Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Seed-Flooding Tolerance Evaluation
2.3. Phenotypic Data Analysis
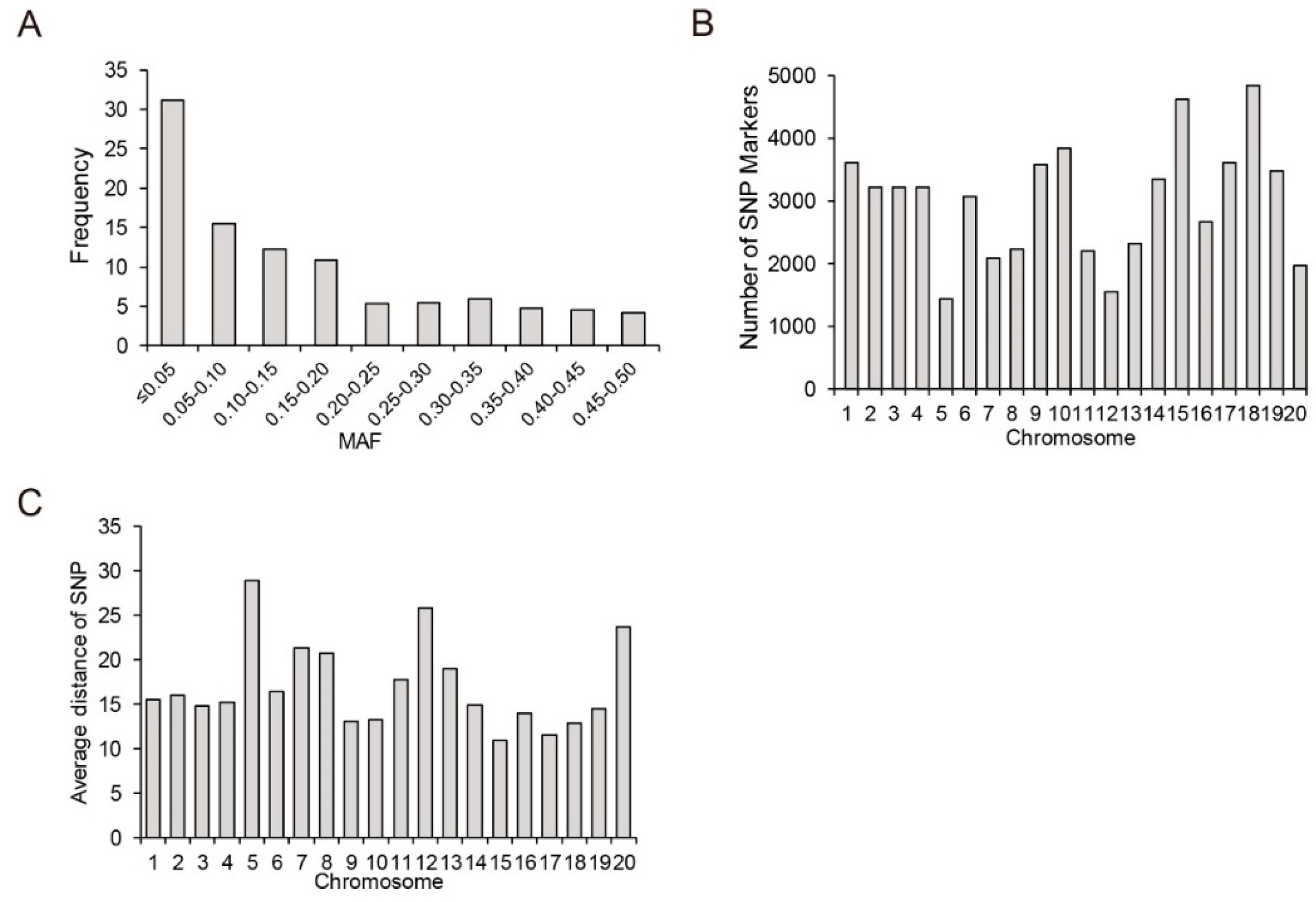
2.4. SNP Data Analysis
2.5. Population Genetic Analysis
2.6. Genome-Wide Association Analysis
2.7. Candidate Gene Predictions and Expression Analysis
2.8. Sequence Analysis of Candidate Genes
2.9. Plasmid Construction and Subcellular Localization
3. Results
3.1. Determination of Optimum Seed-Flooding Treatment Duration
3.2. Phenotypic Evaluation
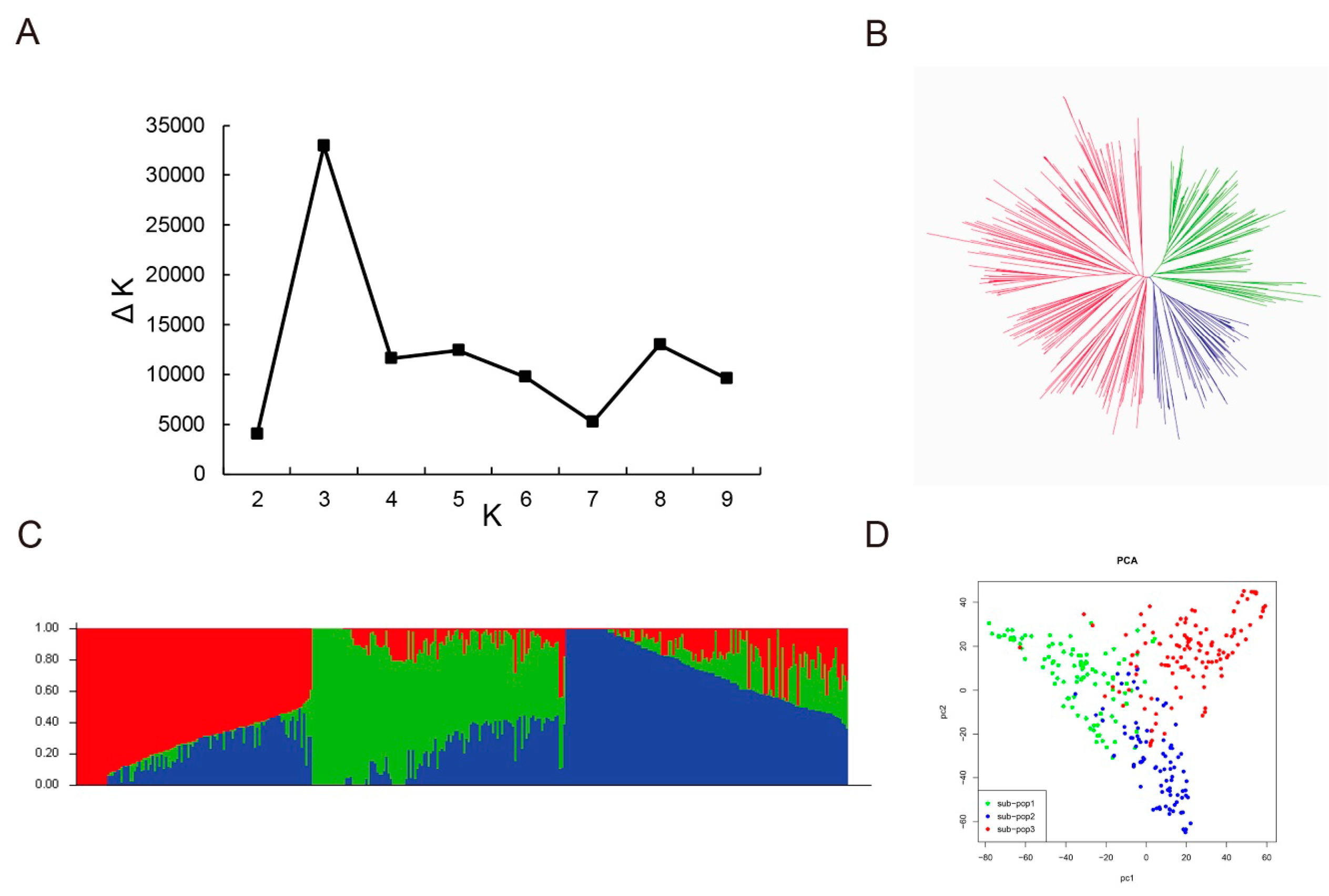
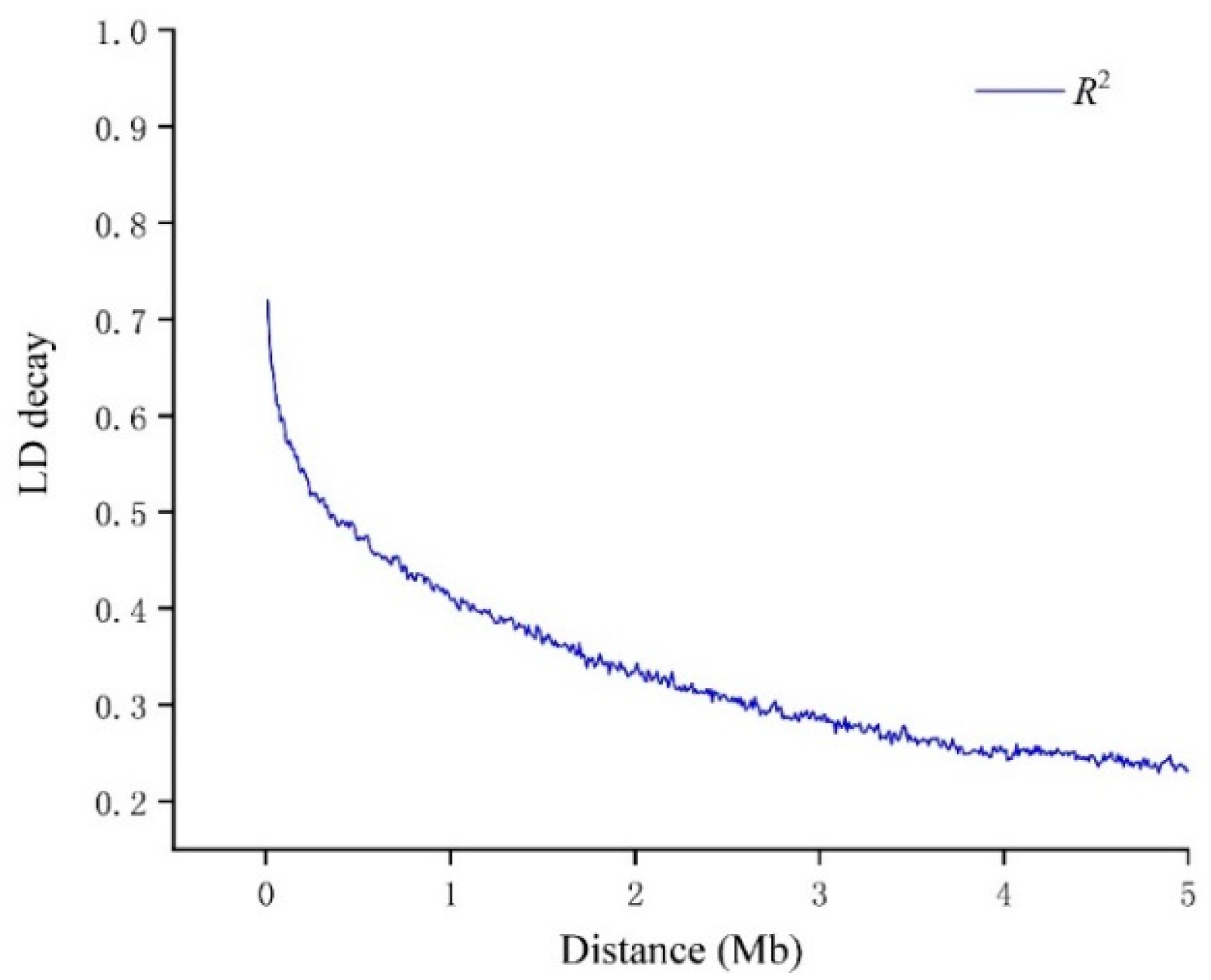
3.3. Genetic Diversity, Population Structure and Linkage Disequilibrium Analysis
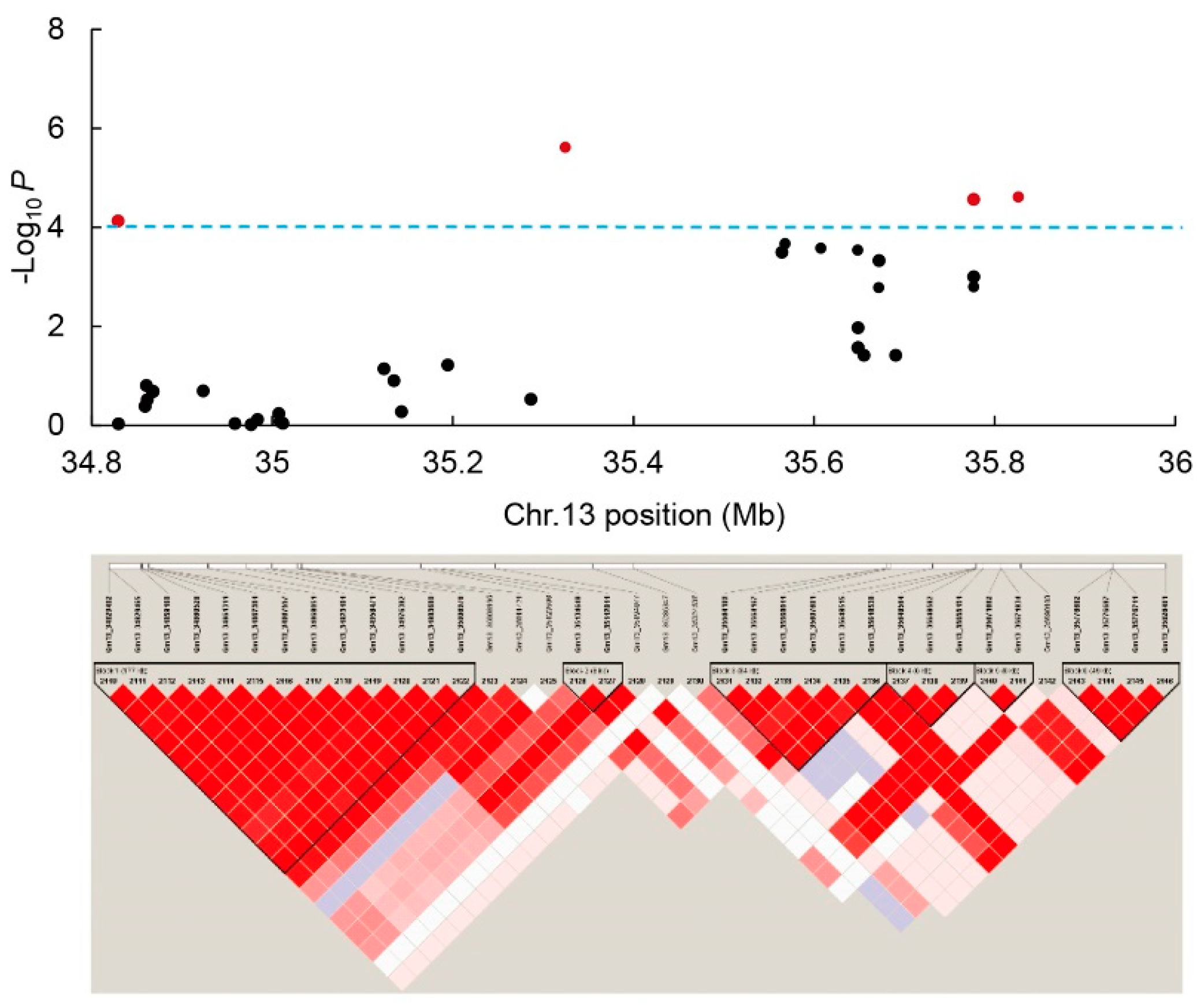
3.4. GWAS Analysis via MLM
3.5. GWAS Analysis via mrMLM, and Comparative Analysis of MLM and mrMLM Results
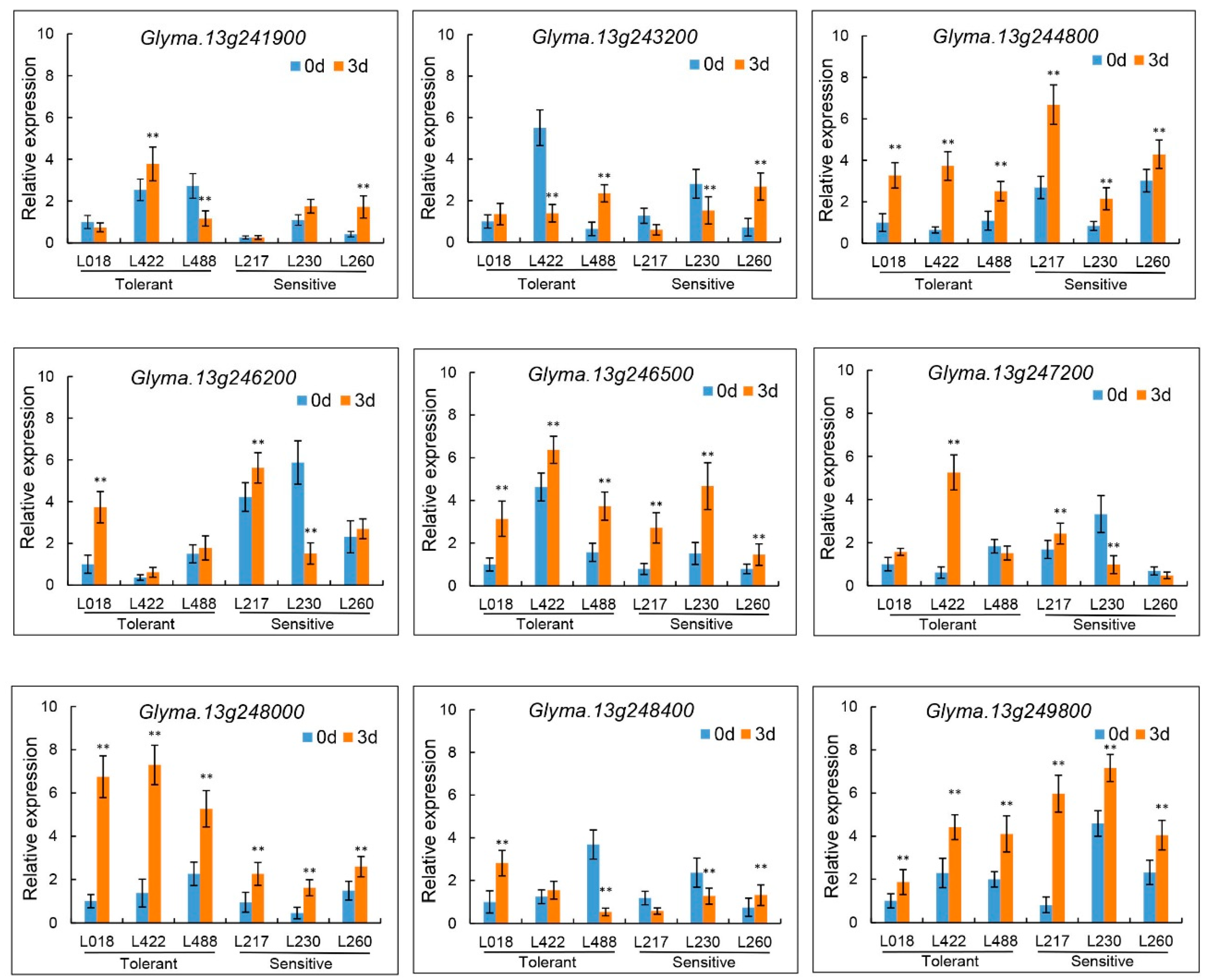
3.6. Candidate Gene Prediction and qRT-PCR Analysis
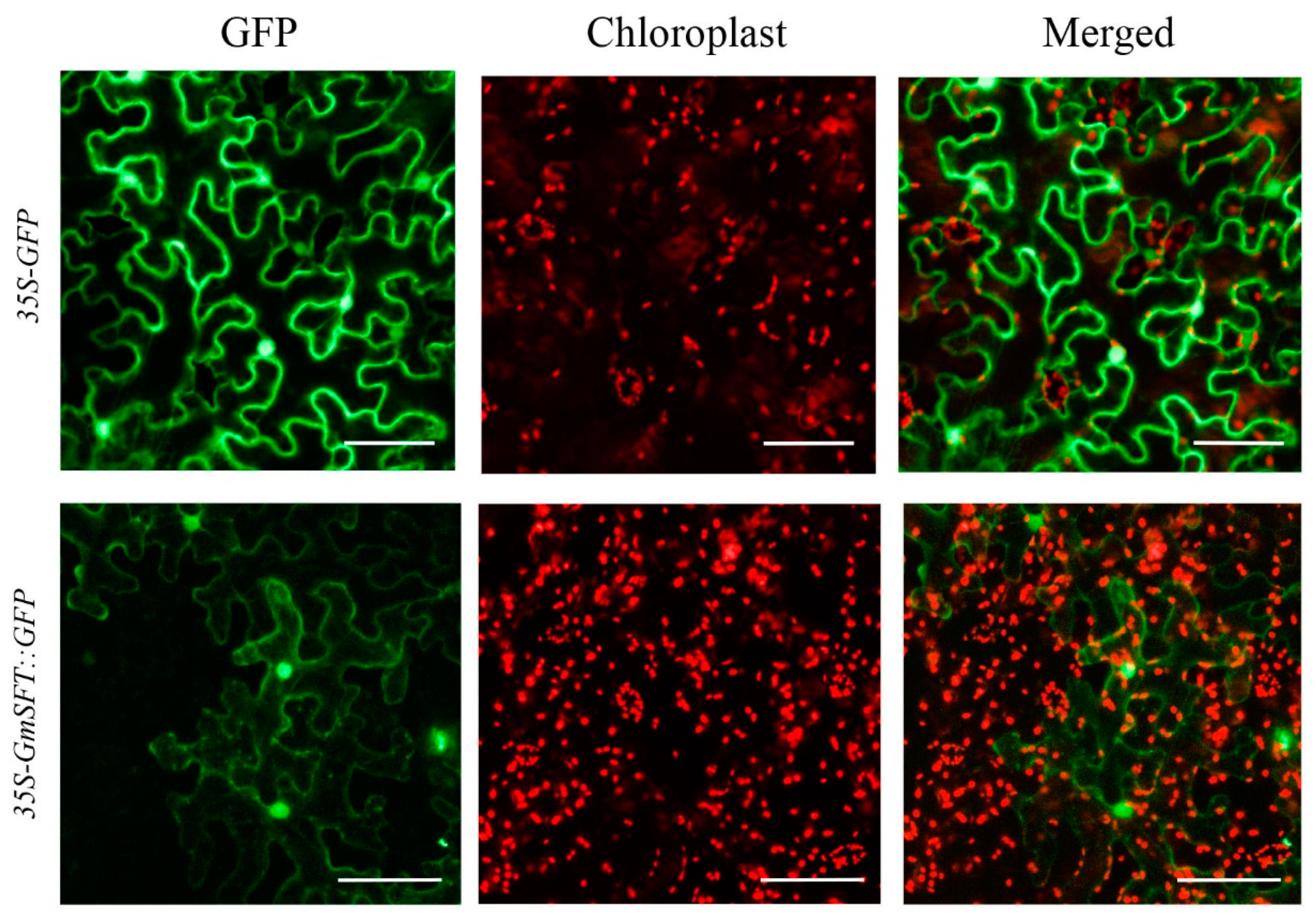
3.7. Sequence Analysis and Subcellular Localization of GmSFT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bailey-Serres, J.; Fukao, T.; Gibbs, D.J.; Holdsworth, M.J.; Lee, S.C.; Licausi, F.; Perata, P.; Voesenek, L.A.; van Dongen, J.T. Making sense of low oxygen sensing. Trends Plant Sci. 2012, 17, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Broughton, S.; Zhou, G.; Teakle, N.; Matsuda, R.; Zhou, M.; O’Leary, R.; Colmer, T.; Li, C. Waterlogging tolerance is associated with root porosity in barley (Hordeum vulgare L.). Mol. Breed. 2015, 35, 27. [Google Scholar] [CrossRef]
- Dong, H. Physiological and molecular adjustment of cotton to waterlogging atpeak-flowering in relation to growth and yield. Field Crop Res. 2015, 179, 164–172. [Google Scholar]
- Yin, D.; Chen, S.; Chen, F.; Guan, Z.; Fang, W. Morphological and physiological responses of two chrysanthemum cultivars differing in their tolerance to waterlogging. Env. Exp. Bot. 2009, 67, 87–93. [Google Scholar] [CrossRef]
- Hou, F.F.; Thseng, F.S. Studies on the flooding tolerance of soybean seed: Varietal differences. Euphytica 1991, 57, 169–173. [Google Scholar] [CrossRef]
- Oosterhuis, D.; Scott, H.D.; Hampton, R.E.; Wullschleger, S.D. Physiological responses of two soybean [Glycine max (L.) Merr] cultivars to short-term flooding. Env. Exp. Bot. 1990, 30, 85–92. [Google Scholar] [CrossRef]
- Mancuso, S.; Shabala, S. Waterlogging Signalling and Tolerance in Plants; Springer: Berlin/Heidelberg, Germany, 2010; pp. 1–294. [Google Scholar]
- VanToai, T.T.; St Martin, S.K.; Chase, K.; Boru, G.; Schnipke, V.; Schmitthenner, A.F.; Lark, K.G. Identification of a QTL associated with tolerance of soybean to soil waterlogging. Crop Sci. 2001, 41, 1247–1252. [Google Scholar] [CrossRef]
- Cornelious, B.; Chen, P.; Chen, Y.; de Leon, N.; Shannon, J.G.; Wang, D. Identification of QTLs underlying water-logging tolerance in soybean. Mol. Breed. 2005, 16, 103–112. [Google Scholar] [CrossRef]
- Githiri, S.M.; Watanabe, S.; Harada, K.; Takahashi, R. QTL analysis of flooding tolerance in soybean at an early vegetative growth stage. Plant Breed. 2006, 125, 613–618. [Google Scholar] [CrossRef]
- Nguyen, V.T.; Vuong, T.D.; VanToai, T.; Lee, J.D.; Wu, X.; Mian, M.A.; Dorrance, A.E.; Shannon, J.G.; Nguyen, H.T. Mapping of quantitative trait loci associated with resistance to and flooding tolerance in soybean. Crop Sci. 2012, 52, 2481. [Google Scholar] [CrossRef]
- Sayama, T.; Nakazaki, T.; Ishikawa, G.; Yagasaki, K.; Yamada, N.; Hirota, N.; Hirata, K.; Yoshikawa, T.; Saito, H.; Teraishi, M. QTL analysis of seed-flooding tolerance in soybean (Glycine max [L.] Merr.). Plant Sci. 2009, 176, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Kraakman, A.T.W.; Niks, R.E.; Van den Berg, P.M.M.M.; Stam, P.; Van Eeuwijk, F.A. Linkage disequilibrium mapping of yield and yield stability in modern spring barley cultivars. Genetics 2004, 168, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Ersoz, E.S.; Yu, J.; Buckler, E.S. Applications of linkage disequilibrium and association mapping in crop plants. In Genomics-Assisted Crop Improvement: Vol. 1: Genomics Approaches and Platforms; Varshney, R.K., Tuberosa, R., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 97–119. [Google Scholar]
- Brachi, B.; Morris, G.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef] [PubMed]
- Hyten, D.; Song, Q.; Choi, I.Y.; Yoon, M.S.; Specht, J.; Matukumalli, L.K.; Nelson, R.; Shoemaker, R.; DYoung, N.; Cregan, P.B. High-throughput genotyping with the goldengate assay in the complex genome of soybean. Theor. Appl. Genet. 2008, 116, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.J.; Kim, S.; Zhao, K.; Bakker, E.; Horton, M.; Jakob, K.; Lister, C.; Molitor, J.; Shindo, C.; Tang, C. Genome-wide association mapping in arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet. 2005, 1, e60. [Google Scholar] [CrossRef]
- Huang, X.; Zhao, Y.; Wei, X.; Li, C.; Wang, A.; Zhao, Q.; Li, W.; Guo, Y.; Deng, L.; Zhu, C. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm. Nat. Genet. 2011, 44, 32. [Google Scholar] [CrossRef]
- Harjes, C.E.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T. Natural genetic variation in Lycopene Epsilon Cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef]
- Abdurakhmonov, I.Y.; Kohel, R.J.; Yu, J.Z.; Pepper, A.E.; Abdullaev, A.A.; Kushanov, F.N.; Salakhutdinov, I.B.; Buriev, Z.T.; Saha, S.; Scheffler, B.E.; et al. Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. Germplasm. Genomics 2008, 92, 478–487. [Google Scholar] [CrossRef]
- Wang, J.; McClean, P.E.; Lee, R.; Goos, R.J.; Helms, T. Association mapping of iron deficiency chlorosis loci in soybean (Glycine max [L.] Merr.) advanced breeding lines. Theor. Appl. Genet. 2008, 116, 777–787. [Google Scholar] [CrossRef]
- Yu, J.; Pressoir, G.; Briggs, H.; Vroh, I.; Yamasaki, M.; Doebley, J.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Feng, J.Y.; Wenlong, R.; Huang, B.; Zhou, L.; Wen, Y.J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef] [PubMed]
- Base, S. 9.3 Procedures Guide: Statistical Procedures; SAS Institute Inc.: Cary, NC, USA, 2011. [Google Scholar]
- Baker, R.J. Estimation of heritability and prediction of selection response in plant populations. Crit. Rev. Plant Sci. 1991, 10, 235–322. [Google Scholar]
- Allen, G.; Flores, M.; Krasnyanski, S.; Kumar, S.; Thompson, W. A modified protocol for rapid DNA isolation from plant tissue using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef] [PubMed]
- Andolfatto, P.; Davison, D.; Erezyilmaz, D.; Hu, T.; Mast, J.; Sunayama-Morita, T.; Stern, D.L. Multiplexed shotgun genotyping for rapid and efficient genetic mapping. Genome Res. 2011, 21, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Li, R.; Yu, C.; Li, Y.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef]
- Yi, X.; Liang, Y.; Huerta-Sanchez, E.; Jin, X.; Cuo, Z.X.P.; Pool, J.E.; Xu, X.; Jiang, H.; Vinckenbosch, N.; Korneliussen, T.S.; et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010, 329, 75–78. [Google Scholar] [CrossRef]
- Scheet, P.; Stephens, M. A fast and flexible statistical model for large-scale population genotype data: Applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 2006, 78, 629–644. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, P.; Zhang, Z.; E Kroon, D.; Casstevens, T.; Ramdoss, Y.; Buckler, E. TASSEL: Software for Association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Meng, S.; Zhao, T.; Xing, G.; Yang, S.; Li, Y.; Guan, R.; Lu, J.; Wang, Y.; Xia, Q.; et al. An innovative procedure of genome-wide association analysis fits studies on germplasm population and plant breeding. Theor. Appl. Genet. 2017, 130, 2327–2343. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chu, S.; Zhang, H.; Zhu, Y.; Cheng, H.; Yu, D. Development and application of a novel genome-wide snp array reveals domestication history in soybean. Sci. Rep. 2016, 6, 20728. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jing, J.; Wang, Z.; Li, X.; Zhao, T. Genetic analysis of seed germination characters under submergence stress in two recombinant inbred line populations of Glycine max Glycine soja. Soybean Sci. 2013, 32, 581–586. [Google Scholar]
- Kim, S.; Plagnol, V.; Hu, T.T.; Toomajian, C.; Clark, R.M.; Ossowski, S.; Ecker, J.R.; Weigel, D.; Nordborg, M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1151. [Google Scholar] [CrossRef]
- Mather, K.A.; Caicedo, A.L.; Polato, N.R.; Olsen, K.M.; McCouch, S.; Purugganan, M.D. The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 2007, 177, 2223–2232. [Google Scholar] [CrossRef]
- Yan, J.; Shah, T.; Warburton, M.L.; Buckler, E.S.; McMullen, M.D.; Crouch, J. Genetic characterization and linkage disequilibrium estimation of a global maize collection using snp markers. PLoS ONE 2009, 4, e8451. [Google Scholar] [CrossRef]
- Lam, H.-M.; Xu, X.; Liu, X.; Chen, W.; Yang, G.; Wong, F.-L.; Li, M.-W.; He, W.; Qin, N.; Wang, B.; et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 2010, 42, 1053–1059. [Google Scholar] [CrossRef]
- Vantoai, T.T.; Beuerlein, A.F.; Schmitthenner, S.K.; St. Martin, S.K. Genetic Variability for Flooding Tolerance in Soybeans. Crop Sci. 1994, 34, 1112–1115. [Google Scholar] [CrossRef]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Tung, C.-W.; Eizenga, G.C.; Wright, M.H.; Ali, M.L.; Price, A.H.; Norton, G.J.; Islam, M.R.; Reynolds, A.; Mezey, J.; et al. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza Sativa. Nat. Commun. 2011, 2, 467. [Google Scholar] [CrossRef] [PubMed]
- Riedelsheimer, C.; Lisec, J.; Czedik-Eysenberg, A.; Sulpice, R.; Flis, A.; Grieder, C.; Altmann, T.; Stitt, M.; Willmitzer, L.; Melchinger, A.E. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 8872–8877. [Google Scholar] [CrossRef] [PubMed]
- Stich, B.; Melchinger, A.E.; Frisch, M.; Maurer, H.P.; Heckenberger, M.; Reif, J.C. Linkage disequilibrium in european elite maize germplasm investigated with SSRs. Theor. Appl. Genet. 2005, 111, 723–730. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Gore, M.; Buckler, E.S.; Yu, J. Status and prospects of association mapping in plants. Plant Genome 2008, 1, 5–20. [Google Scholar] [CrossRef]
- Liu, A.; Burke, J.M. Patterns of nucleotide diversity in wild and cultivated sunflower. Genetics 2006, 173, 321–330. [Google Scholar] [CrossRef]
- Zhu, Y.L.; Song, Q.J.; Hyten, D.L.; Van Tassell, C.P.; Matukumalli, L.K.; Grimm, D.R.; Hyatt, S.M.; Fickus, E.W.; Young, N.D.; Cregan, P.B. Single-nucleotide polymorphisms in soybean. Genetics 2003, 163, 1123–1134. [Google Scholar]
- Nordborg, M. Linkage disequilibrium, gene trees and selfing: An ancestral recombination graph with partial self-fertilization. Genetics 2000, 154, 923–929. [Google Scholar]
- Jaiswal, V.; Gupta, S.; Gahlaut, V.; Muthamilarasan, M.; Bandyopadhyay, T.; Ramchiary, N.; Prasad, M. Genome-wide association study of major agronomic traits in foxtail millet (Setaria italica L.) using ddRAD sequencing. Sci. Rep. 2019, 9, 5020. [Google Scholar] [CrossRef]
- Davey, J.; Davey, J.; Blaxter, M.; Blaxter, M. Radseq: Next-generation population genetics. Brief. Funct. Genomics 2010, 9, 416–423. [Google Scholar] [CrossRef] [PubMed]
- Bhat, J.; Ali, S.; Salgotra, R.; Mir, Z.; Dutta, S.; Jadon, V.; Tyagi, A.; Mushtaq, M.; Jain, N.; Singh, P.; et al. Genomic selection in the Era of next generation sequencing for complex traits in plant breeding. Front. Genet. 2016, 7, 221. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.; Weber, J.; Delaney, E.; Fisher, H.; Hoekstra, H. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Mastretta-Yanes, A.; Arrigo, N.; Alvarez, N.; Jorgensen, T.; Piñero, D.; Emerson, B. Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Mol. Ecol. Resour. 2014, 15, 28–41. [Google Scholar] [CrossRef]
- Pandey, D.M.; Kim, S.R. Identification and expression analysis of hypoxia stress inducible ccch-type zinc finger protein genes in rice. J. Plant Biol. 2012, 55, 489–497. [Google Scholar] [CrossRef]
- Min, J.H.; Chung, J.S.; Lee, K.H.; Kim, C.S. The constans-like 4 transcription factor, ATCOL4, positively regulates abiotic stress tolerance through an abscisic acid-dependent manner in Arabidopsis. J. Integr. Plant Biol. 2015, 57, 313–324. [Google Scholar] [CrossRef]
- Kobayashi, M.; Horiuchi, H.; Fujita, K.; Takuhara, Y.; Suzuki, S. Characterization of grape c-repeat-binding factor 2 and b-box-type zinc finger protein in transgenic Arabidopsis plants under stress conditions. Mol. Biol. Rep. 2012, 39, 7933–7939. [Google Scholar] [CrossRef]
- Liu, X.; Li, R.; Dai, Y.; Yuan, L.; Sun, Q.; Zhang, S.; Wang, X. A B-box zinc finger protein, MdBBX10, enhanced salt and drought stresses tolerance in Arabidopsis. Plant Mol. Biol. 2019, 99, 437–447. [Google Scholar] [CrossRef]
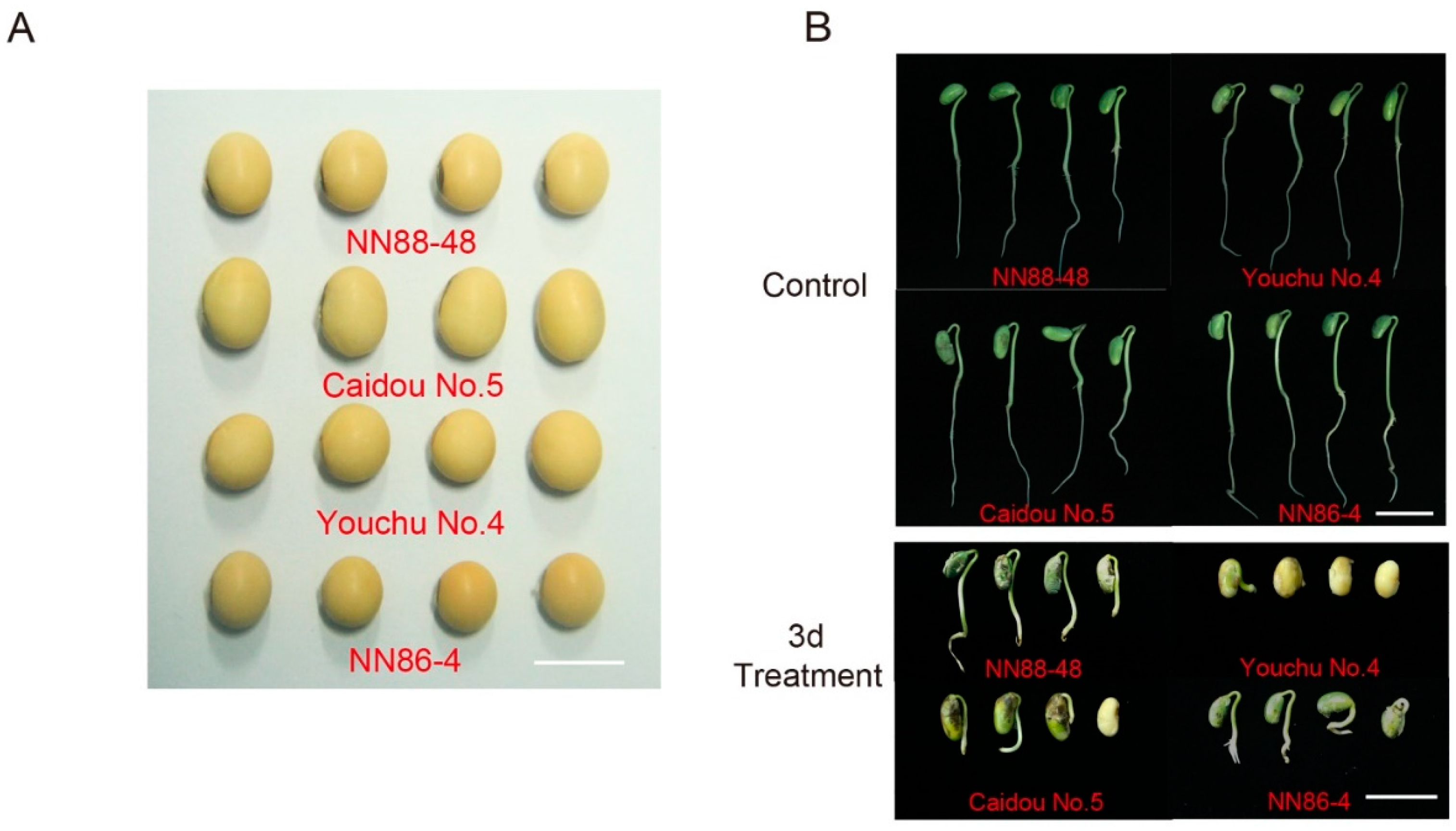


| Trait | Env. | Mean ± SD | Range | h2 | F values from ANOVA | ||
|---|---|---|---|---|---|---|---|
| Line | Env. | Line × Env. | |||||
| GR | JP14 | 0.55 ± 0.29 | 0–1 | 0.74 | 9.84*** | 13.27*** | 0.47ns |
| HY15 | 0.43 ± 0.26 | 0–0.95 | |||||
| NSR | JP14 | 0.40 ± 0.27 | 0–0.94 | 0.69 | 8.11*** | 7.04** | 0.37ns |
| HY15 | 0.27 ± 0.24 | 0–0.93 | |||||
| EC | JP14 | 1248 ± 446 | 154–2840 | 0.77 | 3.58*** | 5.19** | 2.16*** |
| HY15 | 1309 ± 450 | 228–3620 | |||||
| Trait | QTN | Env.a | Chr.b | SNP | Position (bp) | P Value | −log10 Pc | MAF | R2 (%) | Effect | Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|
| GR | qGR-1 | JP14 | 1 | Gm01_380964 | 380964 | 9.83 × 10−5 | 4.01 | 0.07 | 4.49 | -0.027 | T/A |
| qGR-8-1 | JP14 | 8 | Gm08_9896483 | 9896483 | 5.67 × 10−5 | 4.25 | 0.29 | 4.52 | -0.097 | C/T | |
| qGR-8-2 | JP14 | 8 | Gm08_10412475 | 10412475 | 9.37 × 10−5 | 4.03 | 0.25 | 4.26 | -0.003 | A/G | |
| qGR-8-3 | Combined | 8 | Gm08_14242918 | 14242918 | 9.71 × 10−5 | 4.01 | 0.07 | 4.28 | -0.253 | G/T | |
| qGR-13-1 | JP14 | 13 | Gm13_34829465 | 34829465 | 7.42 × 10−5 | 4.13 | 0.09 | 4.48 | -0.018 | C/T | |
| qGR-13-2 | JP14 | 13 | Gm13_35324537 | 35324537 | 4.75 × 10−5 | 4.32 | 0.07 | 4.51 | -0.257 | G/T | |
| (QTN13) | HY15 | 13 | Gm13_35324537 | 35324537 | 2.44 × 10−6 | 5.61 | 0.07 | 6.62 | -0.285 | G/T | |
| Combined | 13 | Gm13_35324537 | 35324537 | 2.65 × 10−5 | 4.58 | 0.06 | 4.97 | 0.197 | C/T | ||
| qGR-13-3 | HY15 | 13 | Gm13_35826401 | 35826401 | 2.43 × 10−5 | 4.61 | 0.05 | 5.28 | -0.267 | A/T | |
| Combined | 13 | Gm13_35776711 | 35776711 | 2.36 × 10−5 | 4.63 | 0.31 | 4.87 | 0.147 | A/G | ||
| qGR-14 | HY15 | 14 | Gm14_46766676 | 46766676 | 3.71 × 10−6 | 5.43 | 0.31 | 6.38 | 0.172 | A/G | |
| Combined | 14 | Gm14_46766676 | 46766676 | 8.28 × 10−5 | 4.08 | 0.13 | 4.35 | -0.175 | C/T | ||
| NSR | qNSR-8 | JP14 | 8 | Gm08_9370630 | 9370630 | 7.92 × 10−5 | 4.1 | 0.12 | 4.38 | -0.11 | A/G |
| Combined | 8 | Gm08_9370743 | 9370743 | 9.72 × 10−5 | 4.01 | 0.08 | 4.52 | 0.229 | A/G | ||
| qNSR-10 | JP14 | 10 | Gm10_4316320 | 4316320 | 2.17 × 10−5 | 4.66 | 0.08 | 5.38 | 0.242 | A/G | |
| HY15 | 10 | Gm10_4314632 | 4314632 | 2.65 × 10−5 | 4.58 | 0.08 | 5.26 | 0.213 | A/T | ||
| Combined | 10 | Gm10_4316320 | 4316320 | 3.73 × 10−5 | 4.43 | 0.07 | 4.81 | -0.264 | G/T | ||
| qNSR-13-1 | JP14 | 13 | Gm13_35324537 | 35324537 | 7.18 × 10−6 | 5.14 | 0.07 | 6.02 | -0.282 | G/T | |
| (QTN13) | HY15 | 13 | Gm13_35324537 | 35324537 | 8.20 × 10−5 | 4.09 | 0.05 | 4.43 | -0.248 | G/T | |
| Combined | 13 | Gm13_35324537 | 35324537 | 2.85 × 10−5 | 4.54 | 0.05 | 4.95 | 0.226 | G/T | ||
| qNSR-13-2 | JP14 | 13 | Gm13_35776682 | 35776682 | 5.02 × 10−5 | 4.3 | 0.06 | 4.69 | -0.213 | C/G | |
| HY15 | 13 | Gm13_35826420 | 35826420 | 9.82 × 10−5 | 4.01 | 0.05 | 4.37 | 0.209 | G/T | ||
| Combined | 13 | Gm13_35826420 | 35826420 | 7.12 × 10−5 | 4.15 | 0.05 | 4.41 | 0.238 | C/T | ||
| qNSR-14 | HY15 | 14 | Gm14_46766676 | 46766676 | 2.45 × 10−5 | 4.61 | 0.31 | 5.31 | 0.146 | A/G | |
| qNSR-20 | HY15 | 20 | Gm20_42414884 | 42414884 | 2.41 × 10−5 | 4.62 | 0.05 | 5.32 | 0.24 | C/T | |
| Combined | 20 | Gm20_42414884 | 42414884 | 6.29 × 10−5 | 4.2 | 0.22 | 4.76 | 272.188 | C/G | ||
| EC | qEC-2-1 | HY15 | 2 | Gm02_5181577 | 5181577 | 3.63 × 10−5 | 4.44 | 0.22 | 5.09 | 285.2 | C/T |
| qEC-2-2 | JP14 | 2 | Gm02_49796905 | 49796905 | 8.91 × 10−5 | 4.05 | 0.47 | 4.46 | -90.75 | C/T | |
| qEC-7-1 | HY15 | 7 | Gm07_2485335 | 2485335 | 3.10 × 10−5 | 4.51 | 0.09 | 5.1 | -519.94 | A/G | |
| qEC-7-2 | JP14 | 7 | Gm07_2942021 | 2942021 | 2.69 × 10−5 | 4.57 | 0.09 | 5.53 | -465.45 | A/G | |
| HY15 | 7 | Gm07_2942021 | 2942021 | 9.74 × 10−6 | 5.01 | 0.09 | 5.87 | -532.98 | A/G | ||
| Combined | 7 | Gm07_2942021 | 2942021 | 6.34 × 10−5 | 4.2 | 0.33 | 4.75 | -106.07 | A/G | ||
| qEC-8 | JP14 | 8 | Gm08_9896483 | 9896483 | 7.20 × 10−5 | 4.14 | 0.29 | 4.62 | 184.03 | C/T | |
| Combined | 8 | Gm08_9752396 | 9752396 | 9.25 × 10−5 | 4.03 | 0.16 | 4.37 | 310.06 | A/G | ||
| qEC-11 | JP14 | 11 | Gm11_4188194 | 4188194 | 7.25 × 10−5 | 4.14 | 0.16 | 4.81 | 326.88 | A/G | |
| Combined | 11 | Gm11_4188194 | 4188194 | 8.70 × 10−5 | 4.06 | 0.13 | 4.76 | 247.19 | A/G | ||
| qEC-13-1 | HY15 | 13 | Gm13_35324537 | 35324537 | 3.47 × 10−5 | 4.46 | 0.08 | 4.95 | 324.5 | G/T | |
| (QTN13) | Combined | 13 | Gm13_35324537 | 35324537 | 7.12 × 10−5 | 4.15 | 0.08 | 4.67 | 333 | G/T | |
| qEC-13-2 | HY15 | 13 | Gm13_35648515 | 35648515 | 4.37 × 10−5 | 4.36 | 0.08 | 4.91 | 400.56 | A/G | |
| qEC-13-3 | HY15 | 13 | Gm13_36352911 | 36352911 | 6.55 × 10−5 | 4.18 | 0.11 | 4.72 | 41.08 | C/T | |
| qEC-18 | JP14 | 18 | Gm18_23932848 | 23932848 | 8.78 × 10−5 | 4.06 | 0.34 | 4.38 | 152.49 | G/T | |
| qEC-19 | HY15 | 19 | Gm19_35610187 | 35610187 | 8.87 × 10−5 | 4.05 | 0.4 | 4.58 | 272.19 | A/G |
| Trait | QTL | Env.a | Chr.b | SNP | Position (bp) | P Value | −log10 Pc | LOD | MAF | R2 (%) | Effect | Allele | CIMd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GR | qGR-2 | HY15 | 2 | Gm02_46786754 | 46786754 | 4.14 × 10−5 | 4.38 | 3.58 | 0.47 | 4.74 | 0.118 | C/T | |
| qGR-7 | HY15 | 7 | Gm07_2485335 | 2485335 | 6.21 × 10−6 | 5.21 | 4.38 | 0.22 | 5.63 | −0.145 | C/G | ||
| qGR-8-1 | JP14 | 8 | Gm08_9896483 | 9896483 | 4.44 × 10−6 | 5.35 | 5.02 | 0.29 | 5.32 | −0.097 | C/T | 1 | |
| qGR-10 | JP14 | 10 | Gm10_4163841 | 4163841 | 1.25 × 10-7 | 6.9 | 6.06 | 0.11 | 5.72 | 0.175 | A/G | ||
| qGR-13-2 | JP14 | 13 | Gm13_35324537 | 35324537 | 1.53 × 10−6 | 5.82 | 5.02 | 0.07 | 5.94 | −0.227 | G/T | 1 | |
| (QTN13) | HY15 | 13 | Gm13_35324537 | 35324537 | 1.87 × 10-7 | 6.73 | 5.8 | 0.07 | 7.22 | −0.265 | G/T | 1 | |
| Joint | 13 | Gm13_35324537 | 35324537 | 4.23 × 10−6 | 5.37 | 4.45 | 0.07 | 5.44 | −0.253 | G/T | 1 | ||
| qGR-15 | HY15 | 15 | Gm15_11267976 | 11267976 | 2.57 × 10-7 | 6.59 | 5.68 | 0.07 | 7.17 | 0.212 | A/T | ||
| qGR-18 | JP14 | 18 | Gm18_1336864 | 1336864 | 2.76 × 10-8 | 7.56 | 6.18 | 0.16 | 7.68 | −0.179 | G/T | ||
| qGR-20 | HY15 | 20 | Gm20_39515872 | 39515872 | 1.69 × 10−5 | 4.77 | 4.01 | 0.32 | 3.96 | −0.115 | A/G | ||
| NSR | qNSR-2 | HY15 | 2 | Gm02_46070310 | 46070310 | 5.28 × 10−5 | 4.28 | 4.01 | 0.14 | 3.89 | −0.144 | C/G | |
| qNSR-7 | JP14 | 7 | Gm07_2584000 | 2584000 | 1.38 × 10-7 | 6.86 | 5.8 | 0.47 | 7.5 | 0.117 | C/T | ||
| qNSR-10 | JP14 | 10 | Gm10_4316320 | 4316320 | 1.57 × 10-7 | 6.8 | 5.97 | 0.08 | 7.11 | 0.242 | A/G | 1 | |
| HY15 | 10 | Gm10_4316320 | 4316320 | 8.89 × 10-8 | 7.05 | 6.08 | 0.08 | 7.61 | 0.219 | A/G | 1 | ||
| qNSR-11 | HY15 | 11 | Gm11_36522210 | 36522210 | 1.69 × 10−5 | 4.77 | 4.11 | 0.08 | 4.28 | −0.161 | A/G | ||
| qNSR-13-1 | JP14 | 13 | Gm13_35324537 | 35324537 | 3.06 × 10-8 | 7.51 | 6.22 | 0.07 | 6.02 | −0.328 | G/T | 1 | |
| (QTN13) | HY15 | 13 | Gm13_35324537 | 35324537 | 4.26 × 10-9 | 8.37 | 7.22 | 0.07 | 7.04 | −0.312 | G/T | 1 | |
| Joint | 13 | Gm13_35324537 | 35324537 | 1.06 × 10-9 | 8.97 | 7.33 | 0.07 | 6.67 | −0.284 | G/T | 1 | ||
| qNSR-18 | HY15 | 18 | Gm18_59484809 | 59484809 | 1.98 × 10−5 | 4.7 | 3.95 | 0.08 | 3.09 | 0.197 | G/T | ||
| qNSR-19 | HY15 | 19 | Gm19_48197396 | 48197396 | 2.16 × 10−6 | 5.67 | 5.31 | 0.1 | 6.83 | −0.147 | C/T | ||
| EC | qEC-3 | JP14 | 3 | Gm03_40169903 | 40169903 | 3.86 × 10−6 | 5.41 | 4.58 | 0.43 | 4.85 | −229.140 | A/G | |
| qEC-7-2 | JP14 | 7 | Gm07_2942021 | 2942021 | 9.56 × 10-7 | 6.02 | 5.19 | 0.09 | 4.91 | −465.450 | A/G | 1 | |
| HY15 | 7 | Gm07_2942021 | 2942021 | 2.07 × 10-7 | 6.68 | 5.76 | 0.09 | 6.81 | −532.980 | A/G | 1 | ||
| qEC-10 | Joint | 10 | Gm10_11026230 | 11026230 | 2.51 × 10−5 | 4.6 | 3.81 | 0.18 | 4.74 | −297.270 | G/T | ||
| qEC-12 | HY15 | 12 | Gm12_990925 | 990925 | 4.90 × 10−5 | 4.31 | 3.72 | 0.46 | 4.55 | −192.640 | A/G | ||
| qEC-13-1 | JP14 | 13 | Gm13_35324537 | 35324537 | 3.06 × 10−5 | 4.51 | 3.69 | 0.07 | 4.66 | 354.477 | G/T | 1 | |
| (QTN13) | HY15 | 13 | Gm13_35324537 | 35324537 | 7.99 × 10-7 | 6.1 | 5.12 | 0.07 | 6.37 | 317.55 | G/T | 1 | |
| Joint | 13 | Gm13_35324537 | 35324537 | 4.10 × 10−6 | 5.39 | 5.05 | 0.07 | 4.98 | 307.325 | G/T | 1 | ||
| qEC-19 | HY15 | 19 | Gm19_35626216 | 35626216 | 1.92 × 10−5 | 4.72 | 4.14 | 0.08 | 5.35 | 400.562 | G/T |
| Gene ID | Positon (bp) | Function annotation |
|---|---|---|
| Glyma.13g241900 | 35181812-35182687 | Dof-type zinc finger DNA-binding family protein |
| Glyma.13g243200 | 35262813-35264688 | NAC domain containing protein |
| Glyma.13g244800 | 35408963-35410102 | Glycine-rich protein |
| Glyma.13g246200 | 35511992-35512643 | F-box family protein |
| Glyma.13g246500 | 35523224-35524550 | transmembrane protein |
| Glyma.13g247200 | 35571708-35573509 | MYB-like DNA-binding protein |
| Glyma.13g248000 | 35620202-35621661 | B-box type zinc finger family protein |
| Glyma.13g248400 | 35651954-35655747 | Leucine-rich Repeat Receptor-like protein kinase |
| Glyma.13g249800 | 35739290-35742405 | Basic helix-loop-helix (bHLH) DNA-binding superfamily protein |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Chang, F.; Lv, W.; Sharmin, R.A.; Wang, Z.; Kong, J.; Bhat, J.A.; Zhao, T. Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS). Genes 2019, 10, 957. https://doi.org/10.3390/genes10120957
Yu Z, Chang F, Lv W, Sharmin RA, Wang Z, Kong J, Bhat JA, Zhao T. Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS). Genes. 2019; 10(12):957. https://doi.org/10.3390/genes10120957
Chicago/Turabian StyleYu, Zheping, Fangguo Chang, Wenhuan Lv, Ripa Akter Sharmin, Zili Wang, Jiejie Kong, Javaid Akhter Bhat, and Tuanjie Zhao. 2019. "Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS)" Genes 10, no. 12: 957. https://doi.org/10.3390/genes10120957
APA StyleYu, Z., Chang, F., Lv, W., Sharmin, R. A., Wang, Z., Kong, J., Bhat, J. A., & Zhao, T. (2019). Identification of QTN and Candidate Gene for Seed-flooding Tolerance in Soybean [Glycine max (L.) Merr.] using Genome-Wide Association Study (GWAS). Genes, 10(12), 957. https://doi.org/10.3390/genes10120957





