Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease
Abstract
1. Introduction
2. Eukaryotic m5C RNA Methyltransferases and Their Catalytic Mechanisms
3. Cellular Functions of m5C RNA Methyltransferases and the Modifications They Install
3.1. NSUN1 and NSUN5 Modify Cytoplasmic Ribosomal RNAs
3.2. Cytoplasmic Transfer RNAs are Methylated by NSUN2, NSUN6 and DNMT2
3.3. NSUN3 and NSUN4 Install m5Cs in Mitochondrial RNAs
3.4. m5C Marks in Messenger RNAs
3.5. Modification of Other RNA Species by m5C Methyltransferases
4. Substrate Recognition by m5C RNA Methyltransferases and Regulation of Their Activity
5. Roles of m5C RNA Methyltransferases in Development and Disease
6. Conclusions and Outlook
Author Contributions
Funding
Conflicts of Interest
References
- Boccaletto, P.; Machnicka, M.A.; Purta, E.; Piatkowski, P.; Baginski, B.; Wirecki, T.K.; de Crecy-Lagard, V.; Ross, R.; Limbach, P.A.; Kotter, A.; et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018, 46, D303–D307. [Google Scholar] [CrossRef]
- Breiling, A.; Lyko, F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015, 8, 24. [Google Scholar] [CrossRef]
- Trixl, L.; Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. Rev. RNA 2019, 10, e1510. [Google Scholar] [CrossRef]
- Lyko, F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Bourgeois, G.; Ney, M.; Gaspar, I.; Aigueperse, C.; Schaefer, M.; Kellner, S.; Helm, M.; Motorin, Y. Eukaryotic rRNA modification by yeast 5- methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLoS One 2015, 10, 1–16. [Google Scholar] [CrossRef]
- Brzezicha, B.; Schmidt, M.; Makałowska, I.; Jarmołowski, A.; Pieńkowska, J.; Szweykowska-Kulińska, Z. Identification of human tRNA: m5C methyltransferase catalysing intron-dependent m5C formation in the first position of the anticodon of the pre-tRNA(CAA)Leu. Nucleic Acids Res. 2006, 34, 6034–6043. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Tuorto, F.; Liebers, R.; Musch, T.; Schaefer, M.; Hofmann, S.; Kellner, S.; Frye, M.; Helm, M.; Stoecklin, G.; Lyko, F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012, 19, 900–905. [Google Scholar] [CrossRef]
- Hussain, S.; Sajini, A.A.; Blanco, S.; Dietmann, S.; Lombard, P.; Sugimoto, Y.; Paramor, M.; Gleeson, J.G.; Odom, D.T.; Ule, J.; et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013, 4, 255–261. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.-F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Tang, H.; Fan, X.; Xing, J.; Liu, Z.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSun2 delays replicative senescence by repressing p27 (KIP1) translation and elevating CDK1 translation. Aging (Albany. NY). 2015, 7, 1143–1158. [Google Scholar] [CrossRef]
- Xing, J.; Yi, J.; Cai, X.; Tang, H.; Liu, Z.; Zhang, X.; Martindale, J.L.; Yang, X.; Jiang, B.; Gorospe, M.; et al. NSun2 Promotes Cell Growth via Elevating Cyclin-Dependent Kinase 1 Translation. Mol. Cell. Biol. 2015, 35, 4043–4052. [Google Scholar] [CrossRef]
- Li, Q.; Li, X.; Tang, H.; Jiang, B.; Dou, Y.; Gorospe, M.; Wang, W. NSUN2-Mediated m5C Methylation and METTL3/METTL14-Mediated m6A Methylation Cooperatively Enhance p21 Translation. J. Cell. Biochem. 2017, 118, 2587–2598. [Google Scholar] [CrossRef]
- Van Haute, L.; Dietmann, S.; Kremer, L.; Hussain, S.; Pearce, S.F.; Powell, C.A.; Rorbach, J.; Lantaff, R.; Blanco, S.; Sauer, S.; et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016, 7, 12039. [Google Scholar] [CrossRef]
- Haag, S.; Sloan, K.E.; Ranjan, N.; Warda, A.S.; Kretschmer, J.; Blessing, C.; Hübner, B.; Seikowski, J.; Dennerlein, S.; Rehling, P.; et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. Embo J. 2016, 35, 2104–2119. [Google Scholar] [CrossRef]
- Nakano, S.; Suzuki, T.; Kawarada, L.; Iwata, H.; Asano, K.; Suzuki, T. NSUN3 methylase initiates 5-formylcytidine biogenesis in human mitochondrial tRNAMet. Nat. Chem. Biol. 2016, 12, 546–551. [Google Scholar] [CrossRef]
- Metodiev, M.D.; Spahr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.-G.; Ruzzenente, B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef]
- Schosserer, M.; Minois, N.; Angerer, T.B.; Amring, M.; Dellago, H.; Harreither, E.; Calle-Perez, A.; Pircher, A.; Gerstl, M.P.; Pfeifenberger, S.; et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat. Commun. 2015, 6, 6158. [Google Scholar] [CrossRef]
- Gigova, A.; Duggimpudi, S.; Pollex, T.; Pollex, T.I.M.; Schaefer, M. A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability A cluster of methylations in the domain IV of 25S rRNA is required for ribosome stability. RNA 2014, 20, 1632–1644. [Google Scholar] [CrossRef]
- Sharma, S.; Yang, J.; Watzinger, P.; Kötter, P.; Entian, K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013, 41, 9062–9076. [Google Scholar] [CrossRef]
- Liu, R.-J.; Long, T.; Li, J.; Li, H.; Wang, E.-D. Structural basis for substrate binding and catalytic mechanism of a human RNA:m5C methyltransferase NSun6. Nucleic Acids Res. 2017, 45, 6684–6697. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Warda, A.S.; Kretschmer, J.; Günnigmann, M.A.; Höbartner, C.; Bohnsack, M.T. NSUN6 is a human RNA methyltransferase that catalyzes formation of m5C72 in specific tRNAs. RNA 2015, 21, 1532–1543. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Li, S.; Balasubramaniyan, N.; Sancho, A.; Benko, S.; Zhang, F.; Vashisht, A.; Rengasamy, M.; Andino, B.; Chen, C.-H.; et al. Deposition of 5-Methylcytosine on Enhancer RNAs Enables the Coactivator Function of PGC-1alpha. Cell Rep. 2016, 14, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.-L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Reid, R.; Greene, P.J.; Santi, D.V. Exposition of a family of RNA m(5)C methyltransferases from searching genomic and proteomic sequences. Nucleic Acids Res. 1999, 27, 3138–3145. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Santi, D.V. m5C RNA and m5C DNA methyl transferases use different cysteine residues as catalysts. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 8263–8265. [Google Scholar] [CrossRef] [PubMed]
- King, M.Y.; Redman, K.L. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 2002, 41, 11218–11225. [Google Scholar] [CrossRef]
- King, M.; Ton, D.; Redman, K.L. A conserved motif in the yeast nucleolar protein Nop2p contains an essential cysteine residue. Biochem. J. 1999, 337, 29–35. [Google Scholar] [CrossRef]
- Redman, K.L. Assembly of protein-RNA complexes using natural RNA and mutant forms of an RNA cytosine methyltransferase. Biomacromolecules 2006, 7, 3321–3326. [Google Scholar] [CrossRef]
- Hussain, S.; Benavente, S.B.; Nascimento, E.; Dragoni, I.; Kurowski, A.; Gillich, A.; Humphreys, P.; Frye, M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 2009, 186, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Jeltsch, A. Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases. Chembiochem 2002, 3, 274–293. [Google Scholar] [CrossRef]
- Cheng, X. Structure and function of DNA methyltransferases. Annu. Rev. Biophys. Biomol. Struct. 1995, 24, 293–318. [Google Scholar] [CrossRef] [PubMed]
- Osterman, D.G.; DePillis, G.D.; Wu, J.C.; Matsuda, A.; Santi, D.V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry 1988, 27, 5204–5210. [Google Scholar] [CrossRef] [PubMed]
- Gabbara, S.; Bhagwat, A.S. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem. J. 1995, 307, 87–92. [Google Scholar] [CrossRef]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.-D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Hayrapetyan, A.; Grosjean, H.; Helm, M. Effect of a quaternary pentamine on RNA stabilization and enzymatic methylation. Biol. Chem. 2009, 390, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef]
- Sharma, S.; Lafontaine, D.L.J. “View From A Bridge”: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Taoka, M.; Nobe, Y.; Yamaki, Y.; Yamauchi, Y.; Ishikawa, H.; Takahashi, N.; Nakayama, H.; Isobe, T. The complete chemical structure of Saccharomyces cerevisiae rRNA: Partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res. 2016, 44, 8951–8961. [Google Scholar] [CrossRef]
- Hong, B.; Brockenbrough, J.S.; Wu, P.; Aris, J.P. Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol. 1997, 17, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Bohnsack, M.T.; Watkins, N.J. The 5S RNP Couples p53 Homeostasis to Ribosome Biogenesis and Nucleolar Stress. Cell Rep. 2013, 5, 237–247. [Google Scholar] [CrossRef]
- Meyer, B.; Wurm, J.P.; Kotter, P.; Leisegang, M.S.; Schilling, V.; Buchhaupt, M.; Held, M.; Bahr, U.; Karas, M.; Heckel, A.; et al. The Bowen-Conradi syndrome protein Nep1 (Emg1) has a dual role in eukaryotic ribosome biogenesis, as an essential assembly factor and in the methylation of Psi1191 in yeast 18S rRNA. Nucleic Acids Res. 2011, 39, 1526–1537. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.; Kretschmer, J.; Bohnsack, M.T. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 2015, 21, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, M.; Steringer, J.P.; Lyko, F. The Drosophila cytosine-5 methyltransferase Dnmt2 is associated with the nuclear matrix and can access DNA during mitosis. PLoS One 2008, 3, e1414. [Google Scholar] [CrossRef] [PubMed]
- Pais de Barros, J.P.; Keith, G.; El Adlouni, C.; Glasser, A.L.; Mack, G.; Dirheimer, G.; Desgres, J. 2’-O-methyl-5-formylcytidine (f5Cm), a new modified nucleotide at the “wobble” of two cytoplasmic tRNAs Leu (NAA) from bovine liver. Nucleic Acids Res. 1996, 24, 1489–1496. [Google Scholar] [CrossRef]
- Kawarada, L.; Suzuki, T.; Ohira, T.; Hirata, S.; Miyauchi, K.; Suzuki, T. ALKBH1 is an RNA dioxygenase responsible for cytoplasmic and mitochondrial tRNA modifications. Nucleic Acids Res. 2017, 45, 7401–7415. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.T.Y.; Pang, Y.L.J.; Deng, W.; Babu, I.R.; Dyavaiah, M.; Begley, T.J.; Dedon, P.C. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat. Commun. 2012, 3, 937. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.; Fierer, J.; Kaiser, S.; Helm, M.; Jurkowski, T.P.; Jeltsch, A. Cytosine methylation of tRNA-Asp by DNMT2 has a role in translation of proteins containing poly-Asp sequences. Cell Discov. 2015, 1, 15010. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Herbst, F.; Alerasool, N.; Bender, S.; Popp, O.; Federico, G.; Reitter, S.; Liebers, R.; Stoecklin, G.; Grone, H.-J.; et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015, 34, 2350–2362. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.; Hartmann, M.; Schuster, I.; Bender, S.; Thuring, K.L.; Helm, M.; Katze, J.R.; Nellen, W.; Lyko, F.; Ehrenhofer-Murray, A.E. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res. 2015, 43, 10952–10962. [Google Scholar] [CrossRef] [PubMed]
- Tuorto, F.; Legrand, C.; Cirzi, C.; Federico, G.; Liebers, R.; Muller, M.; Ehrenhofer-Murray, A.E.; Dittmar, G.; Grone, H.-J.; Lyko, F. Queuosine-modified tRNAs confer nutritional control of protein translation. EMBO J. 2018, 37, e99777. [Google Scholar] [CrossRef]
- Ehrenhofer-Murray, A.E. Cross-Talk between Dnmt2-Dependent tRNA Methylation and Queuosine Modification. Biomolecules 2017, 7, 14. [Google Scholar] [CrossRef]
- Levitt, M. Detailed molecular model for transfer ribonucleic acid. Nature 1969, 224, 759–763. [Google Scholar] [CrossRef]
- Vare, V.Y.P.; Eruysal, E.R.; Narendran, A.; Sarachan, K.L.; Agris, P.F. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules 2017, 7, 29. [Google Scholar] [CrossRef]
- Li, J.; Li, H.; Long, T.; Dong, H.; Wang, E.-D.; Liu, R.-J. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
- Cámara, Y.; Asin-Cayuela, J.; Park, C.B.; Metodiev, M.D.; Shi, Y.; Ruzzenente, B.; Kukat, C.; Habermann, B.; Wibom, R.; Hultenby, K.; et al. MTERF4 regulates translation by targeting the methyltransferase NSUN4 to the mammalian mitochondrial ribosome. Cell Metab. 2011, 13, 527–539. [Google Scholar] [CrossRef]
- Dudek, J.; Rehling, P.; van der Laan, M. Mitochondrial protein import: common principles and physiological networks. Biochim. Biophys. Acta 2013, 1833, 274–285. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Hobartner, C.; Bohnsack, M.T. How RNA modification allows non-conventional decoding in mitochondria. Cell Cycle 2017, 16, 145–146. [Google Scholar] [CrossRef]
- Bilbille, Y.; Gustilo, E.M.; Harris, K.A.; Jones, C.N.; Lusic, H.; Kaiser, R.J.; Delaney, M.O.; Spremulli, L.L.; Deiters, A.; Agris, P.F. The human mitochondrial tRNAMet: Structure/function relationship of a unique modification in the decoding of unconventional codons. J. Mol. Biol. 2011, 406, 257–274. [Google Scholar] [CrossRef]
- Cantara, W.A.; Murphy, F.V.; Demirci, H.; Agris, P.F. Expanded use of sense codons is regulated by modified cytidines in tRNA. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 10964–10969. [Google Scholar] [CrossRef] [PubMed]
- Carlile, T.M.; Rojas-Duran, M.F.; Zinshteyn, B.; Shin, H.; Bartoli, K.M.; Gilbert, W.V. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014, 515, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Erlacher, M.; Rossmanith, W.; Stern-Ginossar, N.; Schwartz, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Taylor, R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, X.-Y.; Micura, R.; Lusser, A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017, 18, 1. [Google Scholar] [CrossRef]
- Legrand, C.; Tuorto, F.; Hartmann, M.; Liebers, R.; Jacob, D.; Helm, M.; Lyko, F. Statistically robust methylation calling for whole-transcriptome bisulfite sequencing reveals distinct methylation patterns for mouse RNAs. Genome Res. 2017, 27, 1589–1596. [Google Scholar] [CrossRef]
- David, R.; Burgess, A.; Parker, B.; Li, J.; Pulsford, K.; Sibbritt, T.; Preiss, T.; Searle, I.R. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell 2017, 29, 445–460. [Google Scholar] [CrossRef]
- Edelheit, S.; Schwartz, S.; Mumbach, M.R.; Wurtzel, O.; Sorek, R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet. 2013, 9, e1003602. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarian, H.; Wagner, N.; Polo, B.; Baudouy, D.; Kiani, J.; Michiels, J.-F.; Cuzin, F.; Rassoulzadegan, M.; Wagner, K.-D. Dnmt2/Trdmt1 as Mediator of RNA Polymerase II Transcriptional Activity in Cardiac Growth. PLoS One 2016, 11, e0156953. [Google Scholar] [CrossRef] [PubMed]
- Long, T.; Li, J.; Li, H.; Zhou, M.; Zhou, X.-L.; Liu, R.-J.; Wang, E.-D. Sequence-specific and Shape-selective RNA Recognition by the Human RNA 5-Methylcytosine Methyltransferase NSun6. J. Biol. Chem. 2016, 291, 24293–24303. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Windhof, I.M.; Maximov, V.; Jurkowski, T.; Jeltsch, A.; Forstner, K.U.; Sharma, C.M.; Graf, R.; Nellen, W. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res. 2013, 41, 8615–8627. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, R.; Aklujkar, M.; Schafer, M.; Reinhardt, R.; Nickel, O.; Reuter, G.; Lovley, D.R.; Ehrenhofer-Murray, A.; Nellen, W.; Ankri, S.; et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res. 2014, 42, 6487–6496. [Google Scholar] [CrossRef] [PubMed]
- Spahr, H.; Habermann, B.; Gustafsson, C.M.; Larsson, N.-G.; Hallberg, B.M. Structure of the human MTERF4-NSUN4 protein complex that regulates mitochondrial ribosome biogenesis. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 15253–15258. [Google Scholar] [CrossRef] [PubMed]
- Yakubovskaya, E.; Guja, K.E.; Mejia, E.; Castano, S.; Hambardjieva, E.; Choi, W.S.; Garcia-Diaz, M. Structure of the essential MTERF4:NSUN4 protein complex reveals how an MTERF protein collaborates to facilitate rRNA modification. Structure 2012, 20, 1940–1947. [Google Scholar] [CrossRef] [PubMed]
- Blanco, S.; Frye, M. Role of RNA methyltransferases in tissue renewal and pathology. Curr. Opin. Cell Biol. 2014, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Rafiq, M.A.; Noor, A.; Hussain, S.; Flores, J.V.; Rupp, V.; Vincent, A.K.; Malli, R.; Ali, G.; Khan, F.S.; et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2012, 90, 856–863. [Google Scholar] [CrossRef]
- Martinez, F.J.; Lee, J.H.; Lee, J.E.; Blanco, S.; Nickerson, E.; Gabriel, S.; Frye, M.; Al-Gazali, L.; Gleeson, J.G. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J. Med. Genet. 2012, 49, 380–385. [Google Scholar] [CrossRef]
- Flores, J.V.; Cordero-Espinoza, L.; Oeztuerk-Winder, F.; Andersson-Rolf, A.; Selmi, T.; Blanco, S.; Tailor, J.; Dietmann, S.; Frye, M. Cytosine-5 RNA Methylation Regulates Neural Stem Cell Differentiation and Motility. Stem cell reports 2017, 8, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Trixl, L.; Amort, T.; Wille, A.; Zinni, M.; Ebner, S.; Hechenberger, C.; Eichin, F.; Gabriel, H.; Schoberleitner, I.; Huang, A.; et al. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell. Mol. Life Sci. 2018, 75, 1483–1497. [Google Scholar] [CrossRef] [PubMed]
- Chi, L.; Delgado-Olguin, P. Expression of NOL1/NOP2/sun domain (Nsun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr. Patterns 2013, 13, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.; Marquez, B.; Suarez, S.; Schimenti, J. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the Sun domain-containing family of putative RNA methyltransferases. Biol. Reprod. 2007, 77, 376–382. [Google Scholar] [CrossRef]
- Khosronezhad, N.; Colagar, A.H.; Jorsarayi, S.G.A. T26248G-transversion mutation in exon7 of the putative methyltransferase Nsun7 gene causes a change in protein folding associated with reduced sperm motility in asthenospermic men. Reprod. Fertil. Dev. 2015, 27, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Khosronezhad, N.; Hosseinzadeh Colagar, A.; Mortazavi, S.M. The Nsun7 (A11337)-deletion mutation, causes reduction of its protein rate and associated with sperm motility defect in infertile men. J. Assist. Reprod. Genet. 2015, 32, 807–815. [Google Scholar] [CrossRef] [PubMed]
- Doll, A.; Grzeschik, K.H. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 2001, 95, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Wang, F.; Ngoc, L.V.; Collignon, E.; Bonvin, E.; Deplus, R.; Calonne, E.; Hassabi, B.; Putmans, P.; Awe, S.; et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016, 351, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Tardu, M.; Lin, Q.; Koutmou, K.S. N4-acetylcytidine and 5-formylcytidine are present in Saccharomyces cerevisiae mRNAs. bioRxiv 2018. [Google Scholar]
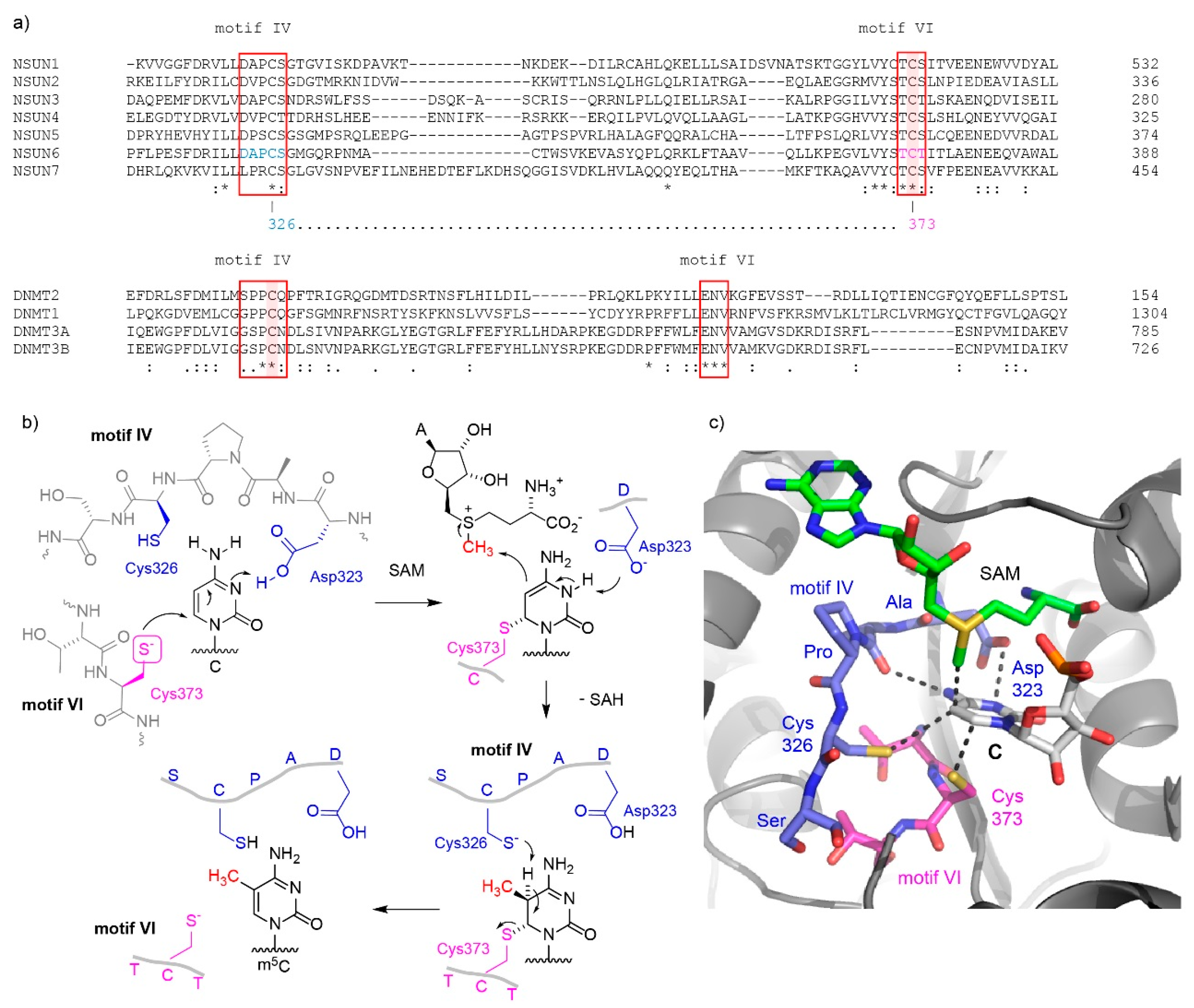
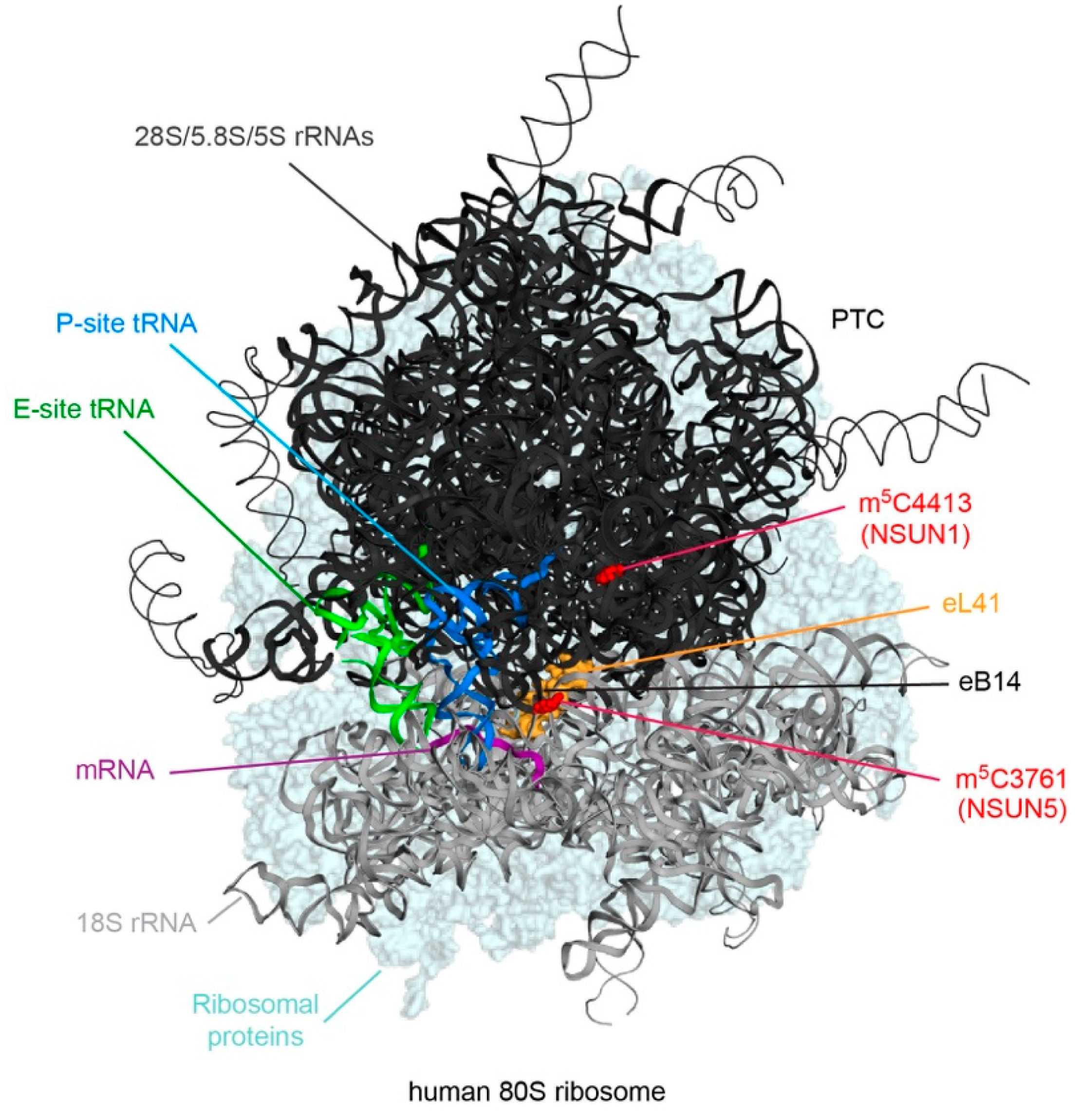
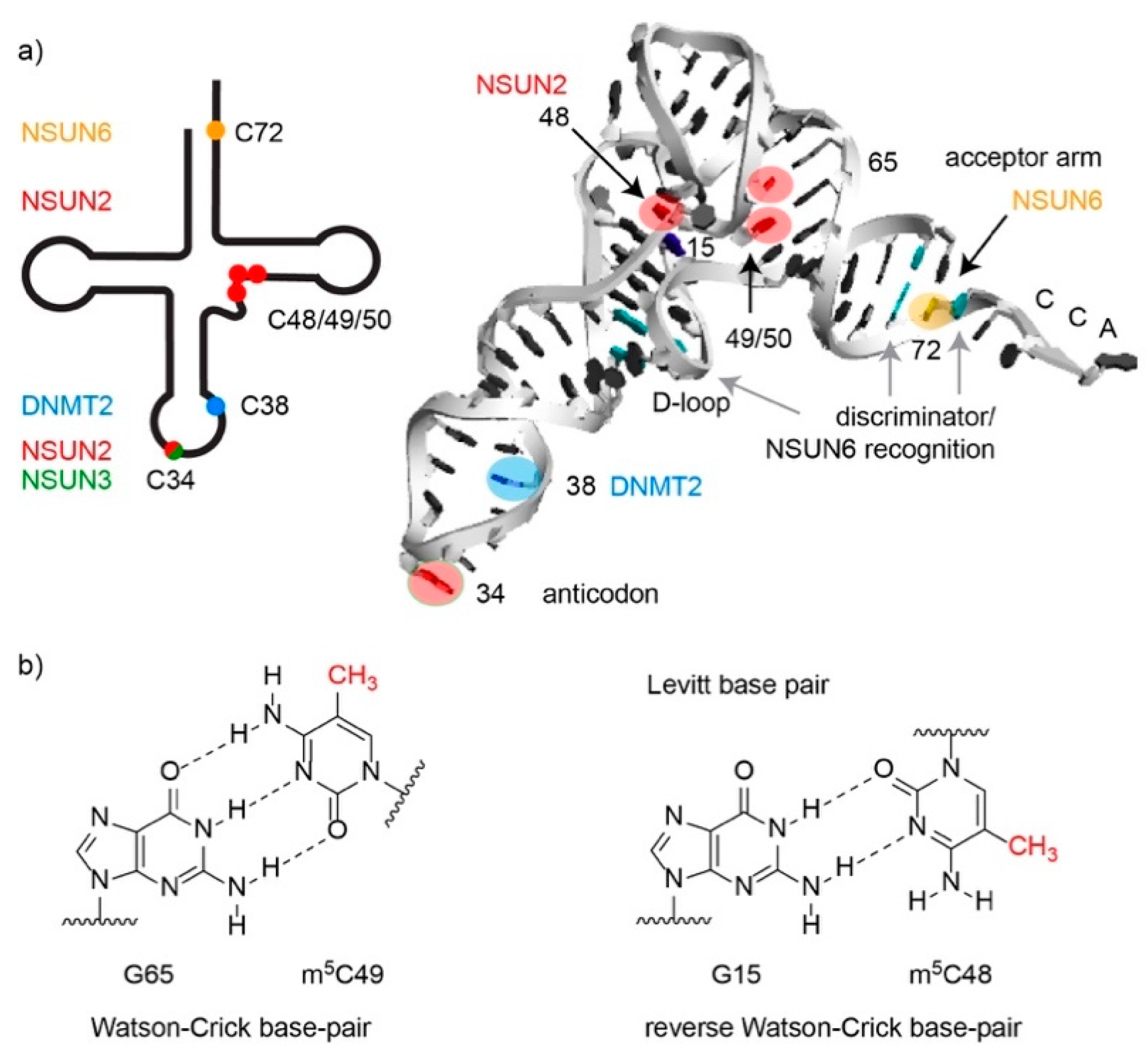
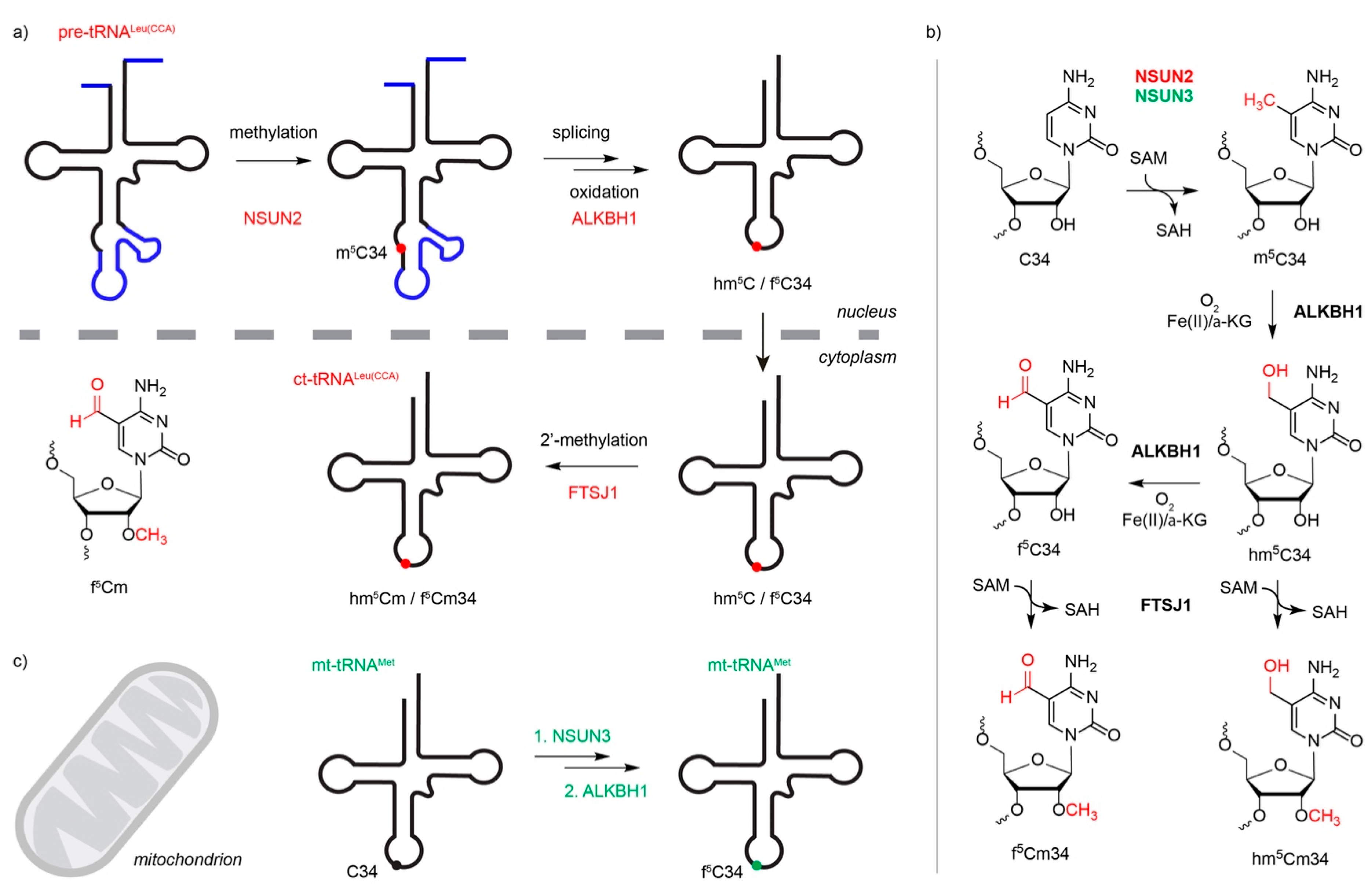
| Methyl- transferase | Subcellular localization | Target RNA(s) | Modification installed | Ref. |
|---|---|---|---|---|
| NSUN1 | Nucleolus | 28S rRNA | m5C4413 | [5] |
| NSUN2 | Nucleus/Nucleolus | Pre-tRNALeu(CAA) | m5C34 | [6] |
| tRNAAla(AGC/CGC/UGC)/His(GUG)/Ile(AAU)/ Leu(CAA/AAG/CAG/UAA/UAG)/Lys(CUU)/ Met(CAU)/Ser(AGA/CGA/GCU/UGA)/Thr(CGT/UGU)/Tyr(GUA) | m5C48 | [7,8] | ||
| tRNAAsp(GUC)/Gln(CUG/UUG)/Lys(UUU)/Phe(GAA)/ Thr(AGU)/Val(AAC/CAC/UAC) | m5C48, 49 | [7,8] | ||
| tRNAGlu(CUC/UUC)/Gly(CCC/GCC/UCC)/Pro(AGG/CGG/UGG) | m5C48, 49, 50 | [7,8] | ||
| vtRNA1.1 | m5C69 | [9] | ||
| vtRNA1.2 | m5C27, 591 | [9] | ||
| vtRNA1.3 | m5C15, 27, 59 | [9] | ||
| mRNA | various | [10,11,12,13] | ||
| NSUN3 | Mitochondria | mt-tRNAMet | m5C34 | [14,15,16] |
| NSUN4 | Mitochondria | mt-12S rRNA | m5C8412 | [17] |
| NSUN5 | Nucleolus | 28S rRNA | m5C3761 | [18,19,20] |
| NSUN6 | Cytoplasm/Golgi | tRNACys/Thr | m5C72 | [21,22] |
| NSUN7 | Nucleus | eRNA (Pfk1/Sirt5/Hmox2/Idh3b) | various | [23] |
| DNMT2 | Cytoplasm/Nucleus | tRNAAsp(GUC)/ Gly(GCC)/ Val(AAC) | m5C38 | [24,25] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohnsack, K.E.; Höbartner, C.; Bohnsack, M.T. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes 2019, 10, 102. https://doi.org/10.3390/genes10020102
Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes. 2019; 10(2):102. https://doi.org/10.3390/genes10020102
Chicago/Turabian StyleBohnsack, Katherine E., Claudia Höbartner, and Markus T. Bohnsack. 2019. "Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease" Genes 10, no. 2: 102. https://doi.org/10.3390/genes10020102
APA StyleBohnsack, K. E., Höbartner, C., & Bohnsack, M. T. (2019). Eukaryotic 5-methylcytosine (m5C) RNA Methyltransferases: Mechanisms, Cellular Functions, and Links to Disease. Genes, 10(2), 102. https://doi.org/10.3390/genes10020102





