Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification of the FAD and FAB2 Gene Families in Maize
2.2. Sequence Alignment and Phylogenetic Tree Construction
2.3. Characteristics Gene Structure and Synteny Analysis
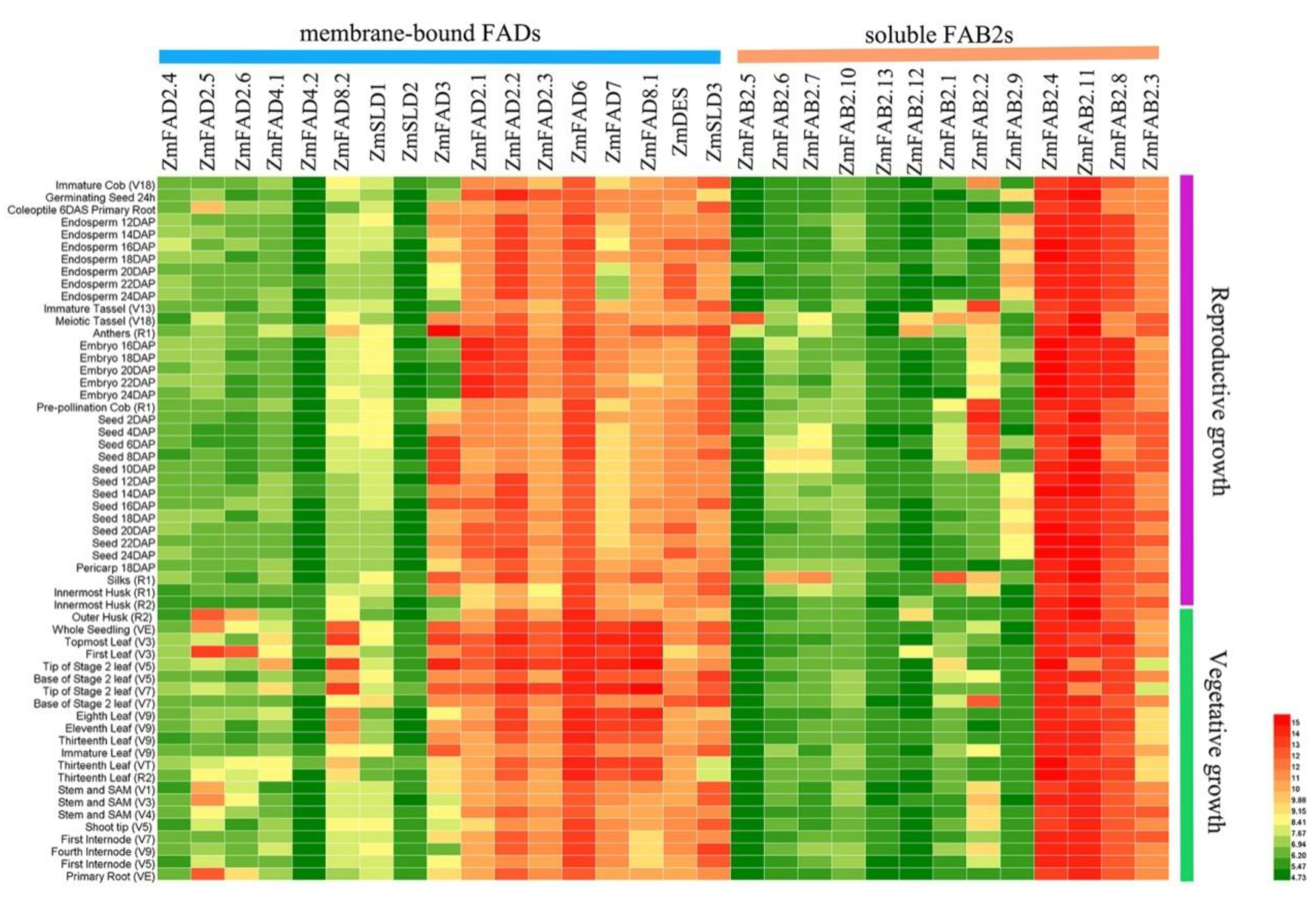
2.4. Expression Profiles of Maize Fatty Acid Desaturase Genes in Diverse Tissues
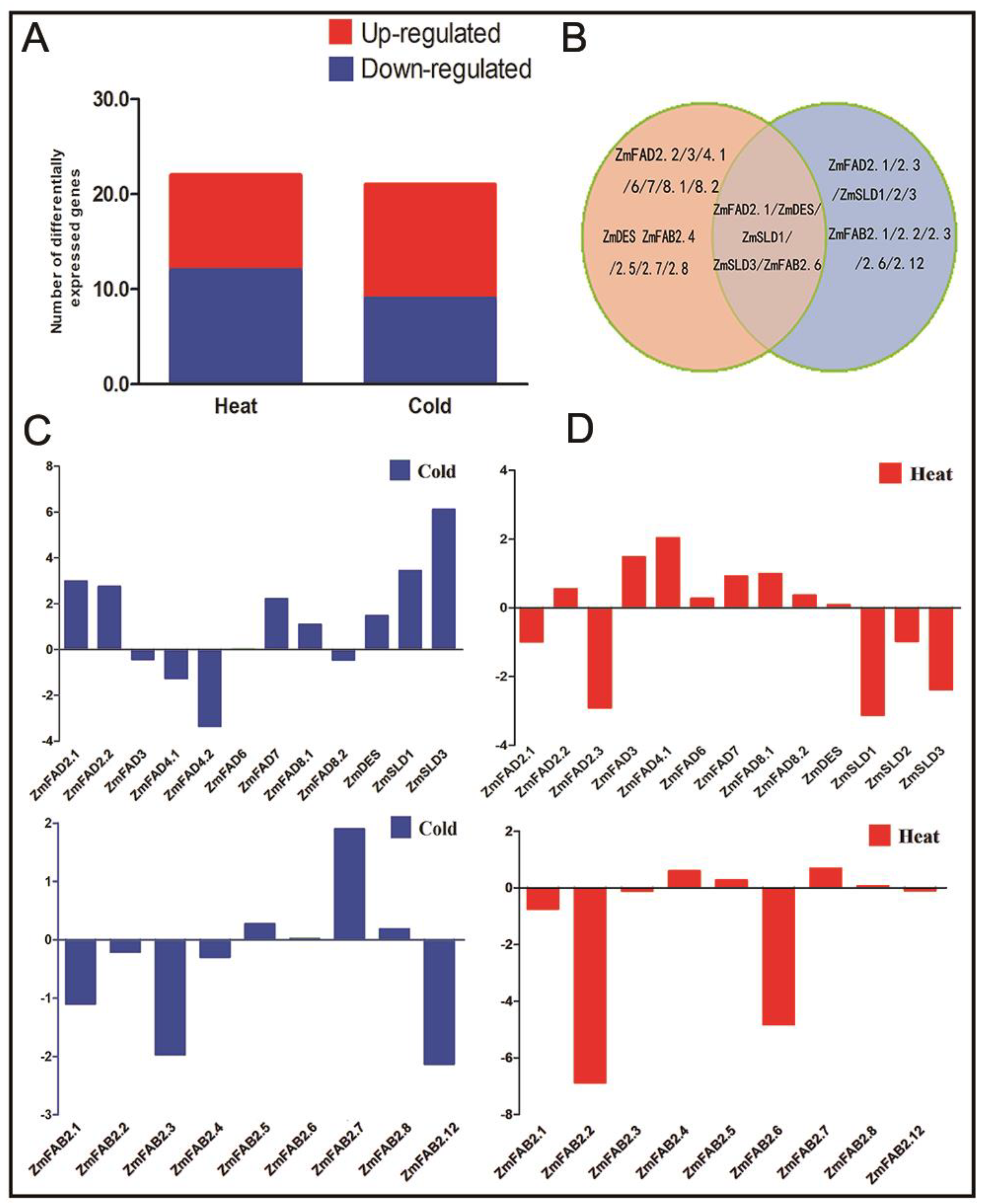
2.5. Expression Profiles of the Maize FAD and FAB2 Genes in Response to Temperature Stresses
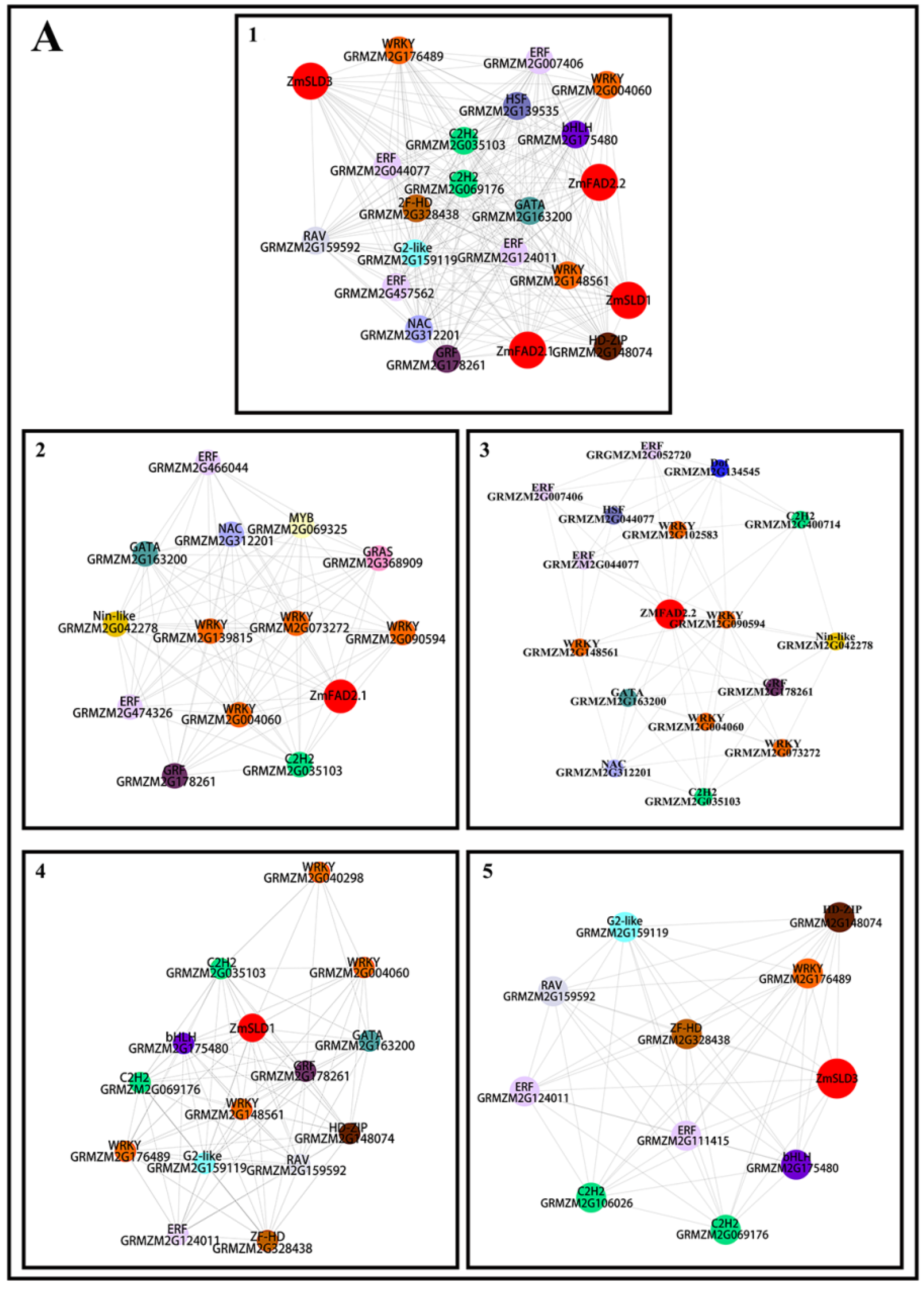
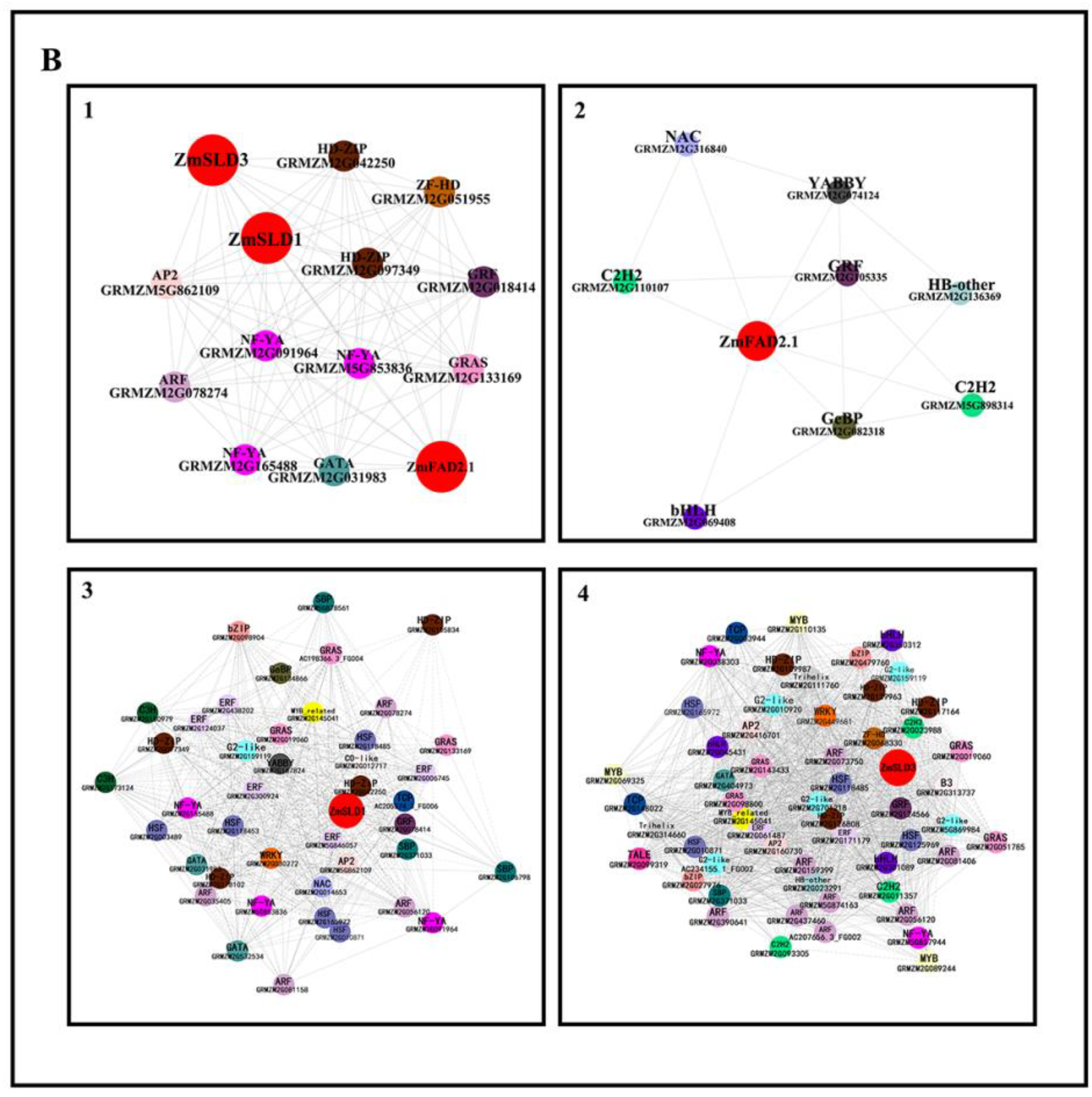
2.6. Co-Expression of FADs and Transcription Factors under Temperature Stresses
2.7. Quantitative Real-Time RT-PCR Analysis
2.8. Statistical Analysis
3. Results
3.1. Characterization of Fatty Acid Desaturase Families in Maize
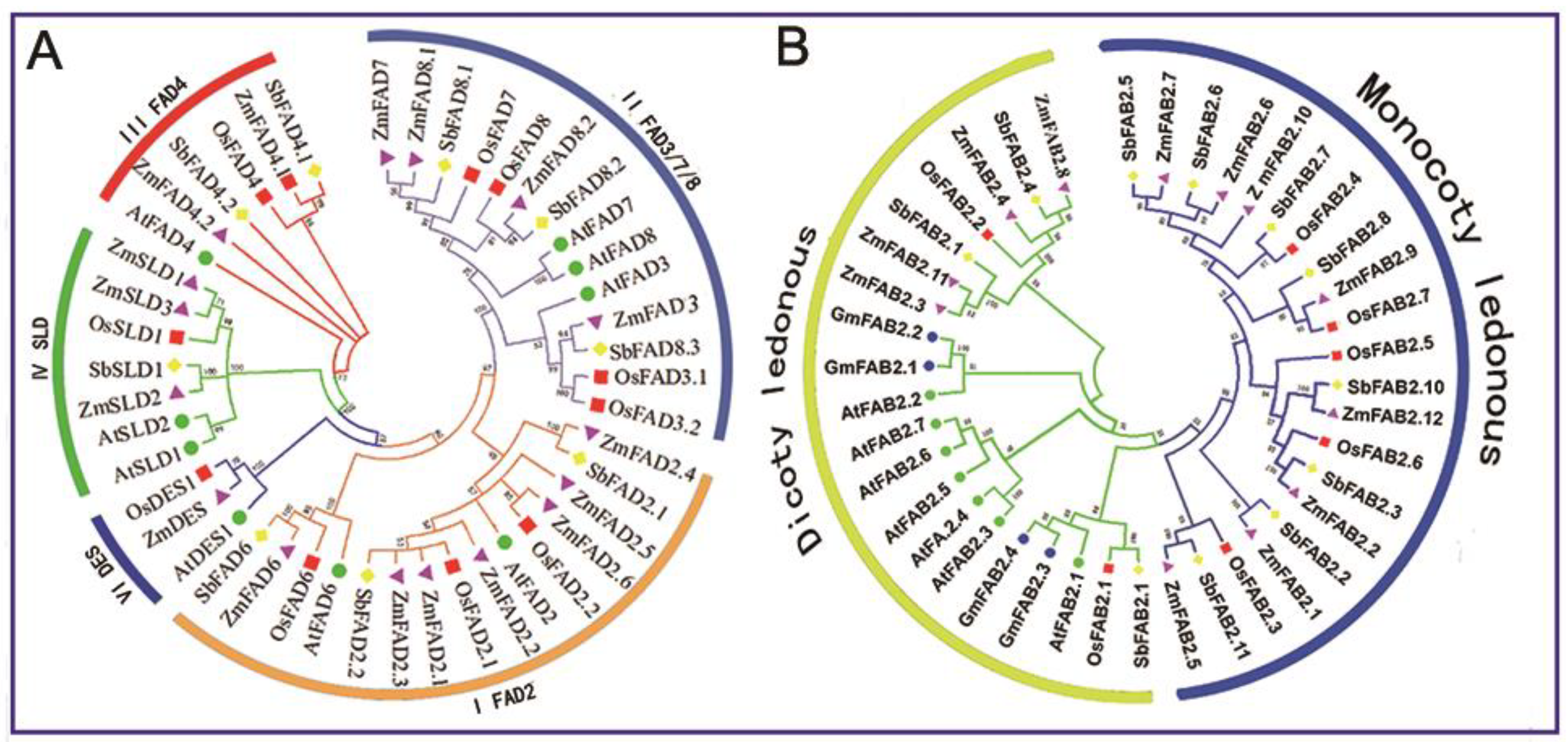
3.2. Phylogenetic Analysis of Maize Fatty Acid Desaturases
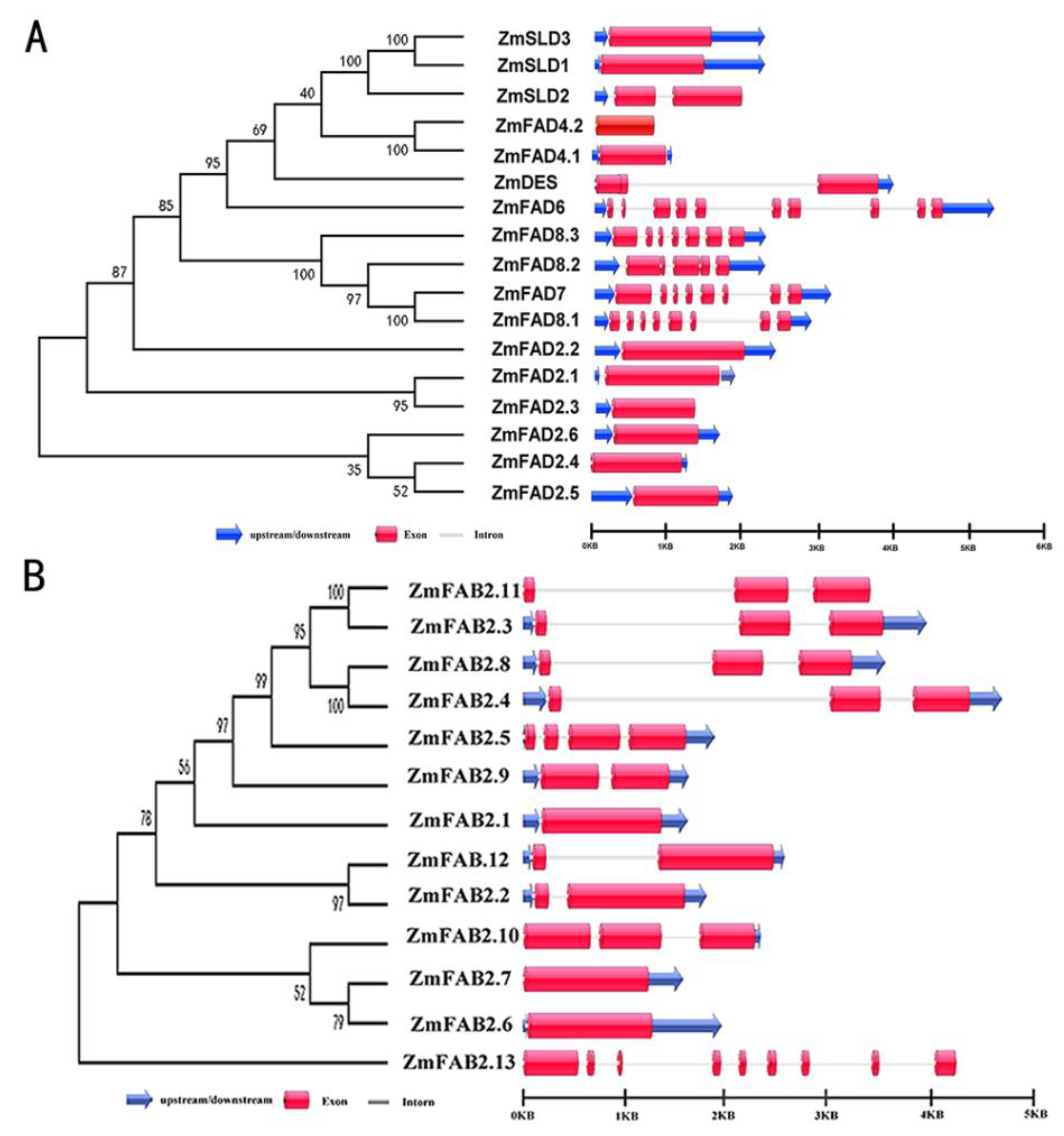
3.3. Exon/Intron Organization of Maize FAD and FAB2 Genes
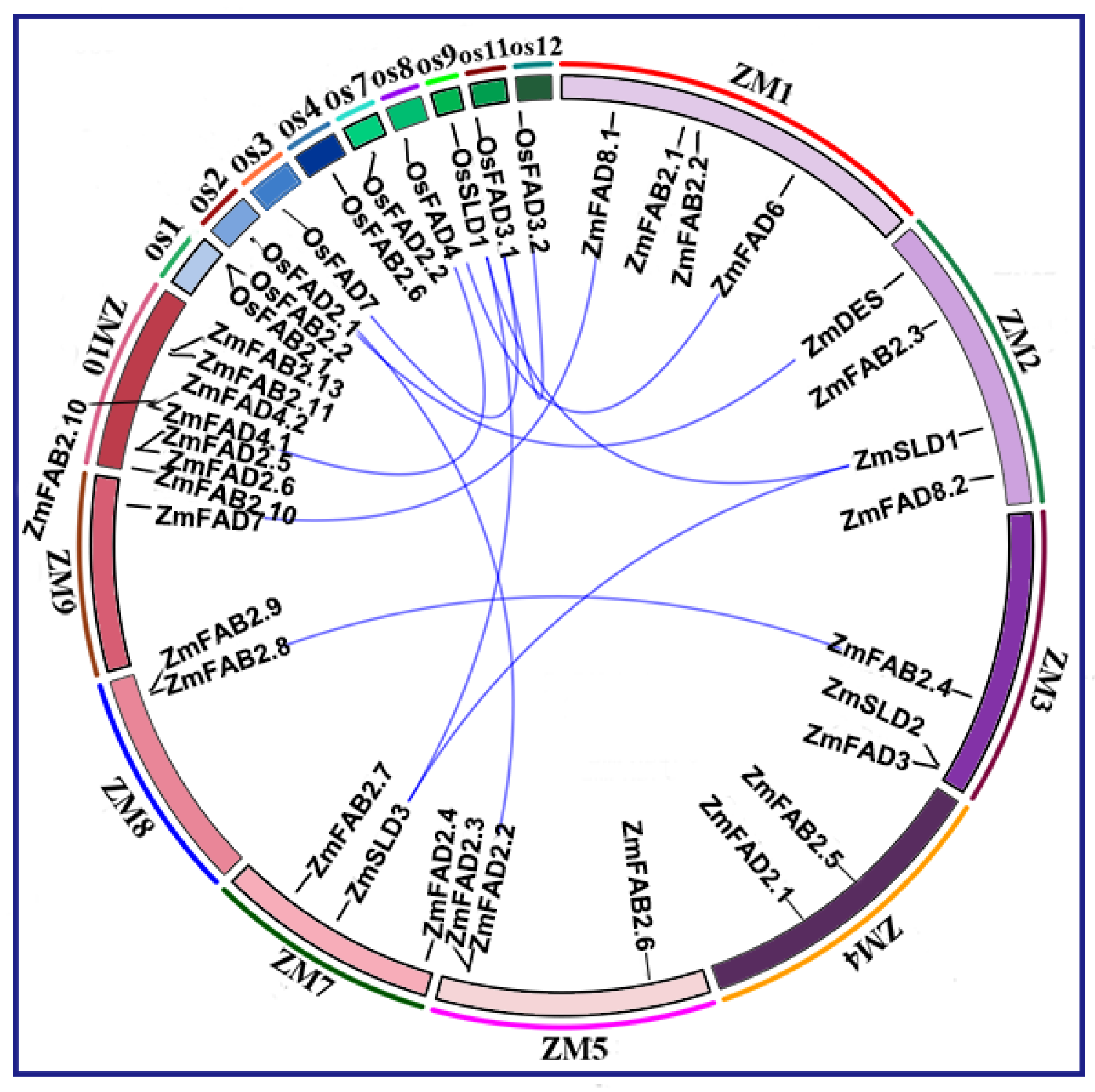
3.4. Syntenic Relations of Fatty Acid Desaturase Genes
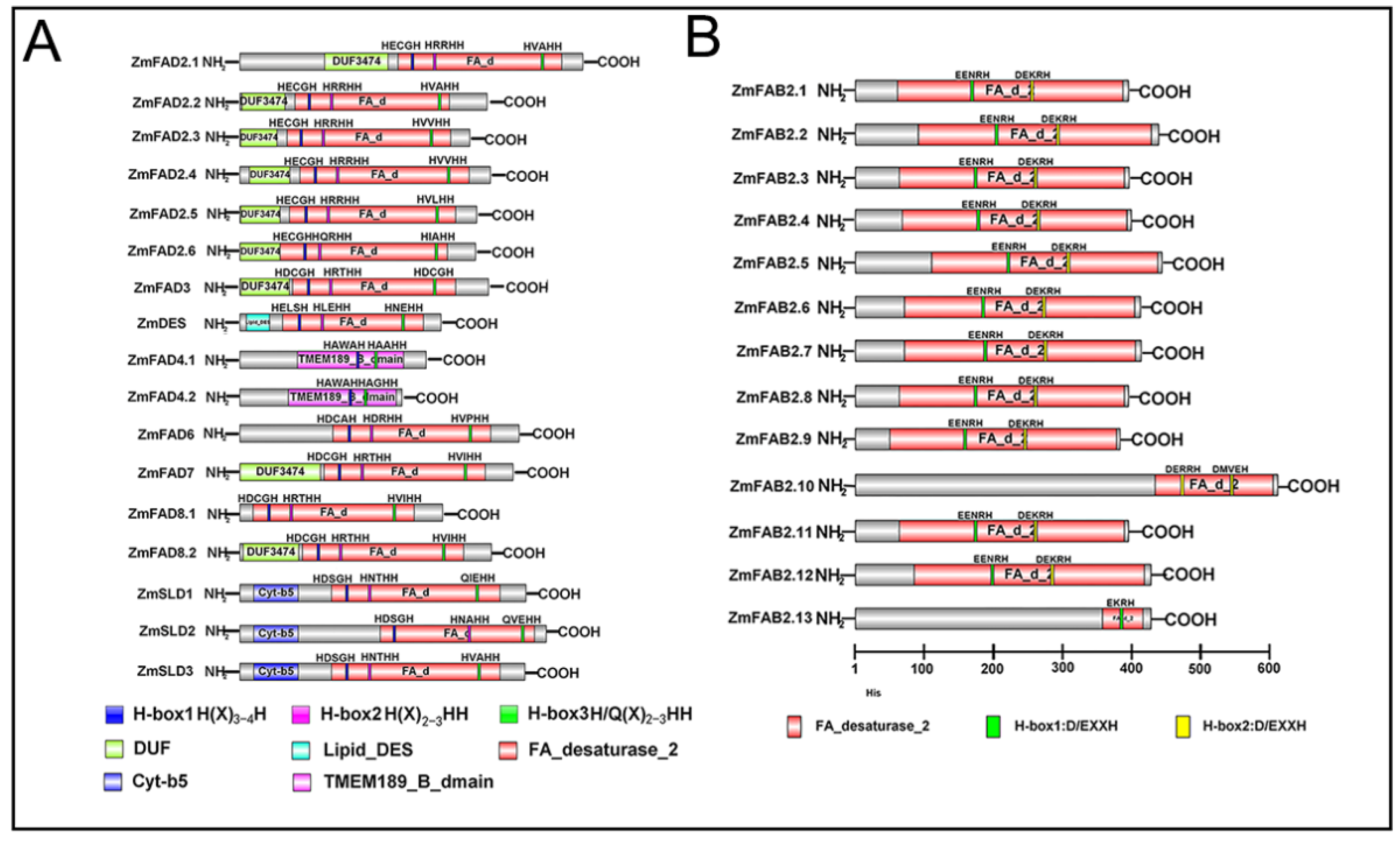
3.5. Conserved Domain Analysis of Maize Fatty Acid Desaturases
3.6. Expression Profiles of Maize Fatty Acid Desaturase Genes in Diverse Tissues
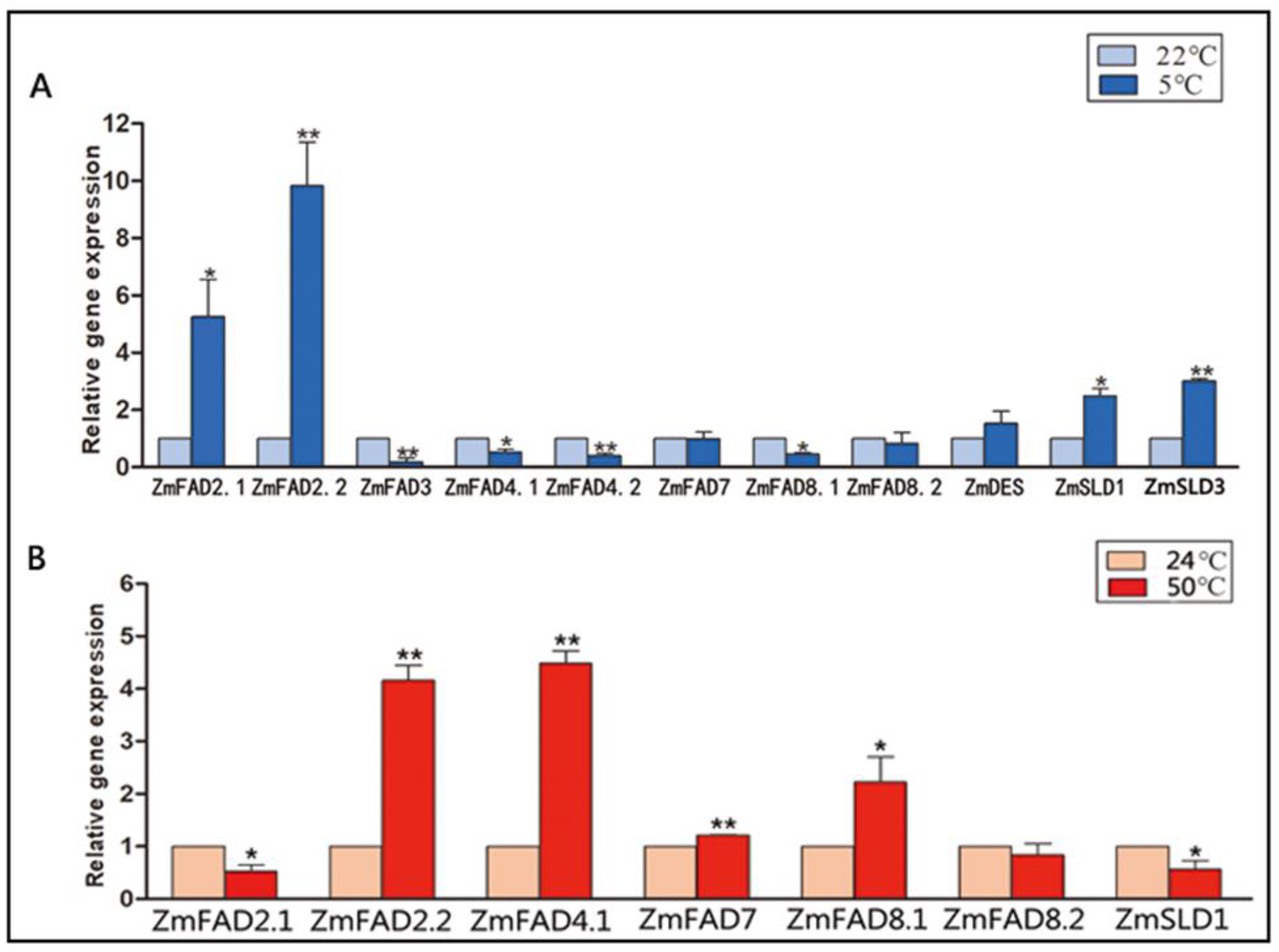
3.7. Expression Profiles of the Maize Fatty Acid Desaturase Genes in Response to Temperature Stresses
3.8. Co-Expression Analysis of Transcription Factors and ZmFADs in Maize
3.9. The qRT-PCR Analysis of the Expression of ZmFAD and ZmTF Genes under Cold and Heat Stresses
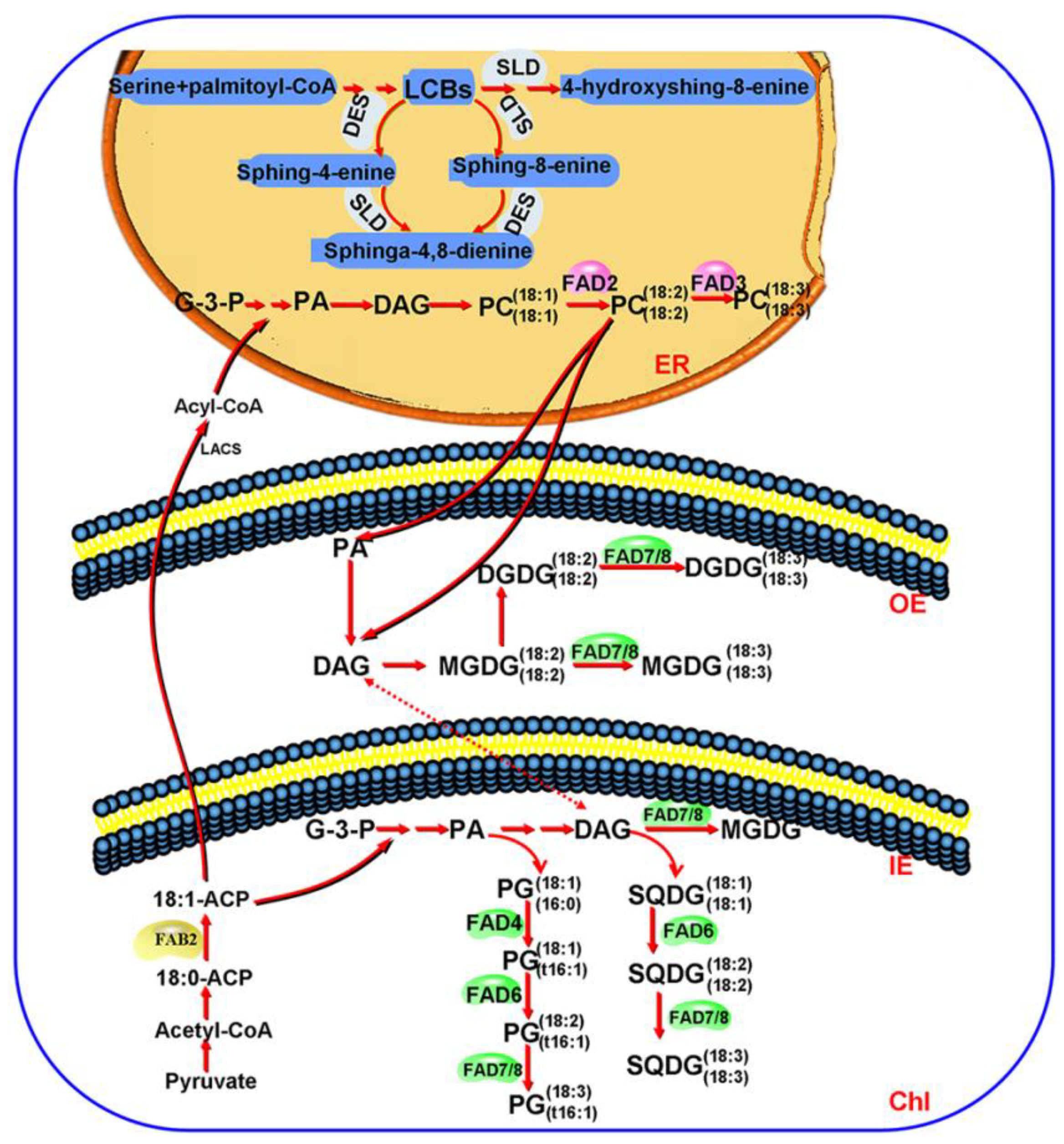
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ohlrogge, J.; Browse, J.J. Lipid biosynthesis. Plant Cell 1995, 7, 957–970. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Xin, Z.; Browse, J. Overexpression of the FAD3 desaturase gene in a mutant of Arabidopsis. Plant Physiol. 1997, 114, 1533–1539. [Google Scholar] [CrossRef] [PubMed]
- Annie, L.; Balk, E.M.; Lichtenstein, A.H. Gender differences in plasma lipid response to dietary fat. Nutr. Rev. 2010, 64, 234–249. [Google Scholar]
- Li, N.; Xu, C.; Li-Beisson, Y.; Philippar, K. Fatty acid and lipid transport in plant cells. Trends Plant Sci. 2016, 21, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wallis, J.G.; John, B. Mutations in the prokaryotic pathway rescue the fatty acid biosynthesis1 mutant in the cold. Plant Physiol. 2015, 169, 442–452. [Google Scholar] [CrossRef]
- Dong, C.; Cao, N.; Zhang, Z.; Shang, Q. Characterization of the fatty acid desaturase genes in cucumber: structure, phylogeny, and expression patterns. PLoS ONE 2016, 11, e0149917. [Google Scholar] [CrossRef]
- Shanklin, J.; Cahoon, E.B. Desaturation and related modifications of fatty acids1. Annu. Rev. Plant Physiol. 1998, 49, 611–641. [Google Scholar] [CrossRef]
- Shanklin, J.; Somerville, C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc. Natl. Acad. Sci. USA 1991, 88, 2510–2514. [Google Scholar] [CrossRef]
- Thiede, M.A.; Ozols, J.; Strittmatter, P. Construction and sequence of cDNA for rat liver stearyl coenzyme a desaturase. J. Biol. Chem. 1986, 261, 13230–13235. [Google Scholar]
- Stukey, J.E.; McDonough, V.M.; Martin, C.E. The OLE1 gene of Saccharomyces cerevisiae encodes the Δ9 fatty acid desaturase and can be functionally replaced by the rat stearoyl-CoA desaturase gene. J. Biol. Chem. 1990, 265, 20144–20149. [Google Scholar]
- Xuan, W.; Zhang, Y.; Liu, Z.; Feng, D.; Luo, M. Molecular cloning and expression analysis of a novel BCCP subunit gene from Aleurites moluccana. Genet. Mol. Res. 2015, 14, 9922–9931. [Google Scholar] [CrossRef] [PubMed]
- González-Thuillier, I.; Venegas-Calerón, M.; Sánchez, R.; Garcés, R.; Wettstein-Knowles, P.V.; Martínez-Force, E. Sunflower (Helianthus annuus) fatty acid synthase complex: β-hydroxyacyl-[acyl carrier protein] dehydratase genes. Planta 2016, 243, 397–410. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.F.R.; Sánchez-García, A.; Salas, J.J.; Garcés, R.; Martínez-Force, E. Characterization of soluble acyl-ACP desaturases from Camelina sativa, Macadamia tetraphylla and Dolichandra unguis-cati. J. Plant Physiol. 2015, 178, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arabidopsis Book 2010, 8, e0133. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; McConn, M.; Douglas, J.J.; Miquel, M. Mutants of Arabidopsis deficient in the synthesis of α-linolenate. biochemical and genetic characterization of the endoplasmic reticulum linoleoyl desaturase. J. Biol. Chem. 1993, 268, 16345–16351. [Google Scholar] [PubMed]
- Wallis, J.G.; Browse, J. Mutants of Arabidopsis reveal many roles for membrane lipids. Prog. Lipid Res. 2002, 41, 254–278. [Google Scholar] [CrossRef]
- Dunn, T.M.; Lynch, D.V.; Michaelson, L.V.; Napier, J.A. A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann. Bot. 2004, 93, 483–497. [Google Scholar] [CrossRef]
- Ming, C.; Markham, J.E.; Cahoon, E.B. Sphingolipid Δ8 unsaturation is important for glucosylceramide biosynthesis and low-temperature performance in Arabidopsis. Plant J. 2012, 69, 769–781. [Google Scholar]
- Narayanan, S.; Prasad, P.V.; Welti, R. Wheat leaf lipids during heat stress: II. lipids experiencing coordinated metabolism are detected by analysis of lipid co-occurrence. Plant Cell Environ. 2016, 39, 608–617. [Google Scholar] [CrossRef]
- Gao, J.; Ajjawi, I.; Manoli, A.; Sawin, A.; Xu, C.; Froehlich, J.E.; Last, R.L.; Benning, C. Fatty acid desaturase4 of Arabidopsis encodes a protein distinct from characterized fatty acid desaturases. Plant J. 2010, 60, 832–839. [Google Scholar] [CrossRef]
- Escandón, M.; Meijón, M.; Valledor, L.; Pascual, J.; Pinto, G.; Cañal, M.J. Metabolome integrated analysis of high-temperature response in Pinus radiata. Front. Plant Sci. 2018, 9, 485. [Google Scholar] [CrossRef] [PubMed]
- Lemieux, B.; Miquel, M.; Somerville, C.; Browse, J. Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theor. Appl. Genet. 1990, 80, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Browse, J.; Kunst, L.; Anderson, S.; Hugly, S.; Somerville, C. A mutant of Arabidopsis deficient in the chloroplast 16:1/18:1 desaturase. Plant Physiol. 1989, 90, 522–529. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, G.L.; Wang, X.M. Role of aminoalcoholphosphotransferases 1 and 2 in phospholipid homeostasis in Arabidopsis. Plant Cell 2015, 27, 1512–1528. [Google Scholar]
- Zhang, M.; Barg, R.; Yin, M.; Gueta-Dahan, Y.; Leikin-Frenkel, A.; Salts, Y.; Shabtai, S.; Ben-Hayyim, G. Modulated fatty acid desaturation via overexpression of two distinct ω-3 desaturases differentially alters tolerance to various abiotic stresses in transgenic tobacco cells and plants. Plant J. 2010, 44, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Tovuu, A.; Zulfugarov, I.S.; Wu, G.X.; Kang, I.S.; Kim, C.; Moon, B.Y.; An, G.; Lee, C.H. Rice mutants deficient in ω−3 fatty acid desaturase (FAD8) fail to acclimate to cold temperatures. Plant Physiol. Biochem. 2016, 109, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-García, M.; Gavilanes-Ruíz, M.; Arce-Cervantes, O. Long-chain bases, phosphatidic acid, MAPKs, and reactive oxygen species as nodal signal transducers in stress responses in Arabidopsis. Front. Plant Sci. 2015, 6, 55. [Google Scholar] [CrossRef]
- Luttgeharm, K.D.; Kimberlin, A.N.; Cahoon, E.B. Plant sphingolipid metabolism and function. Subcell. Biochem. 2016, 86, 249–286. [Google Scholar]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PLoS ONE 2015, 10, e0123281. [Google Scholar] [CrossRef]
- Mikkilineni, V.; Rocheford, T.R. Sequence variation and genomic organization of fatty acid desaturase-2 (fad2) and fatty acid desaturase-6 (fad6) cDNAs in maize. Theor. Appl. Genet. 2003, 106, 1326–1332. [Google Scholar] [CrossRef]
- Tao, F.; Zhu, S.; Fan, J.; Cheng, B. Cloning and sequence analysis of maize FAD2 gene. J. Plant Physiol. 2006, 32, 649–656. [Google Scholar]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Ivica, L.; Tobias, D.; Peer, B. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, 257–260. [Google Scholar]
- Liu, W.; Xie, Y.; Ma, J.; Luo, X.; Nie, P.; Zuo, Z.; Lahrmann, U.; Zhao, Q.; Zheng, Y.; Zhao, Y.; et al. IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 2015, 31, 3359–3361. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. Chapter 2, Unit 2.3. [Google Scholar] [CrossRef] [PubMed]
- Panu, A.; Manohar, J.; Konstantin, A.; Delphine, B.; Gabor, C.; Edouard, D.C.; Séverine, D.; Volker, F.; Arnaud, F.; Elisabeth, G. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef]
- Debbie, W.; Ben, V.; Hardeep, N.; Ron, A.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar]
- Gu, Y.; He, L.; Zhao, C.; Wang, F.; Yan, B.; Gao, Y.; Li, Z.; Yang, K.; Xu, J. Biochemical and transcriptional regulation of membrane lipid metabolism in maize leaves under low temperature. Front. Plant Sci. 2017, 8, 2053. [Google Scholar] [CrossRef]
- Irina, M.; Waters, A.J.; West, P.T.; Michelle, S.; Hirsch, C.N.; Jeffrey, R.I.; Springer, N.M. Transposable elements contribute to activation of maize genes in response to abiotic stress. PLoS Genet. 2015, 11, e1004915. [Google Scholar]
- Michael, S.; Keiichiro, O.; Trey, I.; Steven, M. PiNGO: A Cytoscape plugin to find candidate genes in biological networks. Bioinformatics 2011, 27, 1030–1031. [Google Scholar]
- Zhang, Z.; Zhang, J.; Chen, Y.; Li, R.; Wang, H.; Ding, L.; Wei, J. Isolation, structural analysis, and expression characteristics of the maize (Zea mays L.) hexokinase gene family. Mol. Biol. Rep. 2014, 41, 6157–6166. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Lv, W.; Jiang, S.; Zhang, D.; Cai, G.; Pan, J.; Li, D. Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genom. 2013, 14, 433. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W.R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef]
- Han, Y.; Xu, G.; Du, H.; Hu, J.; Liu, Z.; Li, H.; Li, J.; Yang, X. Natural variations in stearoyl-acp desaturase genes affect the conversion of stearic to oleic acid in maize kernel. Theor. Appl. Genet. 2017, 130, 1–11. [Google Scholar] [CrossRef]
- Michaelson, L.V.; Zäuner, S.; Markham, J.E.; Haslam, R.P.; Desikan, R.; Mugford, S.; Albrecht, S.; Warnecke, D.; Sperling, P.; Heinz, E.; et al. Functional characterization of a higher plant sphingolipid ∆4-desaturase: Defining the role of sphingosine and sphingosine-1-phosphate in Arabidopsis. Plant Physiol. 2009, 149, 487–495. [Google Scholar] [CrossRef]
- Chi, X.; Zhang, X.; Guan, X.; Ding, L.; Li, Y.; Wang, M.; Lin, H.; Qin, S. Fatty acid biosynthesis in eukaryotic photosynthetic microalgae: identification of a microsomal delta 12 desaturase in Chlamydomonas reinhardtii. J. Microbiol. 2008, 46, 189–201. [Google Scholar] [CrossRef]
- Lu, Y.; Chi, X.; Li, Z.; Yang, Q.; Li, F.; Liu, S.; Gan, Q.; Qin, S. Isolation and characterization of a stress-dependent plastidial Δ12 fatty acid desaturase from the Antarctic microalga Chlorella vulgaris NJ-7. Lipids 2010, 45, 179–187. [Google Scholar] [CrossRef]
- Xue, Y.; Chen, B.; Win, A.N.; Fu, C.; Lian, J.; Liu, X.; Wang, R.; Zhang, X.; Chai, Y. Omega-3 fatty acid desaturase gene family from two ω-3 sources, Salvia hispanica and Perilla frutescens: Cloning, characterization and expression. PLoS ONE 2018, 13, e0191432. [Google Scholar] [CrossRef]
- Dörmann, P.; Hoffmann-Benning, S.; Balbo, I.; Benning, C. Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 1995, 7, 1801–1810. [Google Scholar]
- Heinz, E.; Roughan, P.G. Similarities and differences in lipid metabolism of chloroplasts isolated from 18:3 and 16:3 plants. Plant Physiol. 1983, 72, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Löhden, I.; Frentzen, M. Role of plastidial acyl-acyl carrier protein: Glycerol 3-phosphate acyltransferase and acyl-acyl carrier protein hydrolase in channelling the acyl flux through the prokaryotic and eukaryotic pathway. Planta 1988, 176, 506–512. [Google Scholar] [CrossRef] [PubMed]
- Tocher, D.R.; Leaver, M.J.; Hodgson, P.A. Recent advances in the biochemistry and molecular biology of fatty acyl desaturases. Prog. Lipid Res. 1998, 37, 73–117. [Google Scholar] [CrossRef]
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 gene in plants: Occurrence, regulation, and role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed]
- Esteban, A.B.; Sicardo, M.D.; Mancha, M.; Martinez-Rivas, J.M. Growth temperature control of the linoleic acid content in safflower (Carthamus tinctorius) seed oil. J. Agric. Food Chem. 2004, 52, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Matteucci, M.; D’Angeli, S.; Errico, S.; Lamanna, R.; Perrotta, G.; Altamura, M.M. Cold affects the transcription of fatty acid desaturases and oil quality in the fruit of Olea europaea L. genotypes with different cold hardiness. J. Exp. Bot. 2011, 62, 3403–3420. [Google Scholar] [CrossRef]
- Rodriguez-Vargas, S.; Sanchez-Garcia, A.; Martinez-Rivas, J.M.; Prieto, J.A.; Randez-Gil, F. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl. Environ. Microbiol. 2007, 73, 110–116. [Google Scholar] [CrossRef]
- Zhou, Y.; Zeng, L.; Fu, X.; Mei, X.; Cheng, S.; Liao, Y.; Deng, R.; Xu, X.; Jiang, Y.; Duan, X.; et al. The sphingolipid biosynthetic enzyme sphingolipid delta8 desaturase is important for chilling resistance of tomato. Sci. Rep. 2016, 6, 38742. [Google Scholar] [CrossRef]
- Zhang, T.; Song, C.; Song, L.; Shang, Z.; Yang, S.; Zhang, D.; Sun, W.; Shen, Q.; Zhao, D. RNA sequencing and coexpression analysis reveal key genes involved in α-linolenic acid biosynthesis in Perilla frutescens seed. Int. J. Mol. Sci. 2017, 18, 2433. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Wei, J.; He, L.; Zhang, Y.; Zhao, Y.; Xu, X.; Wei, Y.; Ge, S.; Ding, D.; Liu, M.; et al. Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature. Genes 2019, 10, 445. https://doi.org/10.3390/genes10060445
Zhao X, Wei J, He L, Zhang Y, Zhao Y, Xu X, Wei Y, Ge S, Ding D, Liu M, et al. Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature. Genes. 2019; 10(6):445. https://doi.org/10.3390/genes10060445
Chicago/Turabian StyleZhao, Xunchao, Jinpeng Wei, Lin He, Yifei Zhang, Ying Zhao, Xiaoxuan Xu, Yulei Wei, Shengnan Ge, Dong Ding, Meng Liu, and et al. 2019. "Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature" Genes 10, no. 6: 445. https://doi.org/10.3390/genes10060445
APA StyleZhao, X., Wei, J., He, L., Zhang, Y., Zhao, Y., Xu, X., Wei, Y., Ge, S., Ding, D., Liu, M., Gao, S., & Xu, J. (2019). Identification of Fatty Acid Desaturases in Maize and Their Differential Responses to Low and High Temperature. Genes, 10(6), 445. https://doi.org/10.3390/genes10060445




