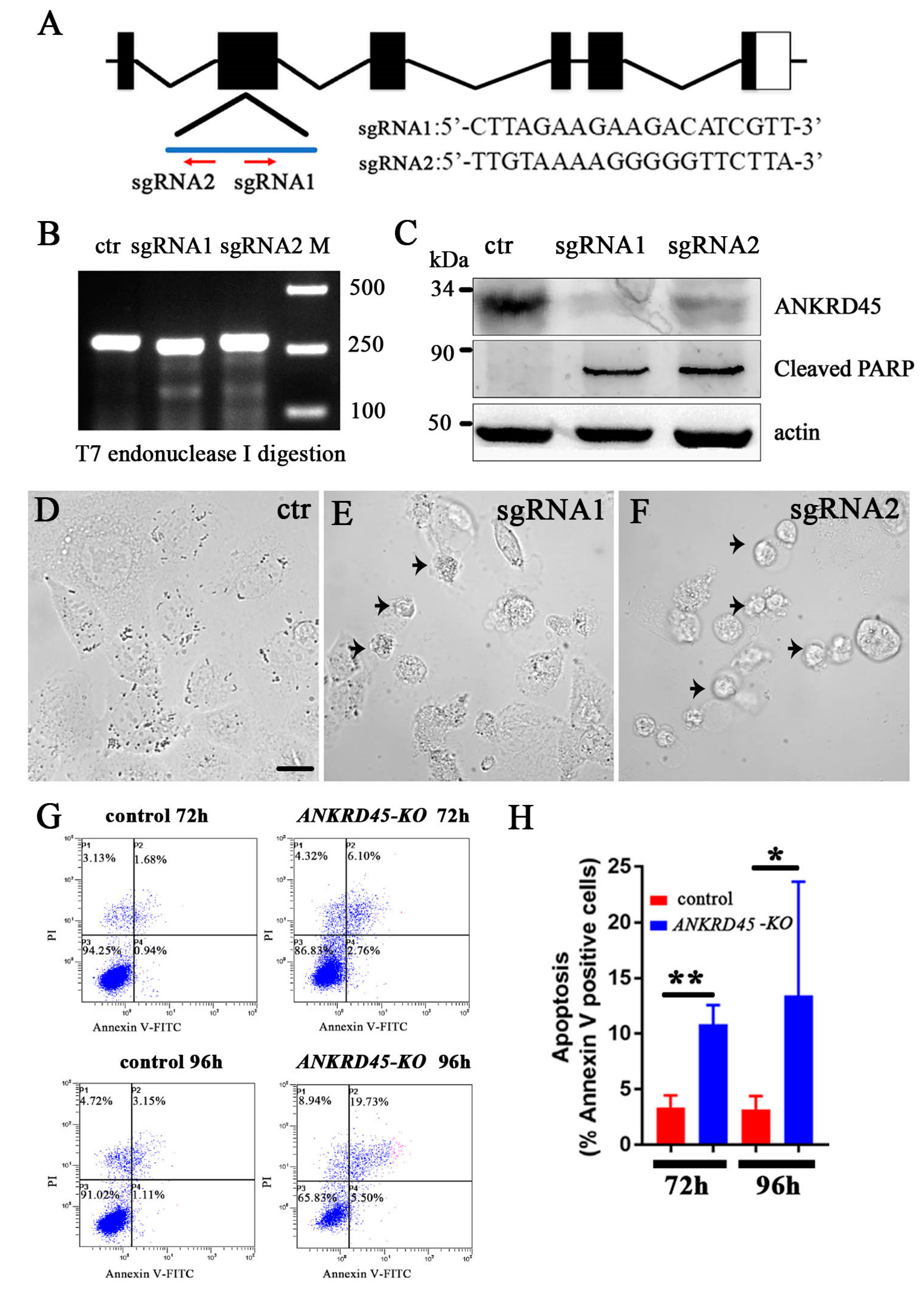
3.6. Loss of ANKRD45 Results in Cell Proliferation Defects in Cultured Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barrick, D.; Ferreiro, D.U.; Komives, E.A. Folding landscapes of ankyrin repeat proteins: Experiments meet theory. Curr. Opin. Struct. Biol. 2008, 18, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Mosavi, L.K.; Cammett, T.J.; Desrosiers, D.C.; Peng, Z.Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. Publ. Protein Soc. 2004, 13, 1435–1448. [Google Scholar] [CrossRef] [PubMed]
- Breeden, L.; Nasmyth, K. Similarity between cell-cycle genes of budding yeast and fission yeast and the notch gene of drosophila. Nature 1987, 329, 651–654. [Google Scholar] [CrossRef] [PubMed]
- Lux, S.E.; John, K.M.; Bennett, V. Analysis of cdna for human erythrocyte ankyrin indicates a repeated structure with homology to tissue-differentiation and cell-cycle control proteins. Nature 1990, 344, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kojic, S.; Radojkovic, D.; Faulkner, G. Muscle ankyrin repeat proteins: Their role in striated muscle function in health and disease. Crit. Rev. Clin. Lab. Sci. 2011, 48, 269–294. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.A.; Tong, L.; Lee, J.O.; Jeffrey, P.D.; Pavletich, N.P. Structural basis for inhibition of the cyclin-dependent kinase cdk6 by the tumour suppressor p16ink4a. Nature 1998, 395, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Ortega, S.; Malumbres, M.; Barbacid, M. Cyclin d-dependent kinases, ink4 inhibitors and cancer. Biochim. Biophys. Acta 2002, 1602, 73–87. [Google Scholar] [CrossRef]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cecillion, M.; Marechal, E.; et al. Notch3 mutations in cadasil, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar] [CrossRef]
- Braune, E.B.; Lendahl, U. Notch—A goldilocks signaling pathway in disease and cancer therapy. Discov. Med. 2016, 21, 189–196. [Google Scholar]
- Baker, R.G.; Hayden, M.S.; Ghosh, S. NF-ĸB, inflammation, and metabolic disease. Cell Metab. 2011, 13, 11–22. [Google Scholar] [CrossRef]
- Kondylis, V.; Kumari, S.; Vlantis, K.; Pasparakis, M. The interplay of IKK, NF-ĸB and RIPK1 signaling in the regulation of cell death, tissue homeostasis and inflammation. Immunol. Rev. 2017, 277, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Di Santi, A.; Cernera, G.; Ombra, M.N.; Castoria, G.; Migliaccio, A. Nuclear receptor-induced transcription is driven by spatially and timely restricted waves of ROS. The role of Akt, IKKα, and DNA damage repair enzymes. Nucleus 2014, 5, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Perillo, B.; Di Santi, A.; Cernera, G.; Ombra, M.N.; Castoria, G.; Migliaccio, A. Phosphorylation of H3 serine 10 by IKKα governs cyclical production of ROS in estrogen-induced transcription and ensures DNA wholeness. Cell Death Differ. 2014, 21, 1503. [Google Scholar] [CrossRef] [PubMed]
- Otto, E.A.; Schermer, B.; Obara, T.; O’Toole, J.F.; Hiller, K.S.; Mueller, A.M.; Ruf, R.G.; Hoefele, J.; Beekmann, F.; Landau, D.; et al. Mutations in invs encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 2003, 34, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Singla, V.; Reiter, J.F. The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science 2006, 313, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Reiter, J.F.; Leroux, M.R. Genes and molecular pathways underpinning ciliopathies. Nat. Rev. Mol. Cell Biol. 2017, 18, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhang, X.; Jia, S.; Yelick, P.C.; Zhao, C. Zebrafish as a model for human ciliopathies. J. Genet. Genom. = Yi Chuan Xue Bao 2016, 43, 107–120. [Google Scholar] [CrossRef]
- Zhang, X.; Jia, S.; Chen, Z.; Chong, Y.L.; Xie, H.; Feng, D.; Wu, X.; Song, D.Z.; Roy, S.; Zhao, C. Cilia-driven cerebrospinal fluid flow directs expression of urotensin neuropeptides to straighten the vertebrate body axis. Nat. Genet. 2018, 50, 1666–1673. [Google Scholar] [CrossRef]
- Li, S.; Balmain, A.; Counter, C.M. A model for ras mutation patterns in cancers: Finding the sweet spot. Nat. Rev. Cancer 2018, 18, 767–777. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. Ras oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef]
- Chew, T.W.; Liu, X.J.; Liu, L.; Spitsbergen, J.M.; Gong, Z.; Low, B.C. Crosstalk of Ras and Rho: Activation of rhoa abates Kras-induced liver tumorigenesis in transgenic zebrafish models. Oncogene 2014, 33, 2717–2727. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Emelyanov, A.; Koh, C.H.; Spitsbergen, J.M.; Lam, S.H.; Mathavan, S.; Parinov, S.; Gong, Z. A high level of liver-specific expression of oncogenic Kras(V12) drives robust liver tumorigenesis in transgenic zebrafish. Dis. Models Mech. 2011, 4, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Chen, Z.; Yang, K.; Miao, S.; Xu, B.; Kang, Y.; Xie, H.; Zhao, C. The cytoplasmic tail of rhodopsin triggers rapid rod degeneration in kinesin-2 mutants. J. Biol. Chem. 2017, 292, 17375–17386. [Google Scholar] [CrossRef] [PubMed]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Gunther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.L.; et al. Genetic compensation triggered by mutant mrna degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Zhu, P.; Shi, H.; Guo, L.; Zhang, Q.; Chen, Y.; Chen, S.; Zhang, Z.; Peng, J.; Chen, J. PTC-bearing mrna elicits a genetic compensation response via Upf3a and COMPASS components. Nature 2019, 568, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Karreth, F.A.; Tuveson, D.A. Modelling oncogenic ras/raf signalling in the mouse. Curr. Opin. Genet. Dev. 2009, 19, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Pohl, C.; Jentsch, S. Final stages of cytokinesis and midbody ring formation are controlled by bruce. Cell 2008, 132, 832–845. [Google Scholar] [CrossRef]
- Sato, N.; Yonemura, S.; Obinata, T.; Tsukita, S.; Tsukita, S. Radixin, a barbed end-capping actin-modulating protein, is concentrated at the cleavage furrow during cytokinesis. J. Cell Biol. 1991, 113, 321–330. [Google Scholar] [CrossRef]
- Sigg, M.A.; Menchen, T.; Lee, C.; Johnson, J.; Jungnickel, M.K.; Choksi, S.P.; Garcia, G., 3rd; Busengdal, H.; Dougherty, G.W.; Pennekamp, P.; et al. Evolutionary proteomics uncovers ancient associations of cilia with signaling pathways. Dev. Cell 2017, 43, 744–762. [Google Scholar] [CrossRef]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef]
- Pelaseyed, T.; Bretscher, A. Regulation of actin-based apical structures on epithelial cells. J. Cell Sci. 2018, 131, jcs221853. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, Y.; Xie, H.; Zhao, C. Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation. Genes 2019, 10, 462. https://doi.org/10.3390/genes10060462
Kang Y, Xie H, Zhao C. Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation. Genes. 2019; 10(6):462. https://doi.org/10.3390/genes10060462
Chicago/Turabian StyleKang, Yunsi, Haibo Xie, and Chengtian Zhao. 2019. "Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation" Genes 10, no. 6: 462. https://doi.org/10.3390/genes10060462
APA StyleKang, Y., Xie, H., & Zhao, C. (2019). Ankrd45 Is a Novel Ankyrin Repeat Protein Required for Cell Proliferation. Genes, 10(6), 462. https://doi.org/10.3390/genes10060462




