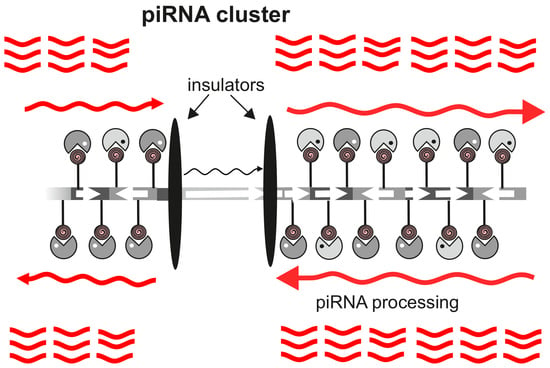
The Integrity of piRNA Clusters is Abolished by Insulators in the Drosophila Germline
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Transgenic Strains
2.2. Small RNA Library Preparation and Analysis
2.3. Chromatin Immunoprecipitation (ChIP)
2.4. RT-PCR
2.5. Motif Finding
3. Results and Discussion
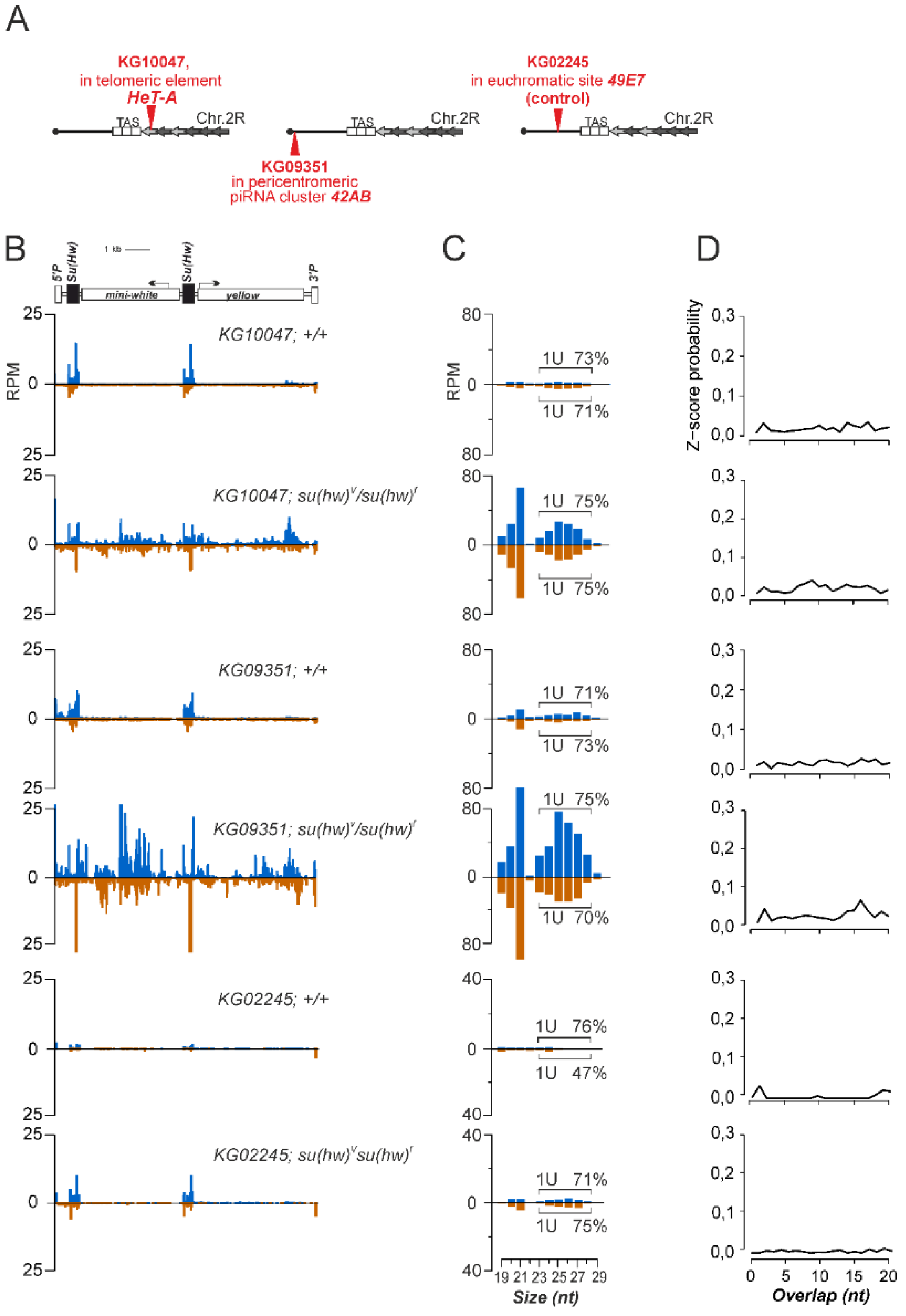
3.1. P{SUPor-P} Transgenic Constructs Inserted into piRNA Clusters Do Not Produce piRNAs
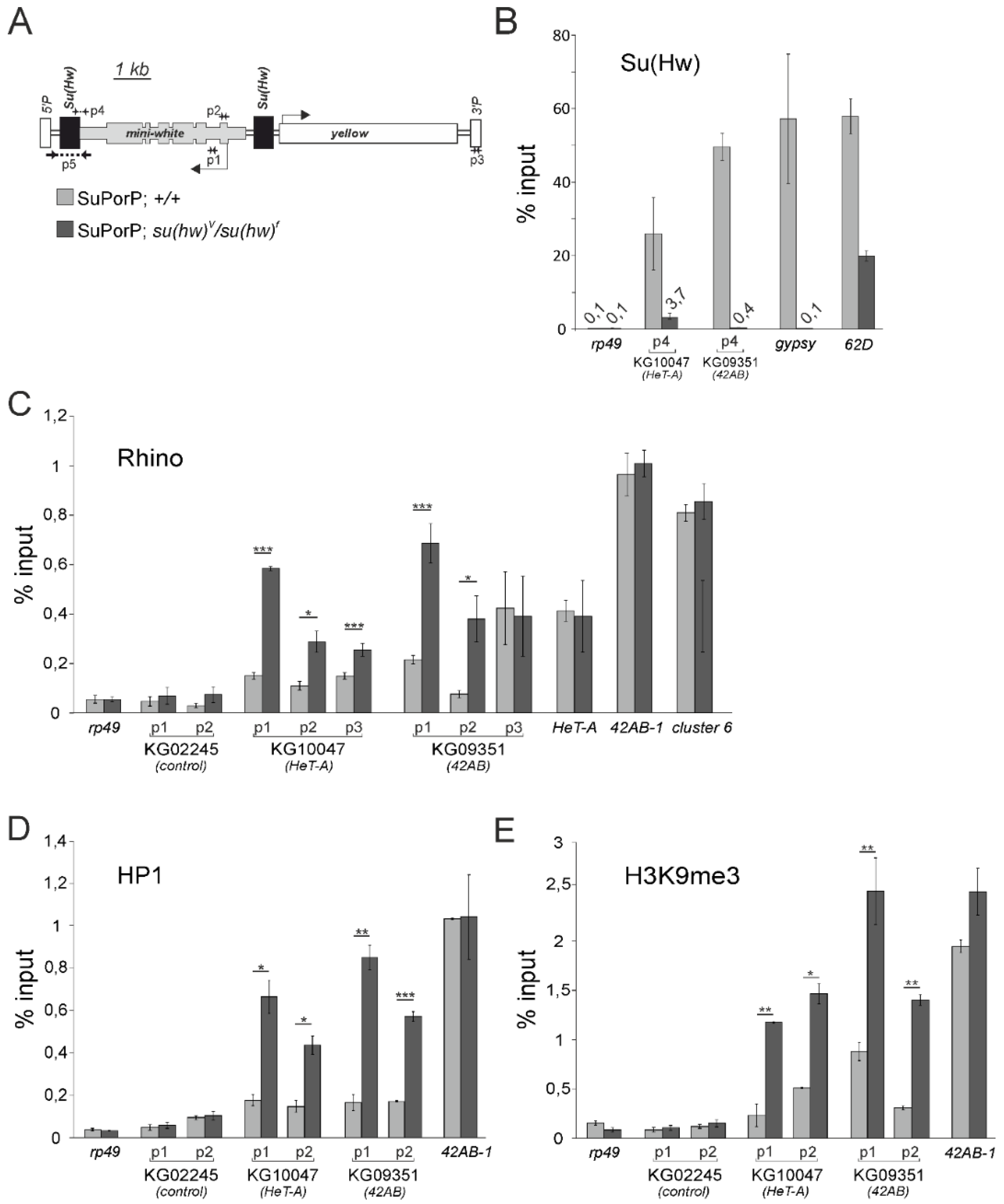
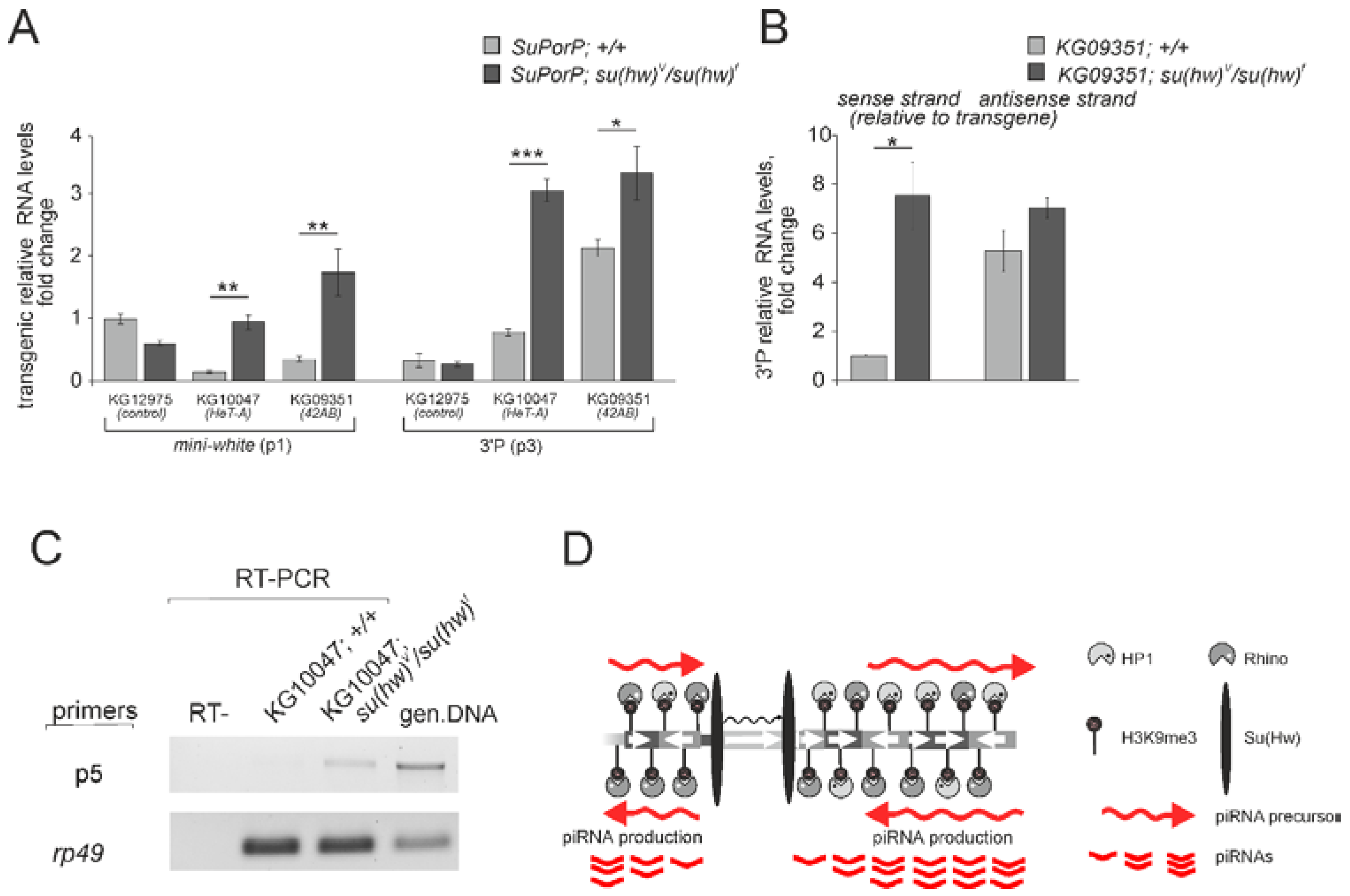
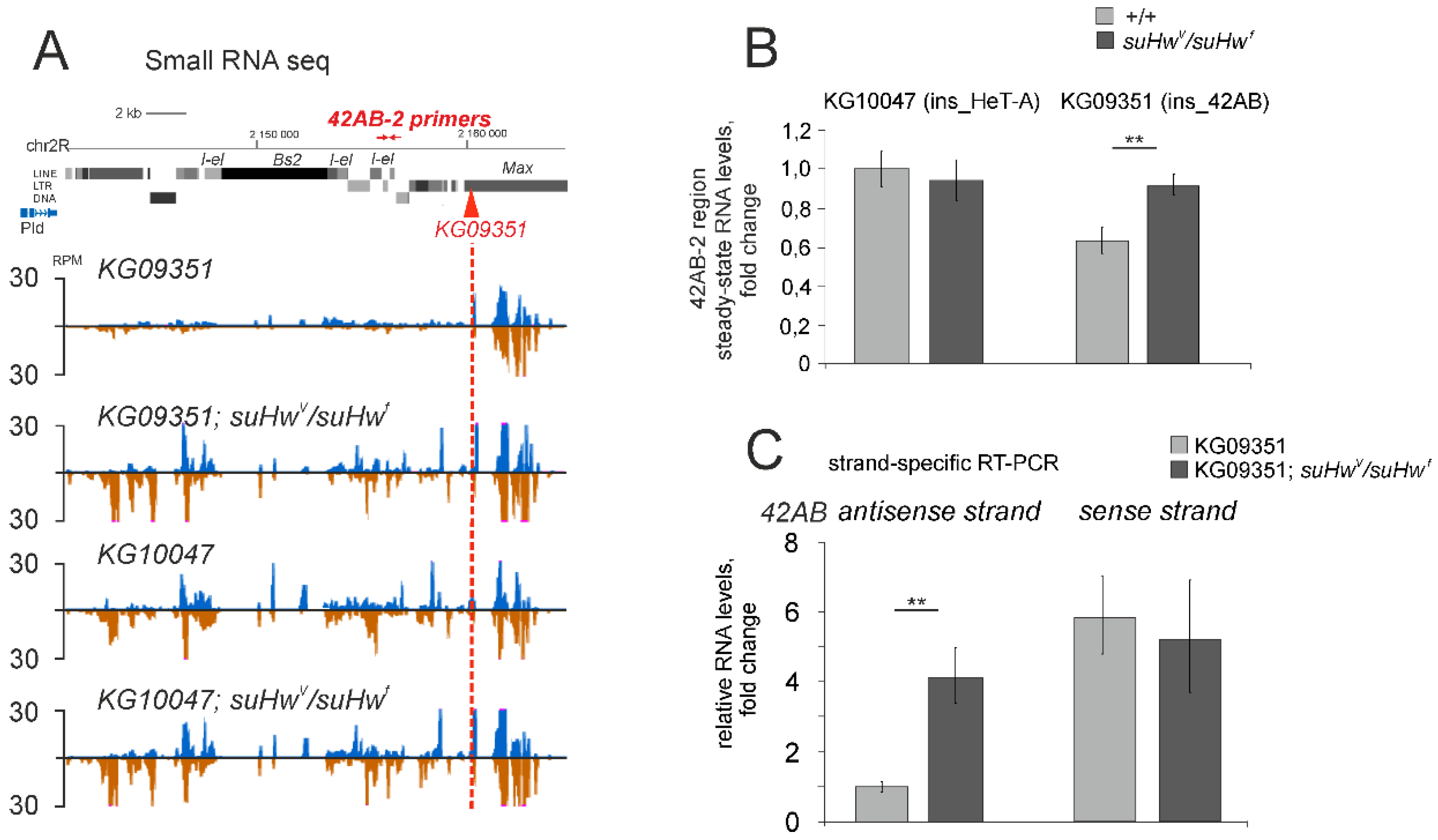
3.2. The Su(Hw) Complex Prevents the Assembly of the Chromatin Structure and Read-Through Transcription Typical of piRNA Clusters
3.3. Su(Hw) Restricts piRNA Production from Telomeric TART Retrotransposons
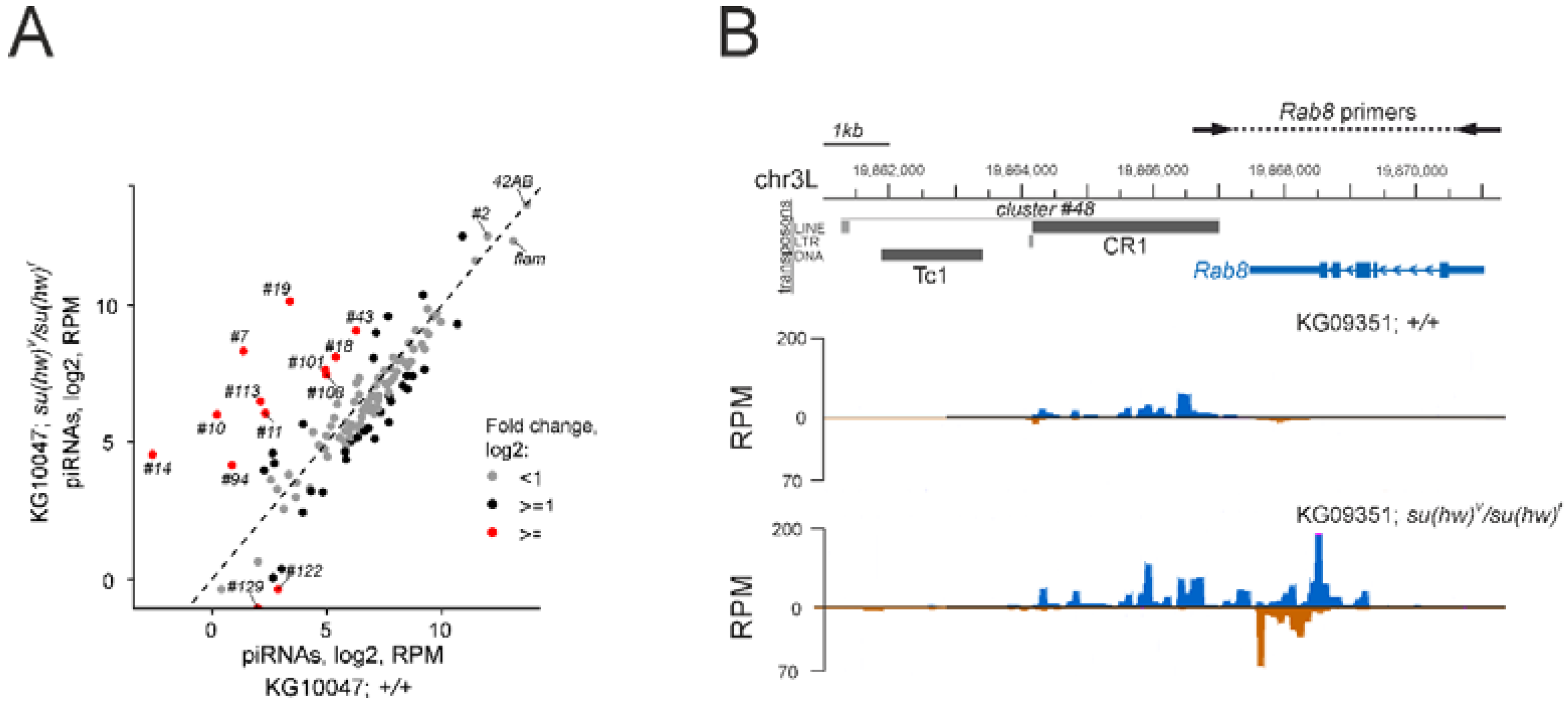
3.4. Su(Hw)-Binding Sites Are Depleted from Dual-Strand piRNA Clusters
3.5. The Su(Hw) Complex Protects Coding Genes from Spurious piRNA Production in the Germline
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Iwasaki, Y.W.; Siomi, M.C.; Siomi, H. PIWI-Interacting RNA: Its Biogenesis and Functions. Annu. Rev. Biochem. 2015, 84, 405–433. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 2014, 157, 1364–1379. [Google Scholar] [CrossRef] [PubMed]
- Pane, A.; Jiang, P.; Zhao, D.Y.; Singh, M.; Schupbach, T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. Embo J. 2011, 30, 4601–4615. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.R.; Tirian, L.; Vunjak, M.; Brennecke, J. A heterochromatin-dependent transcription machinery drives piRNA expression. Nature 2017, 549, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Akulenko, N.; Ryazansky, S.; Morgunova, V.; Komarov, P.A.; Olovnikov, I.; Vaury, C.; Jensen, S.; Kalmykova, A. Transcriptional and chromatin changes accompanying de novo formation of transgenic piRNA clusters. RNA 2018, 24, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Rangan, P.; Malone, C.D.; Navarro, C.; Newbold, S.P.; Hayes, P.S.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. piRNA Production Requires Heterochromatin Formation in Drosophila. Curr. Biol. 2011, 21, 1373–1379. [Google Scholar] [CrossRef] [PubMed]
- Akkouche, A.; Mugat, B.; Barckmann, B.; Varela-Chavez, C.; Li, B.; Raffel, R.; Pelisson, A.; Chambeyron, S. Piwi Is Required during Drosophila Embryogenesis to License Dual-Strand piRNA Clusters for Transposon Repression in Adult Ovaries. Mol. Cell 2017, 66, 411–419 e414. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.H.; Mattei, E.; Gainetdinov, I.; Colpan, C.; Weng, Z.; Zamore, P.D. Maelstrom Represses Canonical Polymerase II Transcription within Bi-directional piRNA Clusters in Drosophila melanogaster. Mol. Cell 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 2014, 157, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Stuwe, E.; Luo, Y.; Ninova, M.; Le Thomas, A.; Rozhavskaya, E.; Li, S.; Vempati, S.; Laver, J.D.; Patel, D.J.; et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol. Cell 2016, 63, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Hur, J.K.; Luo, Y.; Moon, S.; Ninova, M.; Marinov, G.K.; Chung, Y.D.; Aravin, A.A. Splicing-independent loading of TREX on nascent RNA is required for efficient expression of dual-strand piRNA clusters in Drosophila. Genes Dev. 2016, 30, 840–855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, J.; Xu, J.; Zhang, Z.; Koppetsch, B.S.; Schultz, N.; Vreven, T.; Meignin, C.; Davis, I.; Zamore, P.D.; et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell 2012, 151, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Khurana, J.S.; Wang, J.; Xu, J.; Koppetsch, B.S.; Thomson, T.C.; Nowosielska, A.; Li, C.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 2011, 147, 1551–1563. [Google Scholar] [CrossRef] [PubMed]
- Muerdter, F.; Olovnikov, I.; Molaro, A.; Rozhkov, N.V.; Czech, B.; Gordon, A.; Hannon, G.J.; Aravin, A.A. Production of artificial piRNAs in flies and mice. RNA 2012, 18, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Todeschini, A.L.; Teysset, L.; Delmarre, V.; Ronsseray, S. The epigenetic trans-silencing effect in Drosophila involves maternally-transmitted small RNAs whose production depends on the piRNA pathway and HP1. PLoS ONE 2010, 5, e11032. [Google Scholar] [CrossRef] [PubMed]
- Olovnikov, I.; Ryazansky, S.; Shpiz, S.; Lavrov, S.; Abramov, Y.; Vaury, C.; Jensen, S.; Kalmykova, A. De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res. 2013, 41, 5757–5768. [Google Scholar] [CrossRef] [PubMed]
- Kyrchanova, O.; Georgiev, P. Chromatin insulators and long-distance interactions in Drosophila. FEBS Lett. 2014, 588, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Chetverina, D.; Aoki, T.; Erokhin, M.; Georgiev, P.; Schedl, P. Making connections: Insulators organize eukaryotic chromosomes into independent cis-regulatory networks. Bioessays 2014, 36, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Vogelmann, J.; Valeri, A.; Guillou, E.; Cuvier, O.; Nollmann, M. Roles of chromatin insulator proteins in higher-order chromatin organization and transcription regulation. Nucleus 2011, 2, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Brasset, E.; Hermant, C.; Jensen, S.; Vaury, C. The Idefix enhancer-blocking insulator also harbors barrier activity. Gene 2010, 450, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Minervini, C.F.; Ruggieri, S.; Traversa, M.; D’Aiuto, L.; Marsano, R.M.; Leronni, D.; Centomani, I.; De Giovanni, C.; Viggiano, L. Evidences for insulator activity of the 5’UTR of the Drosophila melanogaster LTR-retrotransposon ZAM. Mol. Genet. Genom. 2010, 283, 503–509. [Google Scholar] [CrossRef]
- Geyer, P.K.; Corces, V.G. DNA position-specific repression of transcription by a Drosophila zinc finger protein. Genes Dev. 1992, 6, 1865–1873. [Google Scholar] [CrossRef] [PubMed]
- Gdula, D.A.; Gerasimova, T.I.; Corces, V.G. Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila. Proc. Natl. Acad. Sci. USA 1996, 93, 9378–9383. [Google Scholar] [CrossRef] [PubMed]
- Soshnev, A.A.; Baxley, R.M.; Manak, J.R.; Tan, K.; Geyer, P.K. The insulator protein Suppressor of Hairy-wing is an essential transcriptional repressor in the Drosophila ovary. Development 2013, 140, 3613–3623. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Prasad, S.; Semeshin, V.F.; Andreyeva, E.N.; Nguyen, Q.; Walter, M.F.; Mason, J.M. Two distinct domains in Drosophila melanogaster telomeres. Genetics 2005, 171, 1767–1777. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Radion, E.; Ryazansky, S.; Akulenko, N.; Rozovsky, Y.; Kwon, D.; Morgunova, V.; Olovnikov, I.; Kalmykova, A. Telomeric Retrotransposon HeT-A Contains a Bidirectional Promoter that Initiates Divergent Transcription of piRNA Precursors in Drosophila Germline. J. Mol. Biol. 2017, 429, 3280–3289. [Google Scholar] [CrossRef] [PubMed]
- Golovnin, A.; Volkov, I.; Georgiev, P. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J. Cell Sci. 2012, 125, 2064–2074. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Radion, E.; Morgunova, V.; Ryazansky, S.; Akulenko, N.; Lavrov, S.; Abramov, Y.; Komarov, P.A.; Glukhov, S.I.; Olovnikov, I.; Kalmykova, A. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin 2018, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Melnikova, L.; Kostyuchenko, M.; Parshikov, A.; Georgiev, P.; Golovnin, A. Role of Su(Hw) zinc finger 10 and interaction with CP190 and Mod(mdg4) proteins in recruiting the Su(Hw) complex to chromatin sites in Drosophila. PLoS ONE 2018, 13, e0193497. [Google Scholar] [CrossRef] [PubMed]
- Baxley, R.M.; Soshnev, A.A.; Koryakov, D.E.; Zhimulev, I.F.; Geyer, P.K. The role of the Suppressor of Hairy-wing insulator protein in Drosophila oogenesis. Dev. Biol. 2011, 356, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [PubMed]
- Czech, B.; Malone, C.D.; Zhou, R.; Stark, A.; Schlingeheyde, C.; Dus, M.; Perrimon, N.; Kellis, M.; Wohlschlegel, J.A.; Sachidanandam, R.; et al. An endogenous small interfering RNA pathway in Drosophila. Nature 2008, 453, 798–802. [Google Scholar] [CrossRef] [PubMed]
- De Vanssay, A.; Bouge, A.L.; Boivin, A.; Hermant, C.; Teysset, L.; Delmarre, V.; Antoniewski, C.; Ronsseray, S. Paramutation in Drosophila linked to emergence of a piRNA-producing locus. Nature 2012, 490, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Soshnev, A.A.; He, B.; Baxley, R.M.; Jiang, N.; Hart, C.M.; Tan, K.; Geyer, P.K. Genome-wide studies of the multi-zinc finger Drosophila Suppressor of Hairy-wing protein in the ovary. Nucleic Acids Res. 2012, 40, 5415–5431. [Google Scholar] [CrossRef] [PubMed]
- Georgiev, P.; Kozycina, M. Interaction between mutations in the suppressor of Hairy wing and modifier of mdg4 genes of Drosophila melanogaster affecting the phenotype of gypsy-induced mutations. Genetics 1996, 142, 425–436. [Google Scholar] [PubMed]
- Pai, C.Y.; Lei, E.P.; Ghosh, D.; Corces, V.G. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol. Cell 2004, 16, 737–748. [Google Scholar] [CrossRef] [PubMed]
- Ryazansky, S.; Radion, E.; Mironova, A.; Akulenko, N.; Abramov, Y.; Morgunova, V.; Kordyukova, M.Y.; Olovnikov, I.; Kalmykova, A. Natural variation of piRNA expression affects immunity to transposable elements. PLoS Genet. 2017, 13, e1006731. [Google Scholar] [CrossRef] [PubMed]
- Grentzinger, T.; Armenise, C.; Brun, C.; Mugat, B.; Serrano, V.; Pelisson, A.; Chambeyron, S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res. 2012, 22, 1877–1888. [Google Scholar] [CrossRef] [PubMed]
- Desset, S.; Meignin, C.; Dastugue, B.; Vaury, C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 2003, 164, 501–509. [Google Scholar] [PubMed]
- Sarot, E.; Payen-Groschene, G.; Bucheton, A.; Pelisson, A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 2004, 166, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Malone, C.D.; Aravin, A.A.; Sachidanandam, R.; Stark, A.; Hannon, G.J. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008, 322, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Shpiz, S.; Ryazansky, S.; Olovnikov, I.; Abramov, Y.; Kalmykova, A. Euchromatic transposon insertions trigger production of novel Pi- and endo-siRNAs at the target sites in the drosophila germline. PLoS Genet. 2014, 10, e1004138. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radion, E.; Sokolova, O.; Ryazansky, S.; Komarov, P.A.; Abramov, Y.; Kalmykova, A. The Integrity of piRNA Clusters is Abolished by Insulators in the Drosophila Germline. Genes 2019, 10, 209. https://doi.org/10.3390/genes10030209
Radion E, Sokolova O, Ryazansky S, Komarov PA, Abramov Y, Kalmykova A. The Integrity of piRNA Clusters is Abolished by Insulators in the Drosophila Germline. Genes. 2019; 10(3):209. https://doi.org/10.3390/genes10030209
Chicago/Turabian StyleRadion, Elizaveta, Olesya Sokolova, Sergei Ryazansky, Pavel A. Komarov, Yuri Abramov, and Alla Kalmykova. 2019. "The Integrity of piRNA Clusters is Abolished by Insulators in the Drosophila Germline" Genes 10, no. 3: 209. https://doi.org/10.3390/genes10030209
APA StyleRadion, E., Sokolova, O., Ryazansky, S., Komarov, P. A., Abramov, Y., & Kalmykova, A. (2019). The Integrity of piRNA Clusters is Abolished by Insulators in the Drosophila Germline. Genes, 10(3), 209. https://doi.org/10.3390/genes10030209