Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
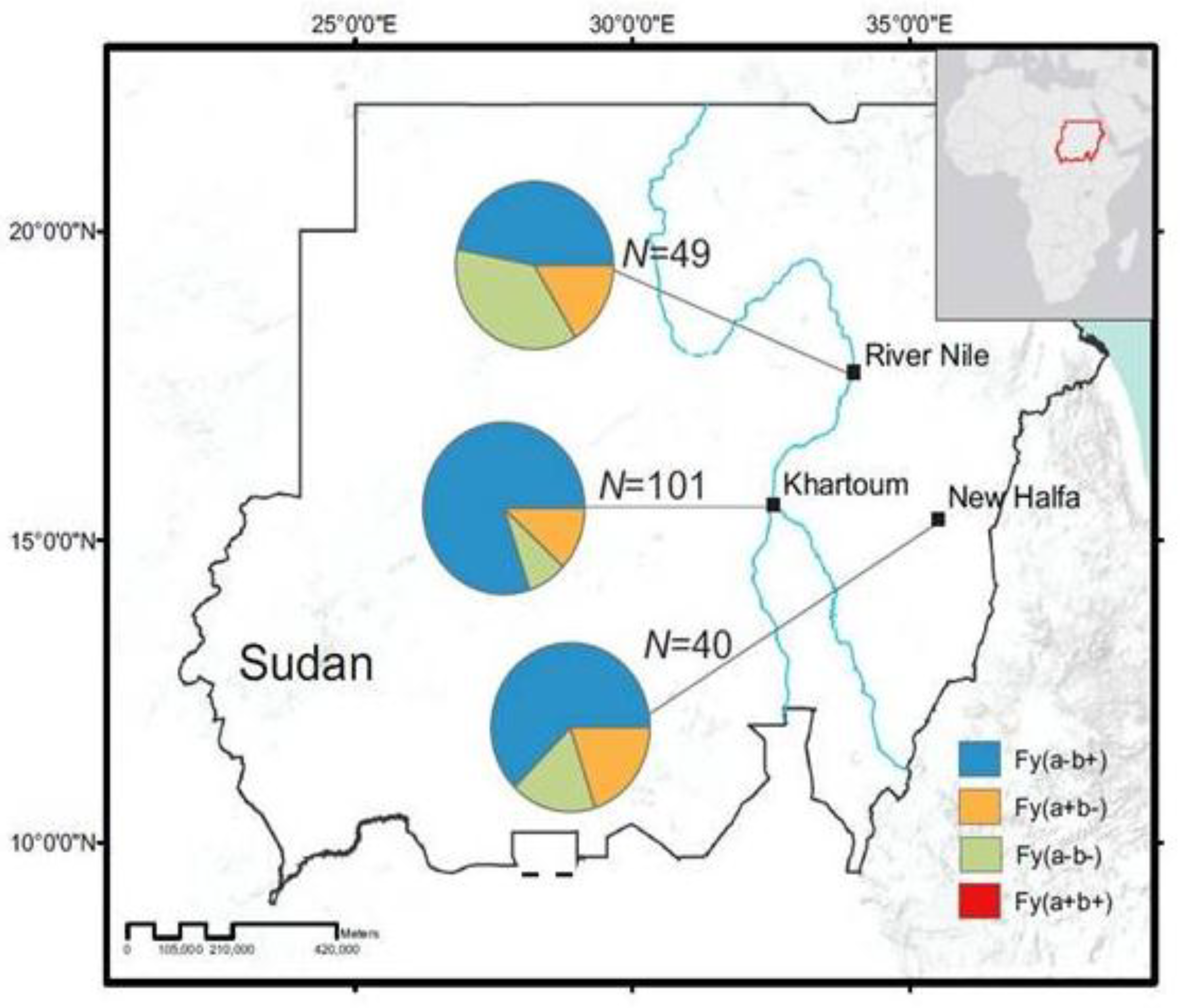
2.2. Study Sites and Subjects
2.3. Sample Collection and DNA Extraction
2.4. Molecular Identification of Plasmodium Species
2.5. Estimation of P. vivax Parasitemia
2.6. Duffy Blood Group Genotyping
2.7. Statistical Analyses
3. Results
3.1. Distribution of Duffy Phenotypes among P. vivax Infections
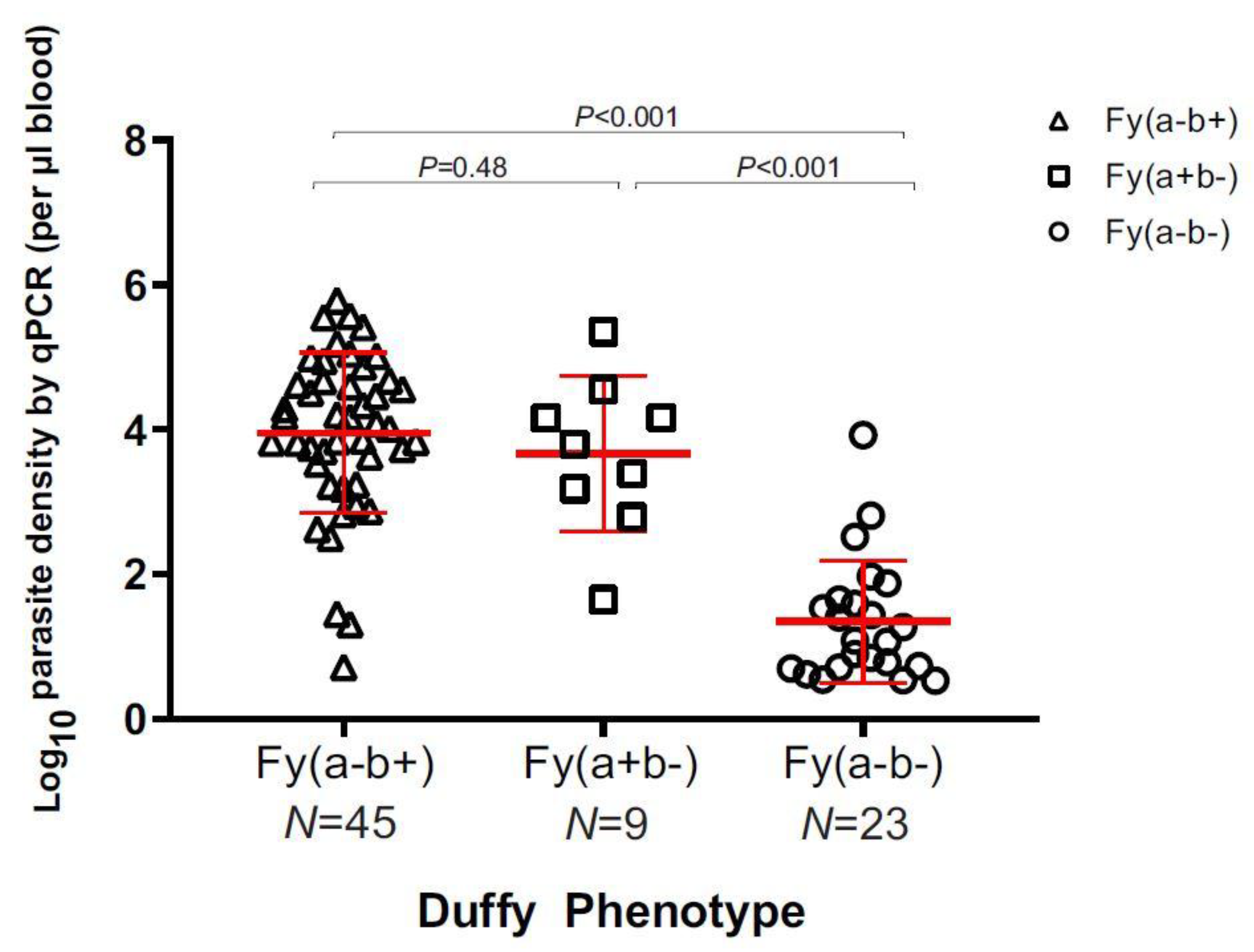
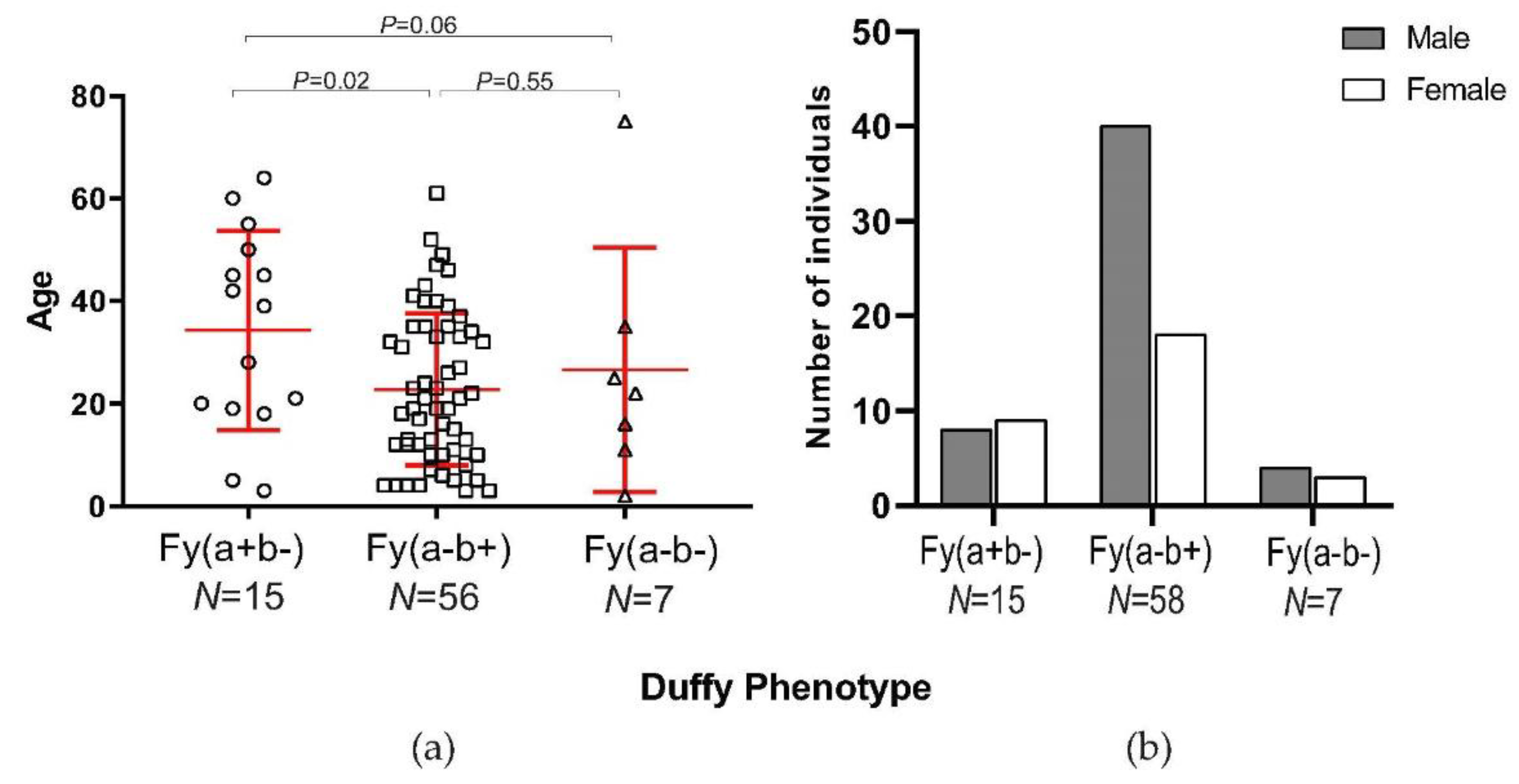
3.2. Parasite Density and Demographic Features in Duffy-Negatives
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. World Malaria Report 2018. Available online: https://www.who.int/malaria/publications/world_malaria_report/en/ (accessed on 19 April 2019).
- Douglas, N.M.; Lampah, D.A.; Kenangalem, E.; Simpson, J.A.; Poespoprodjo, J.R.; Sugiarto, P.; Anstey, N.M.; Price, R.N. Major burden of severe anemia from non-falciiparum malaria species in Southern Papua: a hospital-based surveillance study. PLoS Med. 2013, 10, e1001575. [Google Scholar] [CrossRef] [PubMed]
- Twohig, K.A.; Pfeffer, D.A.; Baird, J.K.; Price, R.N.; Zimmerman, P.A.; Hay, S.I.; Gething, P.W.; Battle, K.E.; Howes, R.E. Growing evidence of Plasmodium vivax across malaria-endemic Africa. PLoS Negl. Trop. Dis. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Abdelraheem, M.H.; Albsheer, M.M.A.; Mohamed, H.S.; Amin, M.; Abdel, H.M.M. Transmission of Plasmodium vivax in Duffy-negative individuals in central Sudan. Trans.R.Soc.Trop.Med.Hyg. 2016, 110, 258–260. [Google Scholar] [CrossRef] [PubMed]
- Suliman, M.M.A.; Hamad, B.M.; Albasheer, M.M.A.; Elhadi, M.; Mustafa, M.A.; Elobied, M.; Hamid, M.M.A. Molecular evidence of high proportion of Plasmodium vivax malaria infection in White Nile area in Sudan. J. Parasitol. Res. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.; Polyakova, J.; Zbrzezna, V.; Williams, K.; Gulati, S.; Pogo, A.O. Cloning of glycoprotein D cDNA, which encodes the major subunit of the Duffy blood group system and the receptor for the Plasmodium vivax malaria parasite. Proc. Natl. Acad. Sci. USA 1993, 90, 10793–10797. [Google Scholar] [CrossRef] [PubMed]
- Miller, L.H.; Mason, S.J.; Clyde, D.F.; McGinniss, M.H. The Resistance Factor to Plasmodium vivax in Blacks. N. Engl. J. Med. 1976, 295, 302–304. [Google Scholar] [CrossRef]
- Chitnis, C.E.; Sharma, A. Targeting the Plasmodium vivax Duffy-binding protein. Trends Parasitol. 2008, 24, 29–34. [Google Scholar] [CrossRef]
- King, C.L.; Adams, J.H.; Zianli, J.; Grimberg, B.T.; McHenry, A.M.; Greenberg, L.J.; Siddiqui, A.; Howes, R.E.; da Silva-Nunes, M.; Ferreira, M.U.; et al. Fya/Fyb antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc. Natl. Acad. Sci. USA 2011, 108. [Google Scholar] [CrossRef]
- De Silva, J.R.; Lau, Y.L.; Fong, M.Y. Genotyping of the Duffy Blood Group among Plasmodium knowlesi-Infected Patients in Malaysia. PLoS ONE 2014, 9, e108951. [Google Scholar] [CrossRef][Green Version]
- Tournamille, C.; Colin, Y.; Cartron, J.P.; Le Van Kim, C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy–negative individuals. Nat. Genet. 1995, 10, 224–228. [Google Scholar] [CrossRef]
- Zimmerman, P.A. Plasmodium vivax infection in Duffy-Negative People in Africa. Am. J. Trop. Med. Hyg. 2017, 97. [Google Scholar] [CrossRef]
- Ménard, D.; Barnadas, C.; Bouchier, C.; Henry-Halldin, C.; Gray, L.R.; Ratsimbasoa, A.; Thonier, V.; Carod, J.F.; Domarle, O.; Colin, Y.; et al. Plasmodium vivax clinical malaria is commonly observed in Duffy-negative Malagasy people. Proc. Natl. Acad. Sci. USA 2010, 107, 5967–5971. [Google Scholar] [CrossRef] [PubMed]
- Wurtz, N.; Lekweiry, K.M.; Bogreau, H.; Pradines, B.; Rogier, C.; Boukhary, A.O.M.S.; Hafid, J.E.; Salem, M.S.O.A.; Trape, J.F.; Basco, L.K.; et al. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. Malar. J. 2011, 10, 336. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Woldearegai, T.G.; Kremsner, P.G.; Kun, J.F.; Mordmüller, B. Plasmodium vivax malaria in duffy-negative individuals from Ethiopia. Trans. R. Soc. Trop. Med. Hyg. 2013, 107, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Russo, G.; Faggioni, G.; Paganotti, G.M.; Dongho, G.B.D.; Pomponi, A.; De Santis, R.; Tebano, G.; Mbida, M.; Sobze, M.S.; Vullo, V.; et al. Molecular evidence of Plasmodium vivax infection in Duffy negative symptomatic individuals from Dschang, West Cameroon. Malar. J. 2017, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Aboud, M.; Makhawi, A.; Verardi, A.; El Raba’a, F.; Elnaiem, D.-E.; Townson, H. A genotypically distinct, melanic variant of Anopheles arabiensis in Sudan is associated with arid environments. Malar. J. 2014, 13, 492. [Google Scholar] [CrossRef] [PubMed]
- Snounou, G.; Viriyakosol, S.; Zhu, X.P.; Jarra, W.; Pinheiro, L.; do Rosario, V.E.; Thaithong, S.; Brown, K.N. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol.Biochem.Parasitol. 1993, 61, 315–320. [Google Scholar] [CrossRef]
- Baum, E.; Sattabongkot, J.; Sirichaisinthop, J.; Kiattibutr, K.; Davies, D.H.; Jain, A.; Lo, E.; Lee, M.C.; Randall, A.Z.; Molina, D.M.; et al. Submicroscopic and asymptomatic Plasmodium falciparum and Plasmodium vivax infections are common in western Thailand—Molecular and serological evidence. Malar. J. 2015, 14, 95. [Google Scholar] [CrossRef]
- FinchTV 1.4. Available online: https://finchtv.software.informer.com/1.4/ (accessed on 7 June 2019).
- NCBI Basic Local Alignment Search Tool. Available online: https://blast.ncbi.nlm.nih.gov/ (accessed on 7 June 2019).
- Kempinska-Podhorodecka, A.; Knap, O.; Drozd, A.; Kaczmarczyk, M.; Parafiniuk, M.; Parczewski, M.; Ciechanowicz, A. Analysis for genotyping Duffy blood group in inhabitants of Sudan, the Fourth Cataract of the Nile. Malar. J. 2012, 11, 115. [Google Scholar] [CrossRef]
- Cavasini, C.E.; de Mattos, L.C.; Couto, Á.A.D.; Bonini-Domingos, C.R.; Valencia, S.H.; Neiras, W.C.; Alves, R.T.; Rossit, A.R.; Castilho, L.; Machado, R.L. Plasmodium vivax infection among Duffy antigen-negative individuals from the Brazilian Amazon region: An exception? Trans. R. Soc. Trop. Med. Hyg. 2007, 101, 1042–1044. [Google Scholar] [CrossRef]
- Mendes, C.; Dias, F.; Figueiredo, J.; Mora, V.G.; Cano, J.; de Sousa, B.; do Rosário, V.E.; Benito, A.; Berzosa, P.; Arez, A.P. Duffy negative antigen is no longer a barrier to Plasmodium vivax—Molecular evidences from the African West Coast (Angola and Equatorial Guinea). PLoS Negl. Trop. Dis. 2011, 5. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.R.; Stoute, J.A.; Amon, J.; Dunton, R.F.; Mtalib, R.; Koros, J.; Owour, B.; Luckhart, S.; Wirtz, R.A.; Barnwell, J.W.; et al. Evidence for transmission of Plasmodium vivax among a Duffy antigen negative population in Western Kenya. Am. J. Trop. Med. Hyg. 2006, 27, 575–581. [Google Scholar] [CrossRef]
- Lo, E.; Yewhalaw, D.; Zhong, D.; Zemene, E.; Degefa, T.; Tushune, K.; Ha, M.; Lee, M.C.; James, A.A.; Yan, G. Molecular epidemiology of Plasmodium vivax and Plasmodium falciparum malaria among Duffy-positive and Duffy-negative populations in Ethiopia. Malar. J. 2015, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Kasehagen, L.J.; Mueller, I.; Kiniboro, B.; Bockarie, M.J.; Reeder, J.C.; Kazura, J.W.; Kastens, W.; McNamara, D.T.; King, C.H.; Whalen, C.C.; et al. Reduced plasmodium vivax Erythrocyte infection in PNG Duffy-negative heterozygotes. PloS ONE 2007, 2. [Google Scholar] [CrossRef] [PubMed]
- Weber, S.S.; Tadei, W.P.; Martins, A.S. Association between Duffy blood group system variants and susceptibility and resistance to malaria in the Brazilian Amazon. Rev. Biol. Neotrop. 2011, 7, 39–44. [Google Scholar] [CrossRef]
| Duffy Phenotype | N | Positive (N = 190) | Negative (N = 67) | p1 | OR (95%CI)2 |
|---|---|---|---|---|---|
| Fy(a+b-) | 29 | 27 (14.2%) | 2 (3%) | 0.0122 | 0.1858 |
| Fy(a-b+) | 149 | 129 (67.9%) | 20 (29.9%) | 0.0001 | 0.2012 |
| Fy(a-b-) | 77 | 34 (17.9%) | 45 (67.1%) | 0.0001 | 1.9567 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albsheer, M.M.A.; Pestana, K.; Ahmed, S.; Elfaki, M.; Gamil, E.; Ahmed, S.M.; Ibrahim, M.E.; Musa, A.M.; Lo, E.; Hamid, M.M.A. Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan. Genes 2019, 10, 437. https://doi.org/10.3390/genes10060437
Albsheer MMA, Pestana K, Ahmed S, Elfaki M, Gamil E, Ahmed SM, Ibrahim ME, Musa AM, Lo E, Hamid MMA. Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan. Genes. 2019; 10(6):437. https://doi.org/10.3390/genes10060437
Chicago/Turabian StyleAlbsheer, Musab M.A., Kareen Pestana, Safaa Ahmed, Mohammed Elfaki, Eiman Gamil, Salma M. Ahmed, Muntaser E. Ibrahim, Ahmed M. Musa, Eugenia Lo, and Muzamil M. Abdel Hamid. 2019. "Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan" Genes 10, no. 6: 437. https://doi.org/10.3390/genes10060437
APA StyleAlbsheer, M. M. A., Pestana, K., Ahmed, S., Elfaki, M., Gamil, E., Ahmed, S. M., Ibrahim, M. E., Musa, A. M., Lo, E., & Hamid, M. M. A. (2019). Distribution of Duffy Phenotypes among Plasmodium vivax Infections in Sudan. Genes, 10(6), 437. https://doi.org/10.3390/genes10060437







