Microhomology Selection for Microhomology Mediated End Joining in Saccharomyces cerevisiae
Abstract
:1. Introduction
2. Material and Methods
2.1. Yeast Strains and Plasmids
2.2. Homothallic Switching Endonuclease Induction
2.3. Analysis of Repair Events
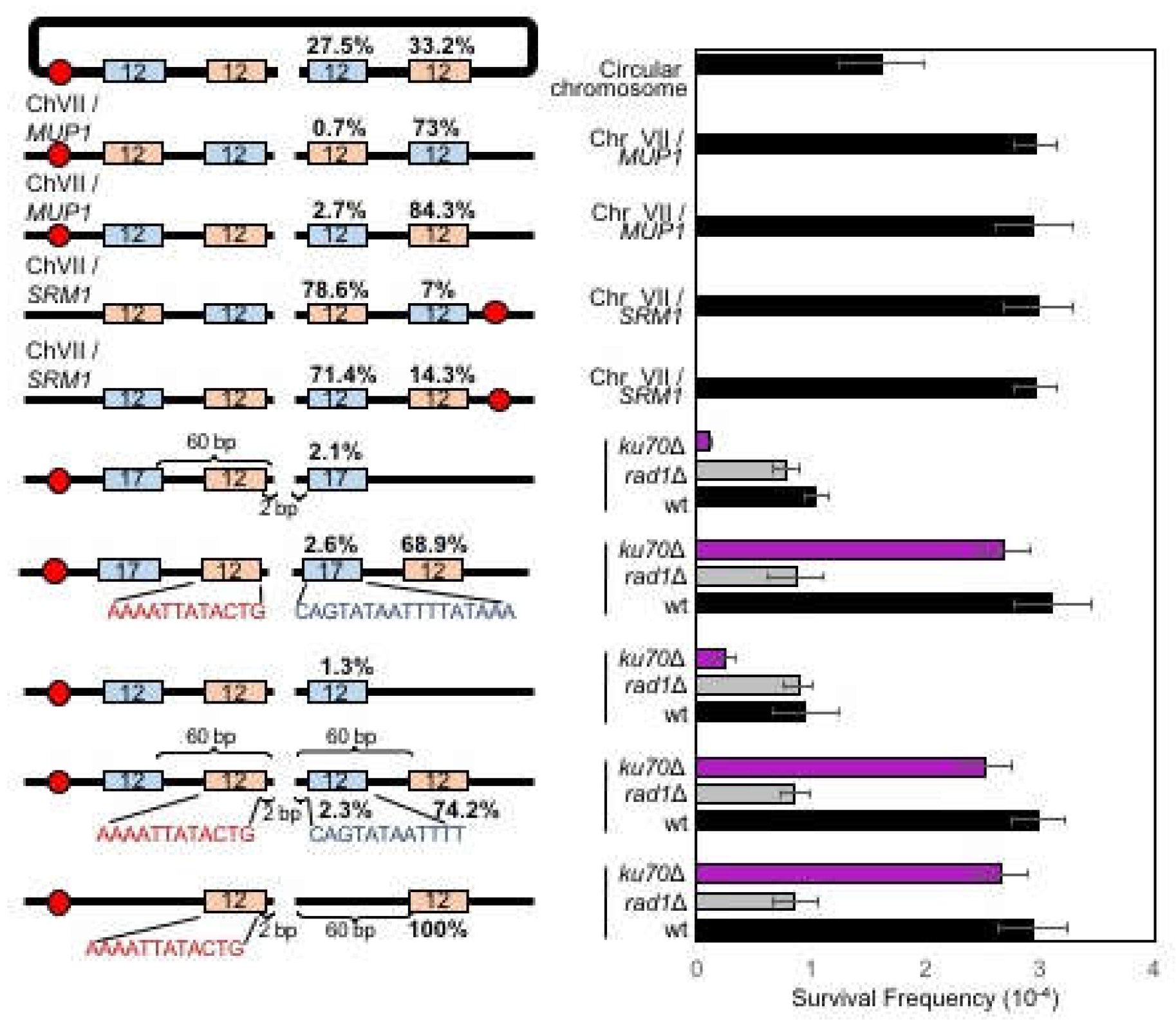
3. Results
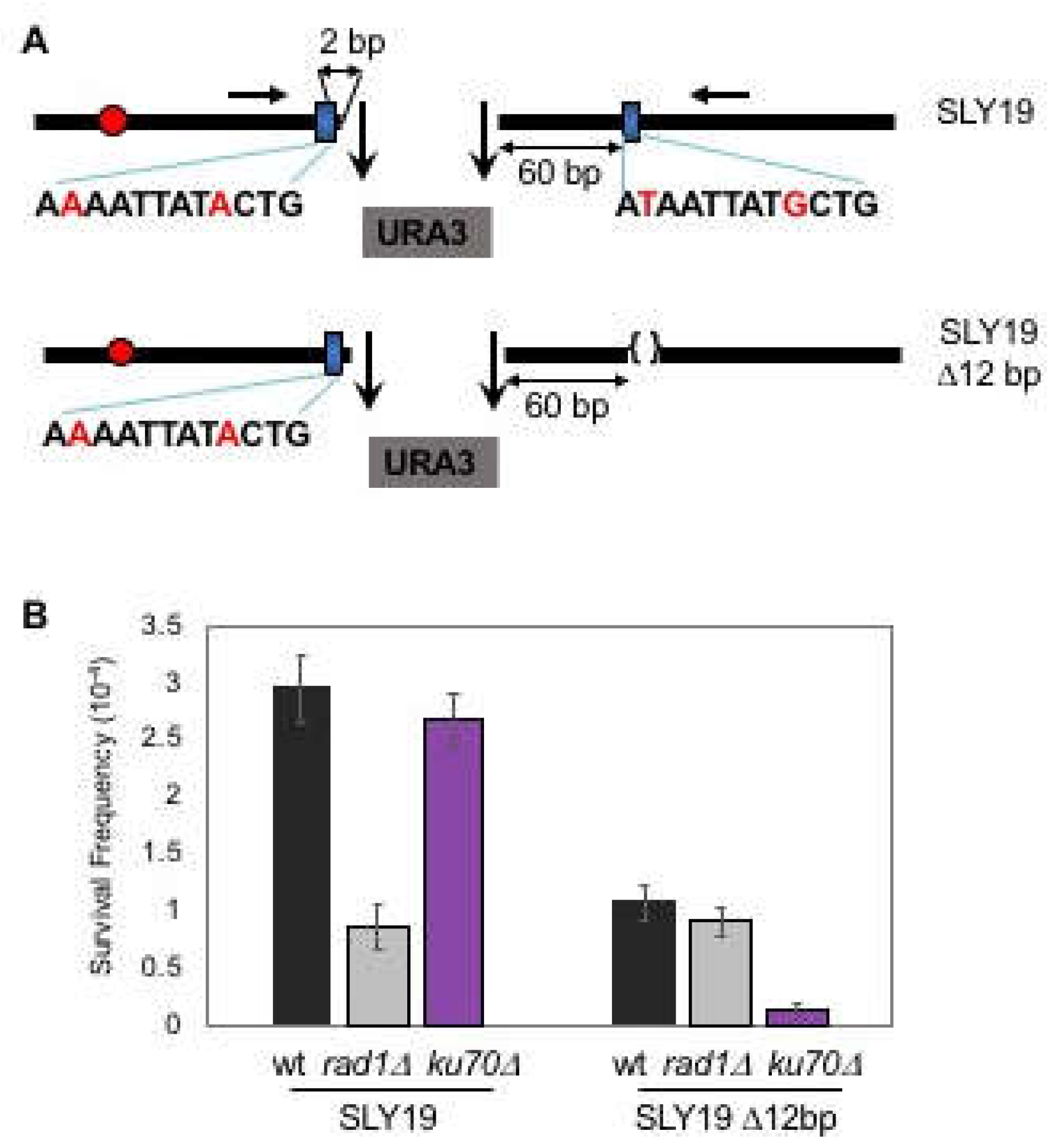
3.1. Microhomology is not Randomly Selected for MMEJ
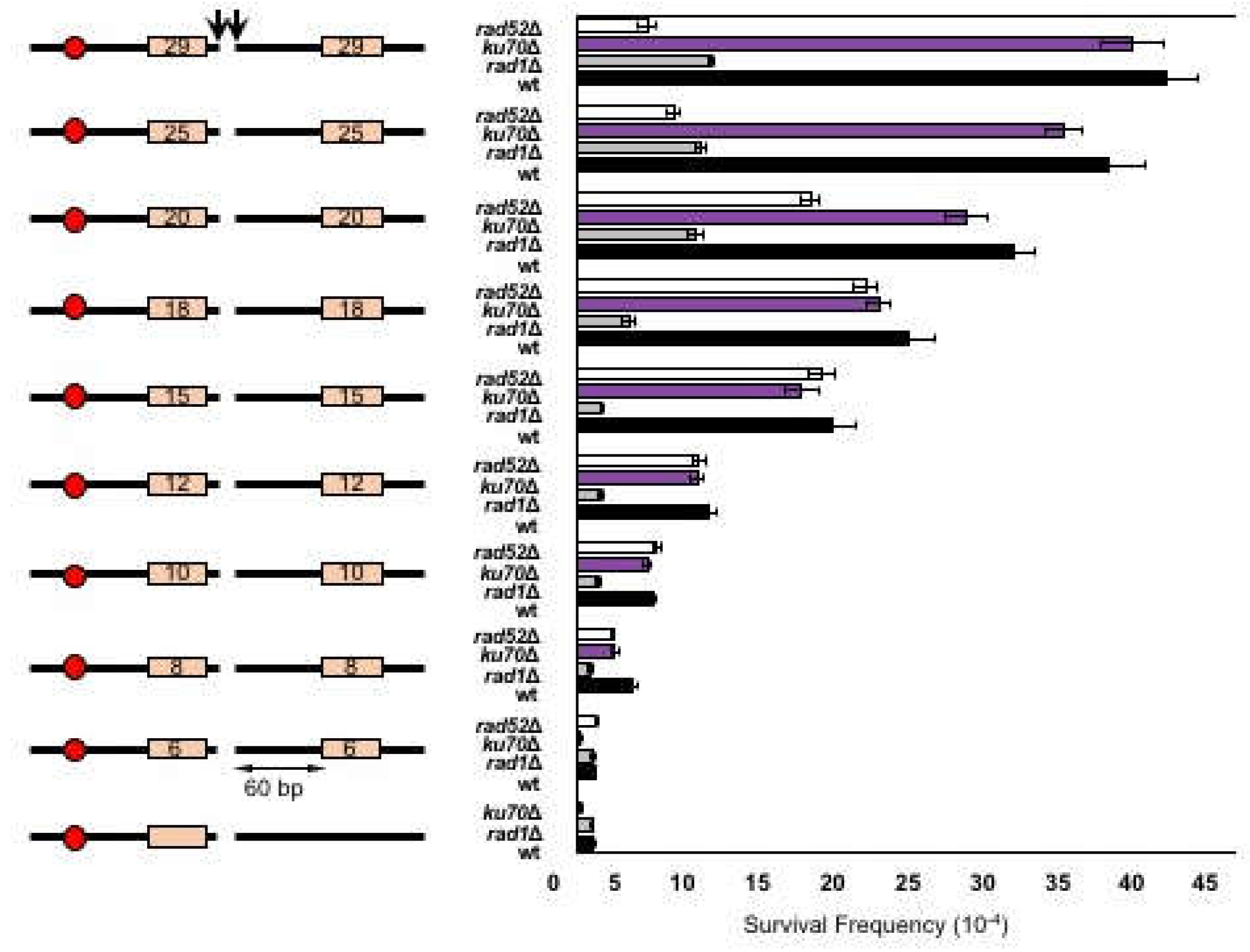
3.2. MMEJ Requires Microhomology of 8- to 20-Nucleotide Long
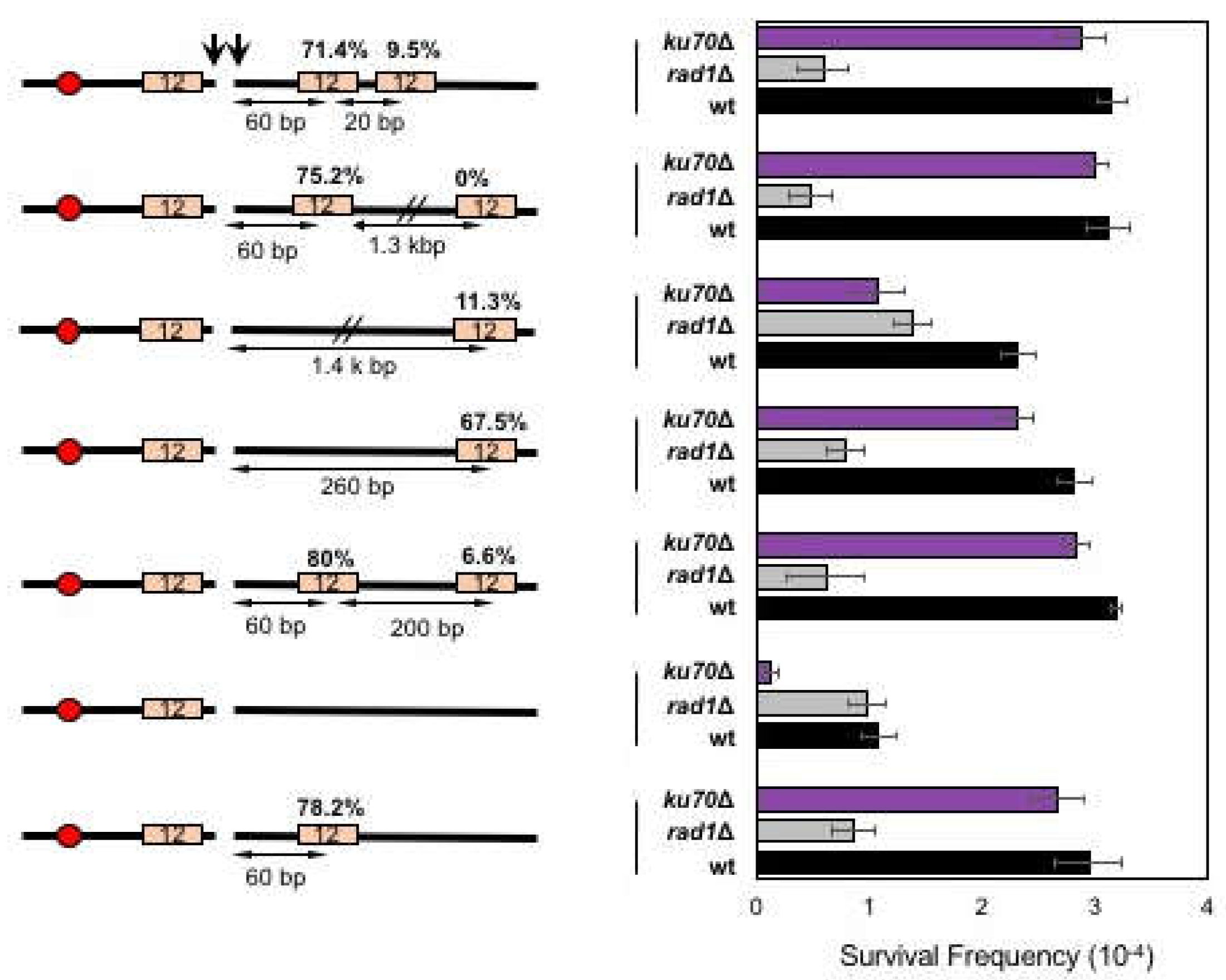
3.3. Effect of Distance between Microhomology and DNA Break on Microhomology Usage
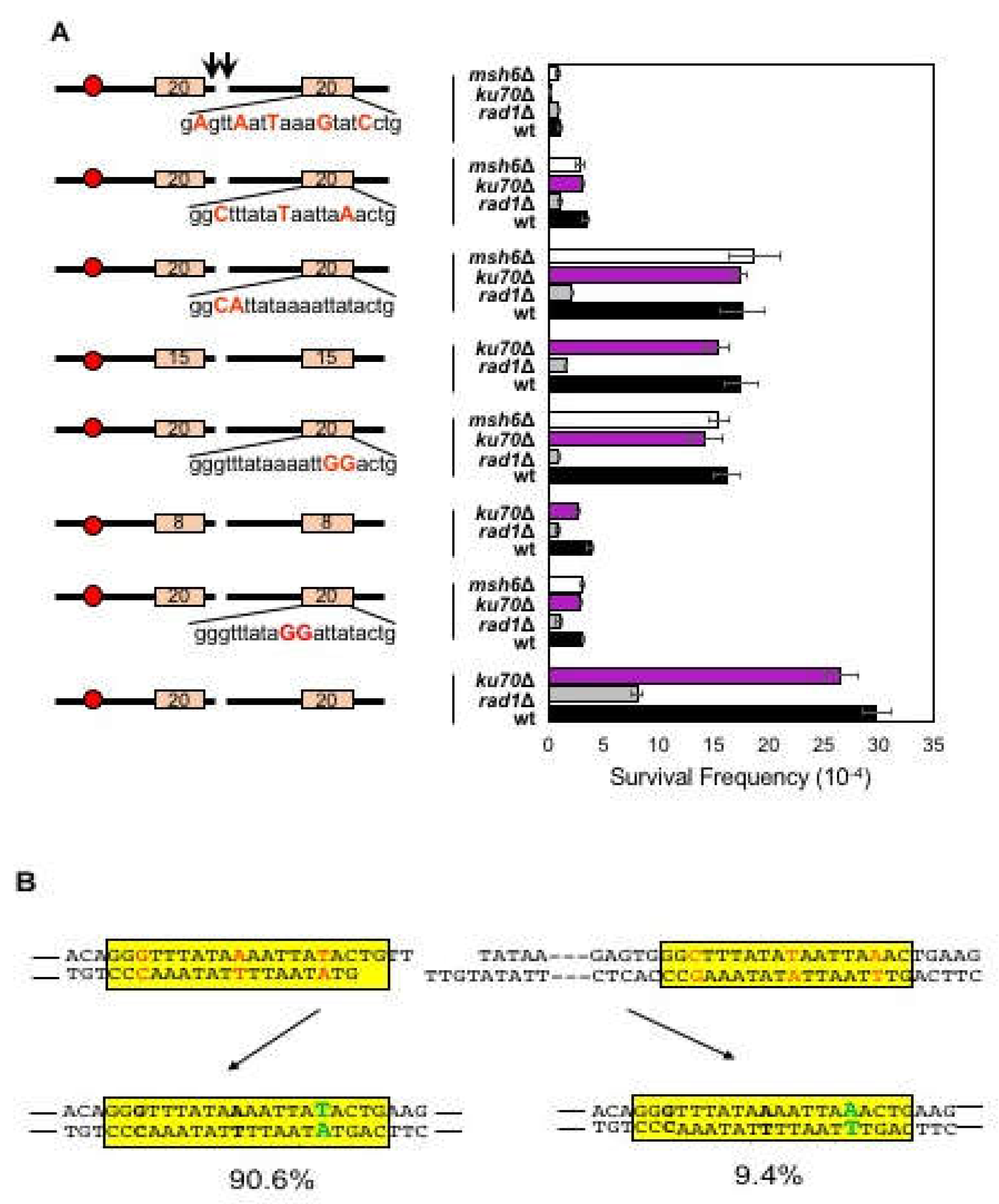
3.4. Effect of Mismatches on Microhomology Selection
3.5. Biased Mismatch Correction in Imperfect Microhomology-Mediated MMEJ
3.6. MMEJ Produces Repair Products Preferentially Deleting the Telomere Proximal Side of the Break
3.7. Chromosome Circularization Disrupts Microhomology Selection Bias in MMEJ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef]
- Seol, J.H.; Shim, E.Y.; Lee, S.E. Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. 2017, 809, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; Symington, L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef]
- Sinha, S.; Villarreal, D.; Shim, E.Y.; Lee, S.E. Risky business: Microhomology-mediated end joining. Mutat. Res. 2016, 788, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Xu, X. Microhomology-mediated end joining: New players join the team. Cell Biosci. 2017, 7, 6. [Google Scholar] [CrossRef]
- Lee-Theilen, M.; Matthews, A.J.; Kelly, D.; Zheng, S.; Chaudhuri, J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 2011, 18, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Lazzerini-Denchi, E.; Sfeir, A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef]
- Simsek, D.; Brunet, E.; Wong, S.Y.; Katyal, S.; Gao, Y.; McKinnon, P.J.; Lou, J.; Zhang, L.; Li, J.; Rebar, E.J.; et al. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011, 7, e1002080. [Google Scholar] [CrossRef]
- Villarreal, D.D.; Lee, K.; Deem, A.; Shim, E.Y.; Malkova, A.; Lee, S.E. Microhomology directs diverse DNA break repair pathways and chromosomal translocations. PLoS Genet. 2012, 8, e1003026. [Google Scholar] [CrossRef]
- Ma, J.L.; Kim, E.M.; Haber, J.E.; Lee, S.E. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol. Cell. Biol. 2003, 23, 8820–8828. [Google Scholar] [CrossRef]
- Lee, K.; Lee, S.E. Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining. Genetics 2007, 176, 2003–2014. [Google Scholar] [CrossRef] [PubMed]
- Decottignies, A. Alternative end-joining mechanisms: A historical perspective. Front. Genet. 2013, 4, 48. [Google Scholar] [CrossRef]
- Liang, L.; Deng, L.; Nguyen, S.C.; Zhao, X.; Maulion, C.D.; Shao, C.; Tischfield, J.A. Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks. Nucleic Acids Res. 2008, 36, 3297–3310. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.T.; Boboila, C.; Souza, E.K.; Franco, S.; Hickernell, T.R.; Murphy, M.; Gumaste, S.; Geyer, M.; Zarrin, A.A.; Manis, J.P.; et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 2007, 449, 478–482. [Google Scholar] [CrossRef] [PubMed]
- Heacock, M.L.; Idol, R.A.; Friesner, J.D.; Britt, A.B.; Shippen, D.E. Telomere dynamics and fusion of critically shortened telomeres in plants lacking DNA ligase IV. Nucleic Acids Res. 2007, 35, 6490–6500. [Google Scholar] [CrossRef]
- McVey, M.; Lee, S.E. MMEJ repair of double-strand breaks (director’s cut): Deleted sequences and alternative endings. Trends Genet. 2008, 24, 529–538. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.M.; McVey, M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res. 2010, 38, 5706–5717. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Nakade, S.; Sakane, Y.; Suzuki, K.T.; Yamamoto, T. MMEJ-assisted gene knock-in using TALENs and CRISPR-Cas9 with the PITCh systems. Nat. Protoc. 2016, 11, 118–133. [Google Scholar] [CrossRef]
- Howard, S.M.; Yanez, D.A.; Stark, J.M. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015, 11, e1004943. [Google Scholar] [CrossRef]
- Deng, S.K.; Gibb, B.; de Almeida, M.J.; Greene, E.C.; Symington, L.S. RPA antagonizes microhomology-mediated repair of DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 405–412. [Google Scholar] [CrossRef]
- McVey, M. RPA puts the brakes on MMEJ. Nat. Struct. Mol. Biol. 2014, 21, 348–349. [Google Scholar] [CrossRef] [PubMed]
- Kent, T.; Chandramouly, G.; McDevitt, S.M.; Ozdemir, A.Y.; Pomerantz, R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta. Nat. Struct. Mol. Biol. 2015, 22, 230–237. [Google Scholar] [CrossRef]
- Soni, A.; Siemann, M.; Grabos, M.; Murmann, T.; Pantelias, G.E.; Iliakis, G. Requirement for Parp-1 and DNA ligases 1 or 3 but not of Xrcc1 in chromosomal translocation formation by backup end joining. Nucleic Acids Res. 2014, 42, 6380–6392. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O’Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Ledermann, J.A. PARP inhibitors in ovarian cancer. Ann. Oncol. 2016, 27 (Suppl. 1), i40–i44. [Google Scholar] [CrossRef]
- Ledermann, J.A.; Drew, Y.; Kristeleit, R.S. Homologous recombination deficiency and ovarian cancer. Eur. J. Cancer 2016, 60, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.W.; Tewari, K.S. New Targeted Agents in Gynecologic Cancers: Synthetic Lethality, Homologous Recombination Deficiency, and PARP Inhibitors. Curr. Treat. Opt. Oncol. 2016, 17, 12. [Google Scholar] [CrossRef]
- Mengwasser, K.E.; Adeyemi, R.O.; Leng, Y.; Choi, M.Y.; Clairmont, C.; D’Andrea, A.D.; Elledge, S.J. Genetic Screens Reveal FEN1 and APEX2 as BRCA2 Synthetic Lethal Targets. Mol. Cell 2019, 73, 885.e6–899.e6. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Moore, J.K.; Holmes, A.; Umezu, K.; Kolodner, R.D.; Haber, J.E. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 1998, 94, 399–409. [Google Scholar] [CrossRef]
- Haber, J.E.; Thorburn, P.C. Healing of broken linear dicentric chromosomes in yeast. Genetics 1984, 106, 207–226. [Google Scholar]
- Zhu, Z.; Chung, W.H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, N.; Goldfarb, T.; Studamire, B.; Alani, E.; Haber, J.E. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc. Natl. Acad. Sci. USA 2004, 101, 9315–9320. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.A.; Baumann, P.; Van Dyck, E.; West, S.C. Precise binding of single-stranded DNA termini by human RAD52 protein. EMBO J. 2000, 19, 4175–4181. [Google Scholar] [CrossRef]
- Stasiak, A.Z.; Larquet, E.; Stasiak, A.; Muller, S.; Engel, A.; Van Dyck, E.; West, S.C.; Egelman, E.H. The human Rad52 protein exists as a heptameric ring. Curr. Biol. 2000, 10, 337–340. [Google Scholar] [CrossRef]
- Anand, R.; Beach, A.; Li, K.; Haber, J. Rad51-mediated double-strand break repair and mismatch correction of divergent substrates. Nature 2017, 544, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Fu, B.X.; Heyer, W.D. DNA polymerases delta and lambda cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2015, 112, E6907–E6916. [Google Scholar] [CrossRef]
- Sinha, S.; Li, F.; Villarreal, D.; Shim, J.H.; Yoon, S.; Myung, K.; Shim, E.Y.; Lee, S.E. Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions. PLoS Genet. 2017, 13, e1006714. [Google Scholar] [CrossRef]
- Kalocsay, M.; Hiller, N.J.; Jentsch, S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol. Cell 2009, 33, 335–343. [Google Scholar] [CrossRef]
| SLY19 | hoΔ MATα∷URA3∷HOcs hmlΔ∷ADE1 hmrΔ∷ADE1 ade1-100 leu2-3,112 lys5 trp1∷hisG ura3-52 ade3∷GAL∷HO |
|---|---|
| YKHL76 | SLY19 ku70Δ::KAN |
| YKHL77 | SLY19 RAD1Δ::KAN |
| YKHL101 | SLY19 12bp microhomology (MH)Δ |
| YKHL102 | JKM179 MAT::URA3::HO::6MH |
| YKHL103 | YKHL102 ku70Δ::KAN |
| YKHL104 | YKHL102 RAD1Δ::KAN |
| YKHL105 | JKM179 MAT::URA3::HO::10MH |
| YKHL106 | YKHL105 ku70Δ::KAN |
| YKHL107 | YKHL105 RAD1Δ::KAN |
| YKHL108 | JKM179 MAT::URA3::HO::12MH |
| YKHL109 | YKHL108 ku70Δ::KAN |
| YKHL111 | YKHL108 RAD1Δ::KAN |
| YKHL112 | JKM179 MAT::URA3::HO::15MH |
| YKHL113 | YKHL112 ku70Δ::KAN |
| YKHL114 | YKHL112 RAD1Δ::KAN |
| YKHL115 | JKM179 MAT::URA3::HO::18MH |
| YKHL116 | YKHL115 ku70Δ::KAN |
| YKHL117 | YKHL115 RAD1Δ::KAN |
| YKHL118 | JKM179 MAT::URA3::HO::20MH |
| YKHL119 | YKHL118 ku70Δ::KAN |
| YKHL121 | YKHL118 RAD1Δ::KAN |
| YKHL122 | YKHL118 rad52Δ::KAN |
| YKHL123 | JKM179 MAT::URA3::HO::25MH |
| YKHL124 | YKHL123 ku70Δ::KAN |
| YKHL125 | YKHL123 rad1Δ::KAN |
| YKHL126 | YKHL123 rad52Δ::KAN |
| YKHL127 | JKM179 MAT::URA3::HO::29MH |
| YKHL128 | YKHL127 ku70Δ::KAN |
| YKHL129 | YKHL127 RAD1Δ::KAN |
| YKHL131 | YKHL127 rad52Δ::KAN |
| YKHL132 | JKM179 MAT::URA3::HO::200bp::12MH |
| YKHL133 | YKHL132 ku70Δ::KAN |
| YKHL134 | YKHL132 RAD1Δ::KAN |
| YKHL142 | JKM179 MAT::URA3::HO::12MH::200bp::12MH |
| YKHL143 | YKHL142 ku70Δ::KAN |
| YKHL144 | YKHL142 RAD1Δ::KAN |
| YKHL145 | JKM179 MAT::URA3::HO::12MH::20bp::12MH |
| YKHL146 | YKHL145 ku70Δ::KAN |
| YKHL147 | YKHL145 RAD1Δ::KAN |
| YKHL148 | JKM179 MAT::URA3::HO::12MH w/ mismatch gggtttataGGattatactg |
| YKHL149 | YKHL148 ku70Δ::KAN |
| YKHL151 | YKHL148 RAD1Δ::KAN |
| YJHJ1 | YKHL148 msh6Δ::KAN |
| YKHL152 | JKM179 MAT::URA3::HO::12MH w/ mismatch gggtttataaaattGGactg |
| YKHL153 | YKHL152 ku70Δ::KAN |
| YKHL154 | YKHL152 RAD1Δ::KAN |
| YJHJ2 | YKHL152 msh6Δ::KAN |
| YKHL155 | JKM179 MAT::URA3::HO::12MH w/ mismatch ggCAttataaaattatactg |
| YKHL156 | YKHL155 ku70Δ::KAN |
| YKHL157 | YKHL155 RAD1Δ::KAN |
| YJHJ3 | YKHL155 msh6Δ::KAN |
| YKHL158 | JKM179 MAT::URA3::HO::12MH w/ mismatch ggCtttataTaattaAactg |
| YKHL159 | YKHL158 ku70Δ::KAN |
| YKHL161 | YKHL158 RAD1Δ::KAN |
| YJHJ4 | YKHL158 msh6Δ::KAN |
| YKHL165 | JKM179 MAT::URA3::HO::12MH w/ mismatch gAgttAatTaaaGtatCctg |
| YKHL166 | YKHL165 ku70Δ::KAN |
| YKHL167 | YKHL165 RAD1Δ::KAN |
| YKHL179 | YKHL152 msh6Δ::KAN |
| YKHL182 | JKM179 MAT::URA3::12MH-1::12MH-2::HO::12MH-1::12MH-2 |
| YKHL183 | YKHL182 ku70Δ::KAN |
| YKHL184 | YKHL182 RAD1Δ::KAN |
| YKHL185 | JKM179 MAT::URA3::12MH::12MH::HO::12MH |
| YKHL186 | YKHL185 ku70Δ::KAN |
| YKHL187 | YKHL185 RAD1Δ::KAN |
| YKHL188 | JKM179 MAT::URA3::17MH::12MH::HO::17MH::12MH |
| YKHL189 | YKHL188 ku70Δ::KAN |
| YKHL201 | YKHL188 RAD1Δ::KAN |
| YKHL202 | JKM179 MAT::URA3::17MH::12MH::HO::17MH::12MH |
| YKHL203 | YKHL188 ku70Δ::KAN |
| YKHL204 | YKHL188 RAD1Δ::KAN |
| YKHL208 | JKM179 MATΔ Mup1::12MH-2::12MH-1::HO::12MH-2::12MH-1 |
| YKHL209 | JKM179 MATΔ Mup1::12MH-1::12MH-2::HO::12MH-1::12MH-2 |
| R072 | described at Haber and Thorburn (1984) |
| YKHL211 | R072 circular ChIII MAT::URA3::HO::12MH::20bp::12MH |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, K.; Ji, J.-H.; Yoon, K.; Che, J.; Seol, J.-H.; Lee, S.E.; Shim, E.Y. Microhomology Selection for Microhomology Mediated End Joining in Saccharomyces cerevisiae. Genes 2019, 10, 284. https://doi.org/10.3390/genes10040284
Lee K, Ji J-H, Yoon K, Che J, Seol J-H, Lee SE, Shim EY. Microhomology Selection for Microhomology Mediated End Joining in Saccharomyces cerevisiae. Genes. 2019; 10(4):284. https://doi.org/10.3390/genes10040284
Chicago/Turabian StyleLee, Kihoon, Jae-Hoon Ji, Kihoon Yoon, Jun Che, Ja-Hwan Seol, Sang Eun Lee, and Eun Yong Shim. 2019. "Microhomology Selection for Microhomology Mediated End Joining in Saccharomyces cerevisiae" Genes 10, no. 4: 284. https://doi.org/10.3390/genes10040284
APA StyleLee, K., Ji, J.-H., Yoon, K., Che, J., Seol, J.-H., Lee, S. E., & Shim, E. Y. (2019). Microhomology Selection for Microhomology Mediated End Joining in Saccharomyces cerevisiae. Genes, 10(4), 284. https://doi.org/10.3390/genes10040284





