Transcriptome Sequencing Reveals the Traits of Spermatogenesis and Testicular Development in Large Yellow Croaker (Larimichthys crocea)
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
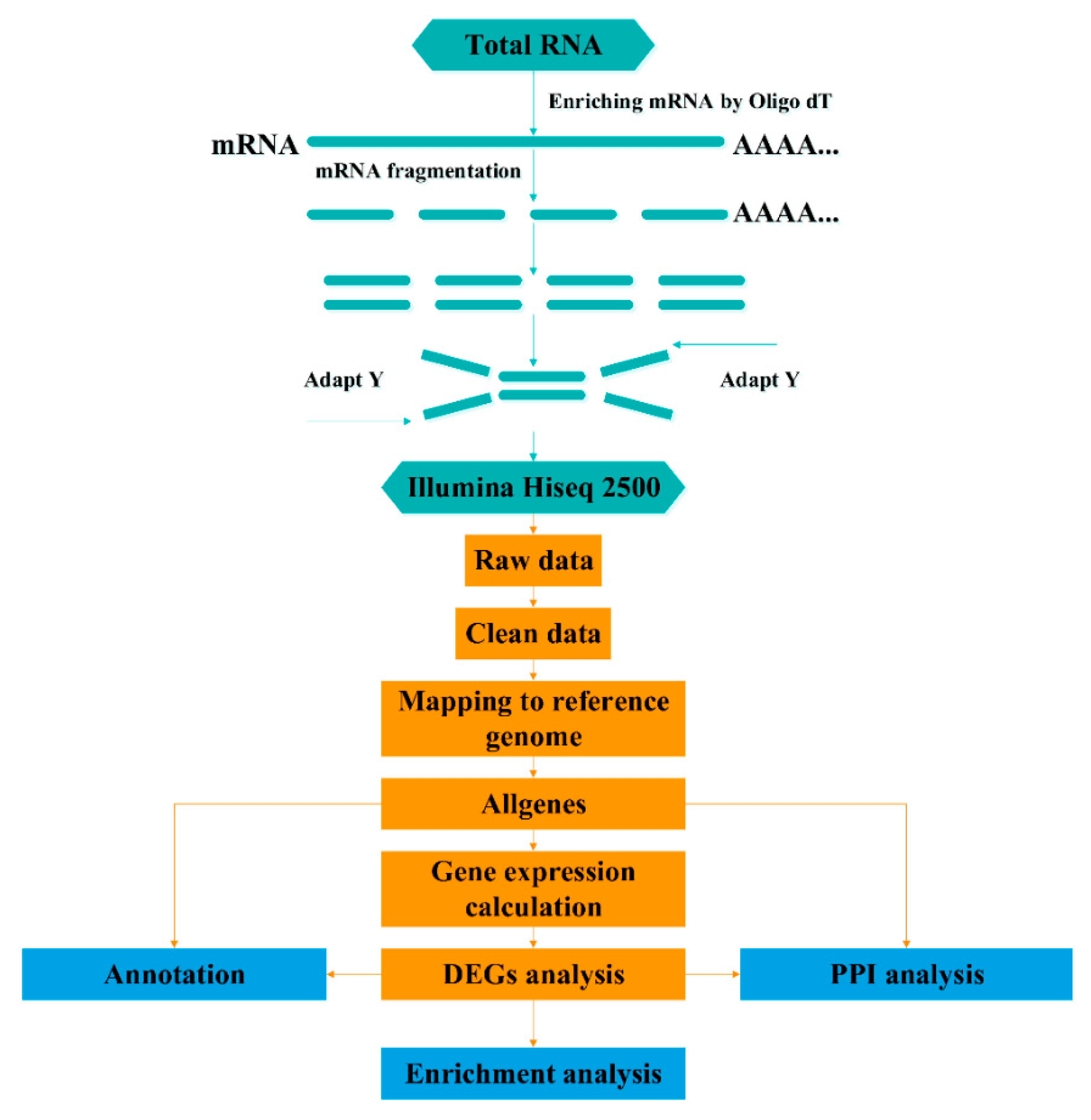
2.2. Sample Collection, Histological Identification, RNA Isolation, and cDNA Library Preparation
2.3. Sequencing and Mapping to the Reference Genome
2.4. Differential Expression Analysis
2.5. Functional Annotation and Pathway Enrichment Analysis
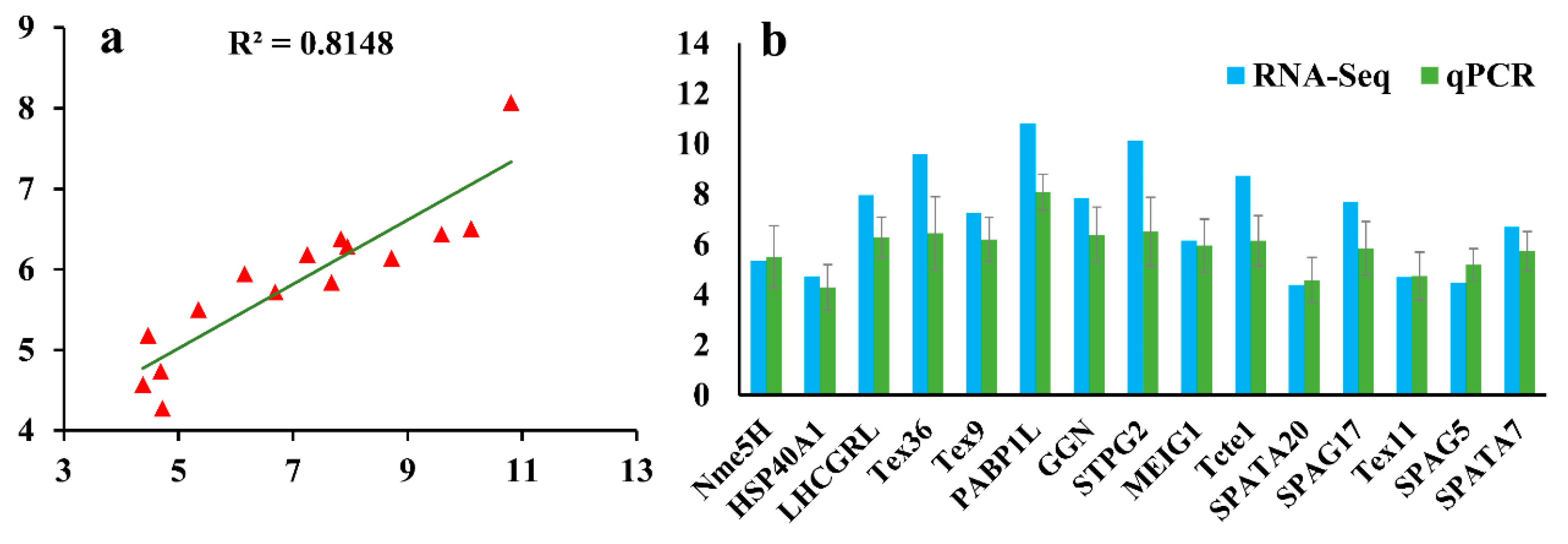
2.6. Quantitative Real-Time PCR (qPCR) Validation
3. Results
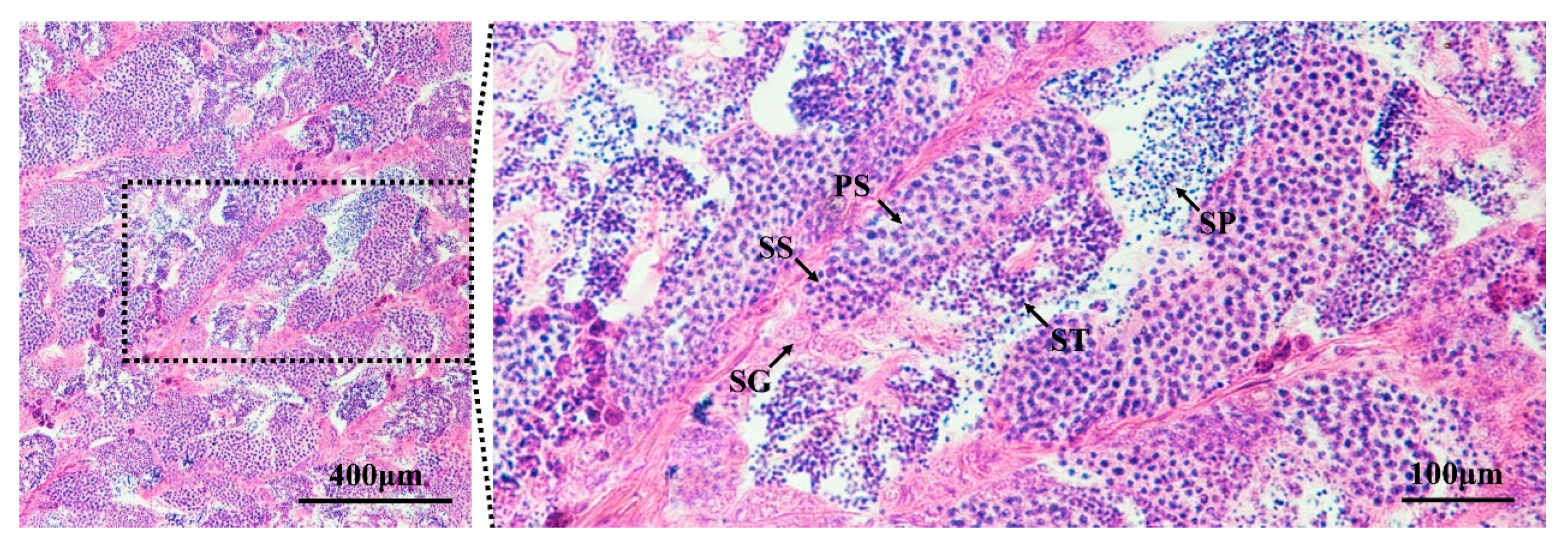
3.1. Identification of Testes in Developmental Stage IV
3.2. Overview of the Transcriptome Profiles
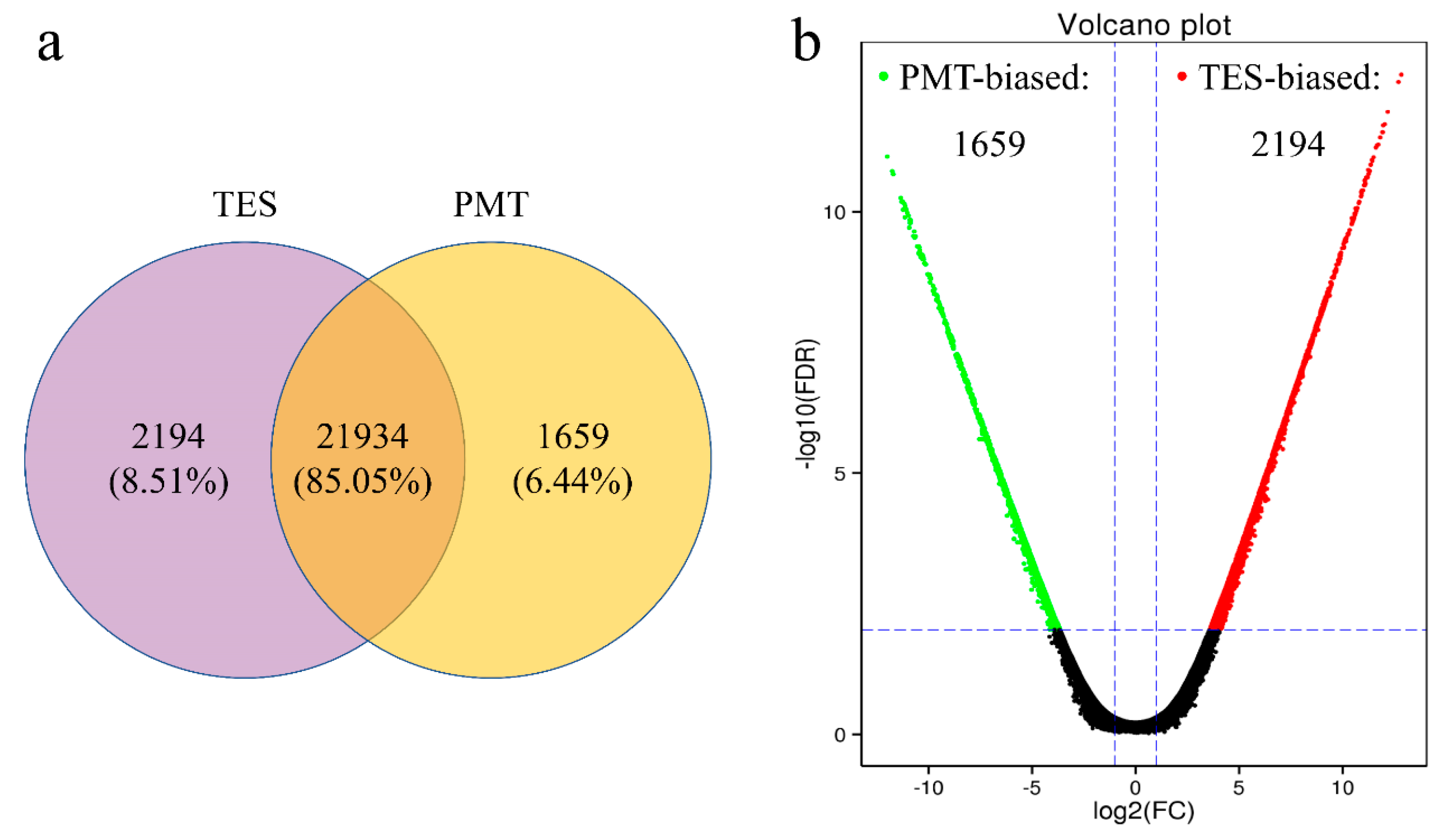
3.3. Differentially Expressed Genes (DEGs) of the Two Libraries
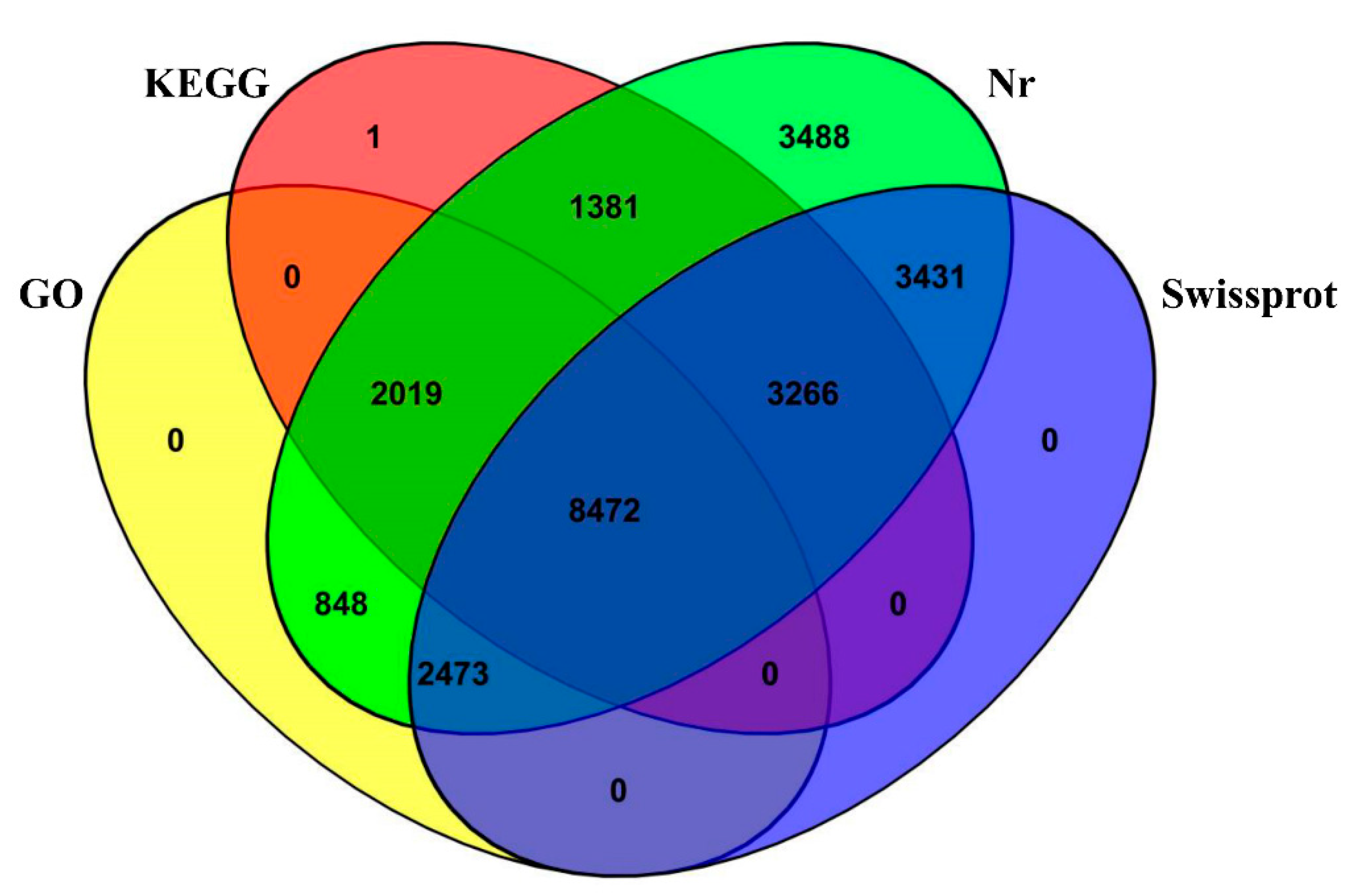
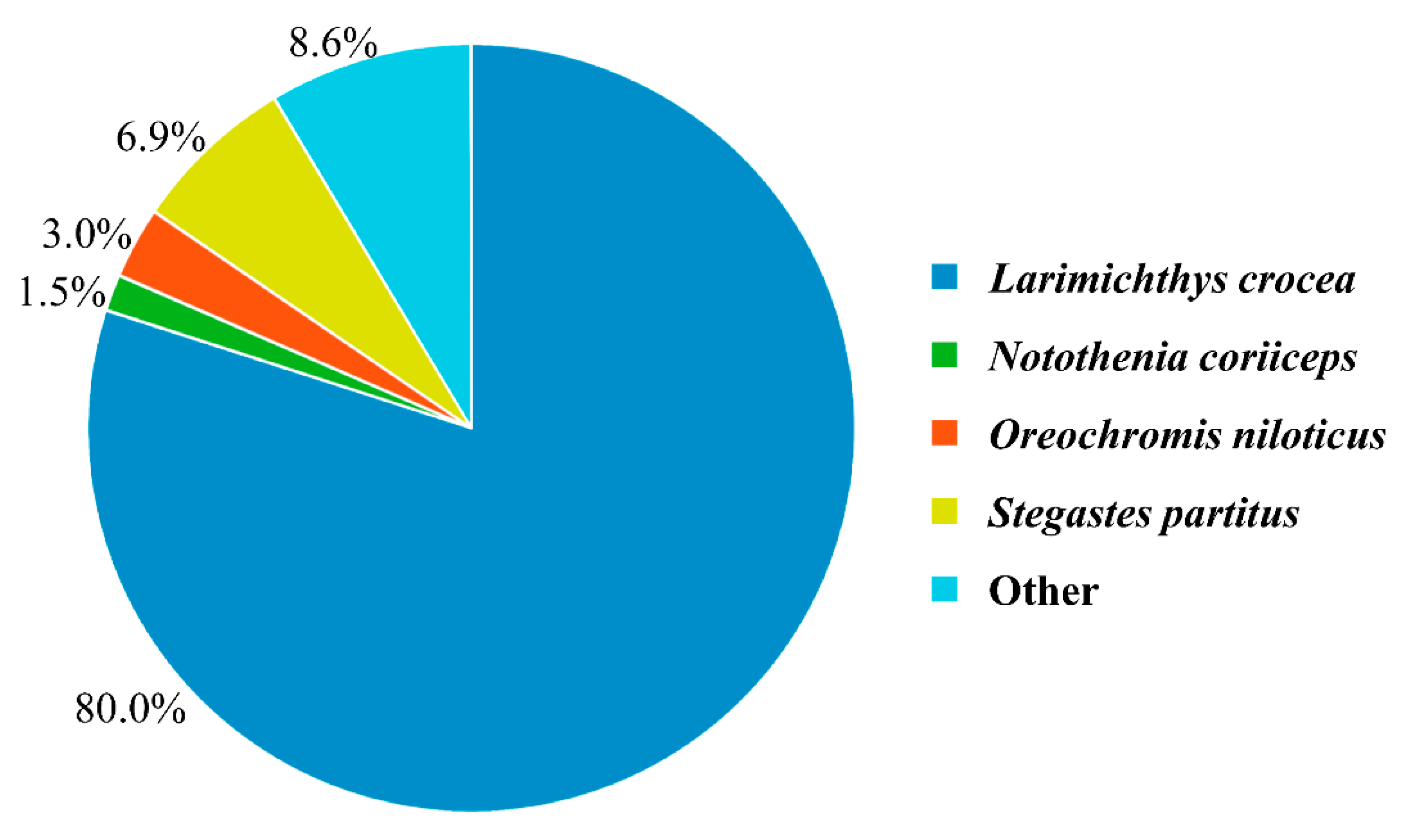
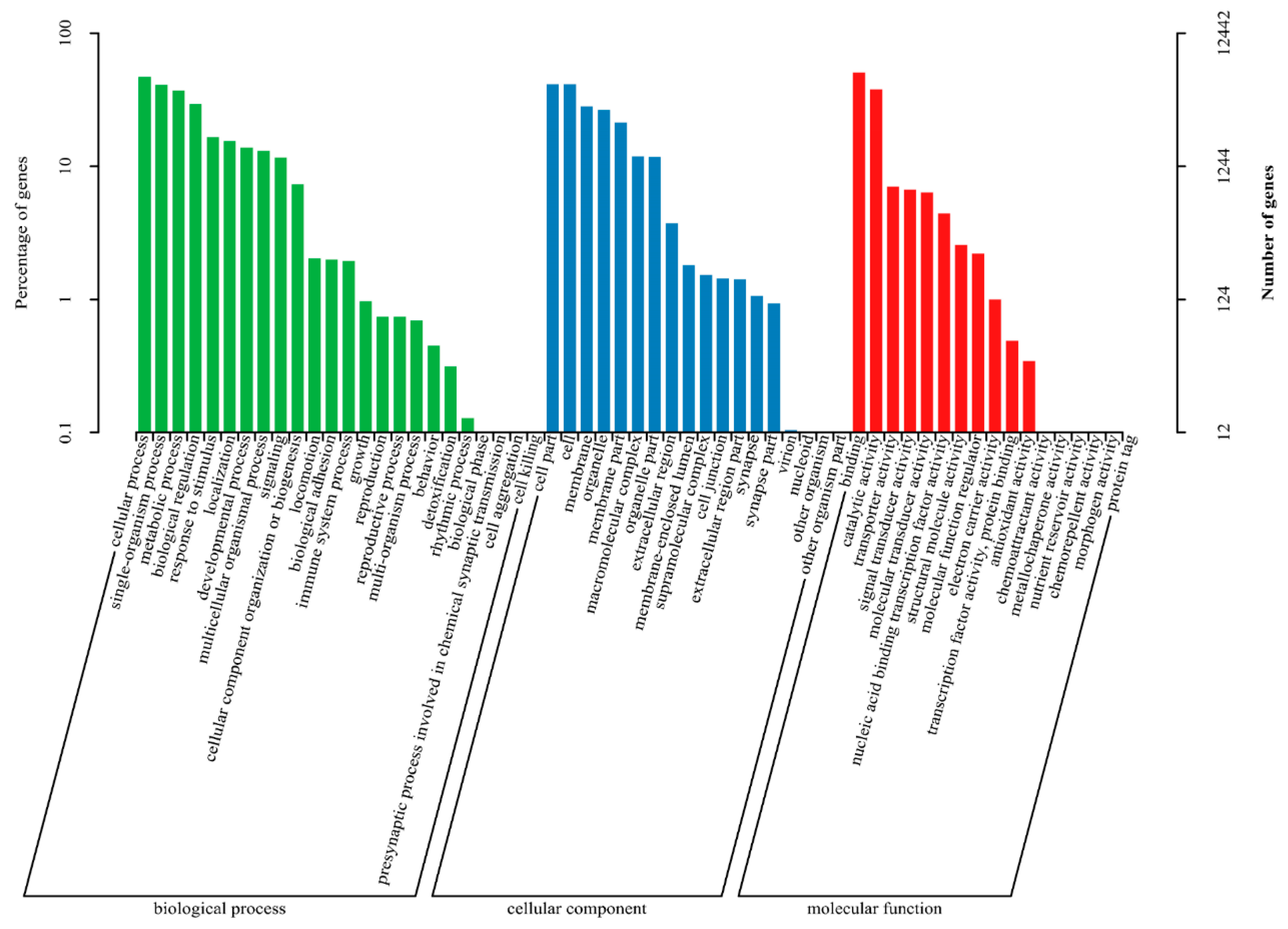
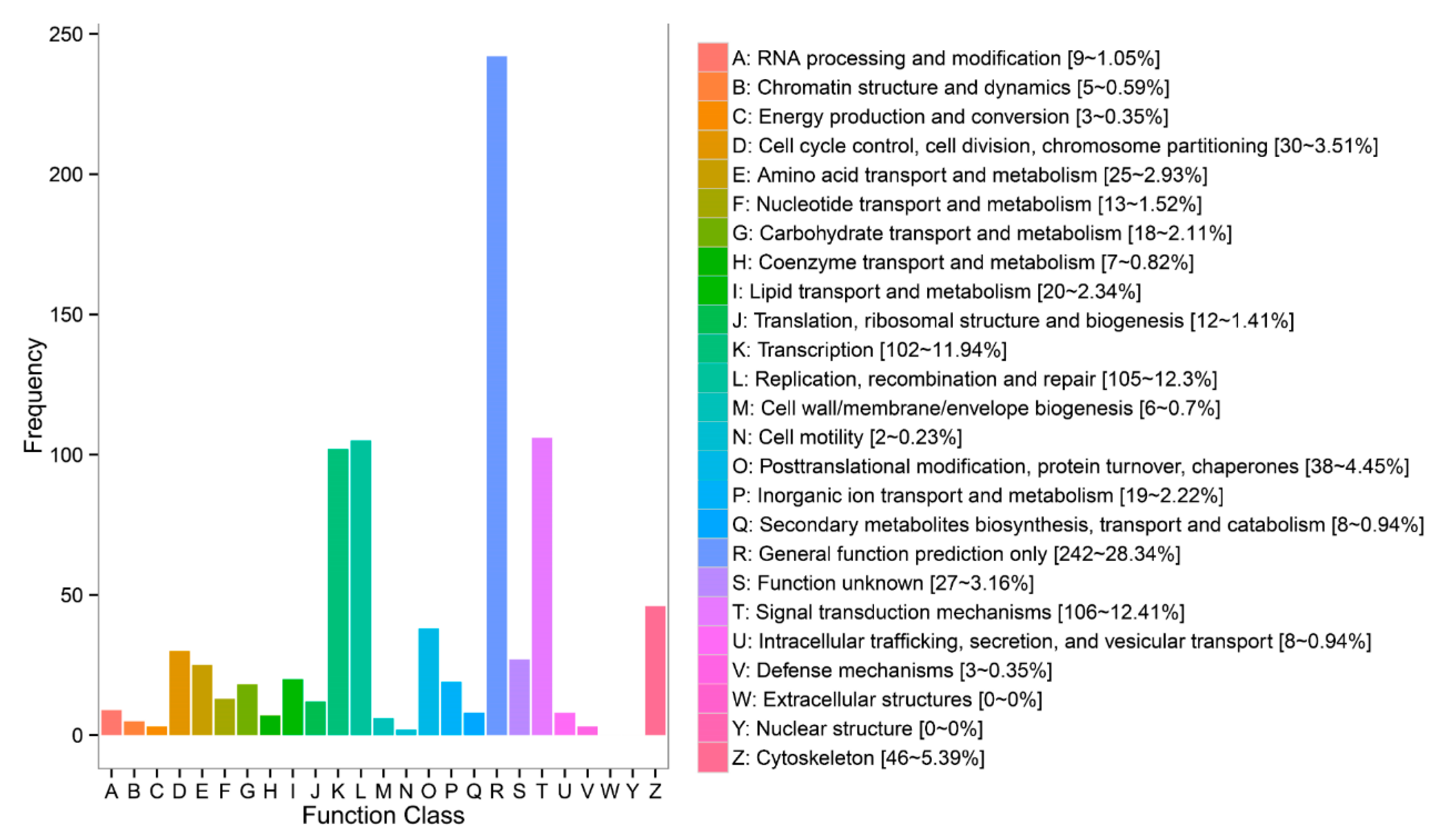
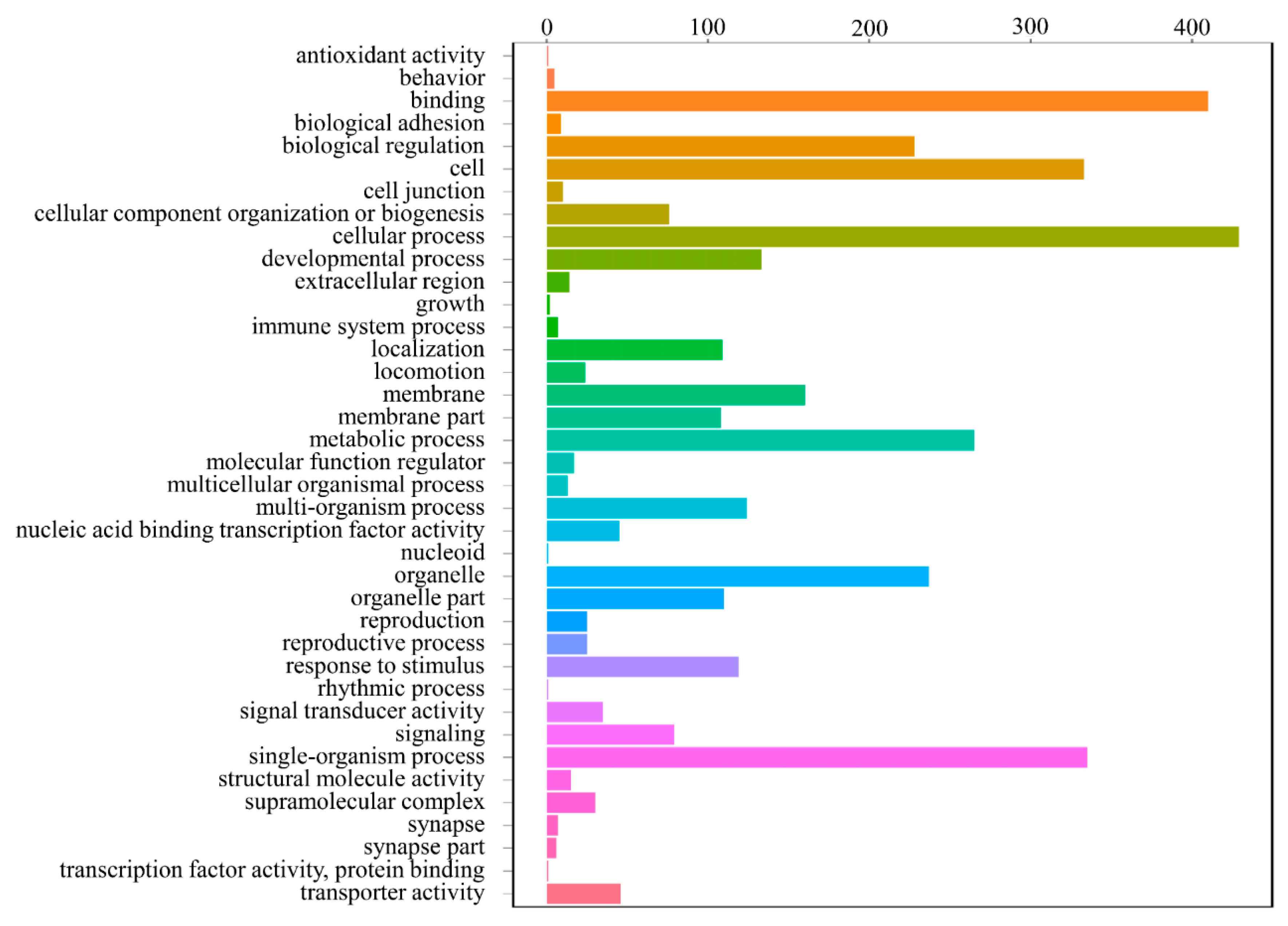
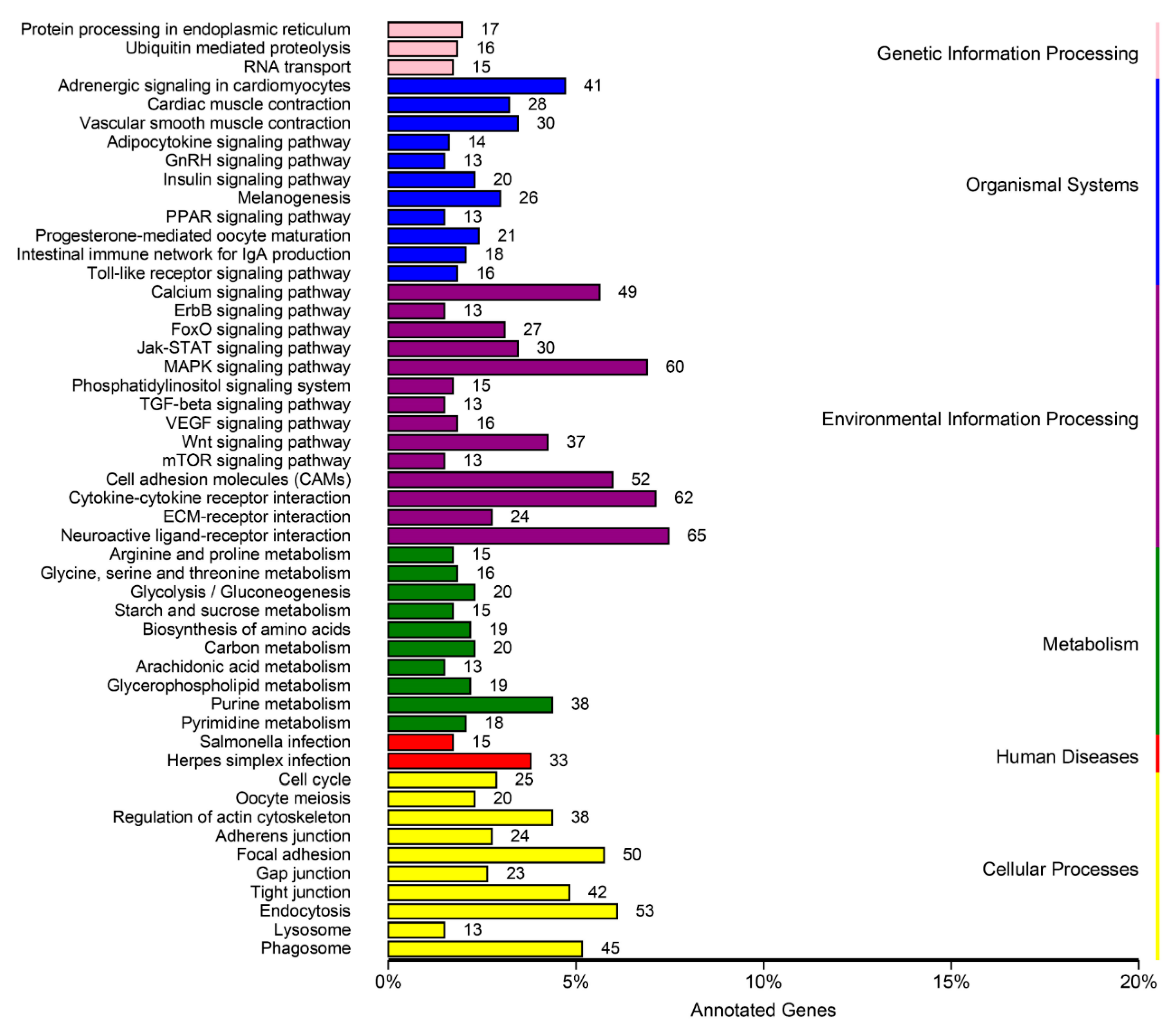
3.4. DEGs Annotation and Pathway Analysis
3.5. Validation of RNA-Seq Data by qPCR
4. Discussion
4.1. Neuroactive Ligand–Receptor Interaction Pathway
4.2. GnRH and MAPK Signaling Pathway
4.3. Cell Cycle Pathway
4.4. Genes Encoding Microtubule-Based Motor Proteins
4.5. Actin Cytoskeleton and Myosins
4.6. Heat Shock Protein Genes (HSPs)
4.7. Synaptonemal Complex Protein 2 Gene (Sycp2)
4.8. Doublesex- and Mab-3-Related Transcription Factor 1 (Dmrt1)
4.9. Spermatogenesis-Associated Genes (Spatas)
4.10. DEAD-Box Helicases (Ddxs)
4.11. Tudor Domain-Containing Protein Genes (Tdrds) and Piwis
5. Conclusions
6. Data Accessibility
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fisheries Bureau, Department of Agriculture of China. China Fishery Statistical Yearbook; Fisheries Bureau, Department of Agriculture of China: Beijing, China, 2018.
- Zhou, J. Study on endocrine mechanism of earlier gonadal maturation in cultured large yellow croaker, Pseudosciaena crocea. Master’s Thesis, State Oceanic Administration, Xiamen, China, 2001. [Google Scholar]
- Zhang, W.; Wan, H.L.; Jiang, H.; Zhao, Y.L.; Zhang, X.W.; Hu, S.N.; Wang, Q. A transcriptome analysis of mitten crab testes (Eriocheir sinensis). Genet. Mol. Biol. 2011, 34, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Waiho, K.; Fazhan, H.; Shahreza, M.S.; Moh, J.H.Z.; Noorbaiduri, S.; Wong, L.L.; Sinnasamy, S.; Ikhwanuddin, M. Transcriptome Analysis and Differential Gene Expression on the Testis of Orange Mud Crab, Scylla olivacea, during Sexual Maturation. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.-F.; Ramachandra, R.K.; Rexroad, C.E.; Yao, J. Molecular characterization and expression profiles of cyclin B1, B2 and Cdc2 kinase during oogenesis and spermatogenesis in rainbow trout (Oncorhynchus mykiss). Anim. Reprod. Sci. 2008, 105, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhu, J.; Zhou, H.; Yang, W. The formation of zona radiata in Pseudosciaena crocea revealed by light and transmission electron microscopy. Micron 2012, 43, 435–444. [Google Scholar] [CrossRef]
- Liu, J. A study on twice maturity characteristic of cultured large yellow croaker in one year. J. Jimei Univ. Nat. Sci. 2004, 9, 200–204. [Google Scholar]
- Han, K.; Chen, S.; Cai, M.; Jiang, Y.; Zhang, Z.; Wang, Y. Nanos3 not nanos1 and nanos2 is a germ cell marker gene in large yellow croaker during embryogenesis. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2018, 218, 13–22. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, K.; Cai, M.; Wang, Y.; Zhang, Z. Characterization and Spatiotemporal Expression of Klf4 in Large Yellow Croaker Larimichthys Crocea. DNA Cell Biol. 2017, 36, 655–671. [Google Scholar] [CrossRef]
- Zhang, D.L.; Yu, D.H.; Luo, H.Y.; Wang, Z.Y. WD Repeat-containing Protein 73, A Novel Gene Correlated with Gonad Development in Large Yellow Croaker, Larimichthys crocea. J. World Aquacult. Soc. 2016, 47, 268–276. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, X.; Zhao, Y.; Hou, C.; Zhu, J. The C-terminal kinesin motor KIFC1 may participate in nuclear reshaping and flagellum formation during spermiogenesis of Larimichthys crocea. Fish Physiol. Biochem. 2017, 43, 1351–1371. [Google Scholar] [CrossRef]
- Todd, E.V.; Liu, H.; Lamm, M.S.; Thomas, J.T.; Rutherford, K.; Thompson, K.C.; Godwin, J.R.; Gemmell, N.J. Female Mimicry by Sneaker Males Has a Transcriptomic Signature in Both the Brain and the Gonad in a Sex-Changing Fish. Mol. Biol. Evol. 2018, 35, 225–241. [Google Scholar] [CrossRef]
- Wang, Z.; Qiu, X.; Kong, D.; Zhou, X.; Guo, Z.; Gao, C.; Ma, S.; Hao, W.; Jiang, Z.; Liu, S.; et al. Comparative RNA-Seq analysis of differentially expressed genes in the testis and ovary of Takifugu rubripes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.F.; Liu, S.K.; Yao, J.; Zhang, Y.; Yuan, Z.H.; Jiang, C.; Chen, A.L.; Fu, Q.; Su, B.F.; Dunham, R.; et al. Transcriptome Display During Testicular Differentiation of Channel Catfish (Ictalurus punctatus) as Revealed by RNA-Seq Analysis. Biol. Reprod. 2016, 95, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Culture and Biology of Large Yellow Croaker; Xiamen University Press: Xiamen, China, 2013. [Google Scholar]
- Zhan, S.; Zhao, W.; Song, T.; Dong, Y.; Guo, J.; Cao, J.; Zhong, T.; Wang, L.; Li, L.; Zhang, H. Dynamic transcriptomic analysis in hircine longissimus dorsi muscle from fetal to neonatal development stages. Funct. Integr. Genom. 2018, 18, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Hao, Y.; Yin, Z.; Wu, G.; Jiang, X. Transcriptome of wax apple (Syzygium samarangense) provides insights into nitric oxide-induced delays of postharvest cottony softening. Acta Physiol. Plant. 2017, 39, 273. [Google Scholar] [CrossRef]
- Storey, J.D. The Positive False Discovery Rate: A Bayesian Interpretation and the q-Value. Ann. Stat. 2003, 31, 2013–2035. [Google Scholar] [CrossRef]
- Billard, R. Spermatogenesis and spermatology of some teleost fish species. Reprod. Nutr. Dev. 1986, 26, 877–920. [Google Scholar] [CrossRef]
- Du, X.X.; Wang, B.; Liu, X.M.; Liu, X.B.; He, Y.; Zhang, Q.Q.; Wang, X.B. Comparative transcriptome analysis of ovary and testis reveals potential sex-related genes and pathways in spotted knifejaw Oplegnathus punctatus. Gene 2017, 637, 203–210. [Google Scholar] [CrossRef]
- Li, Y.H.; Wang, H.P.; Yao, H.; O’Bryant, P.; Rapp, D.; Guo, L.; Waly, E.A. De novo transcriptome sequencing and analysis of male, pseudo-male and female yellow perch, Perca flavescens. PLoS ONE 2017, 12, e0171187. [Google Scholar] [CrossRef]
- Sun, L.X.; Wang, Y.Y.; Zhao, Y.; Wang, H.; Li, N.; Ji, X.S. Global DNA Methylation Changes in Nile Tilapia Gonads during High Temperature-Induced Masculinization. PLoS ONE 2016, 11, e0158483. [Google Scholar] [CrossRef]
- Liu, Z.H.; Chen, Q.L.; Chen, Q.; Li, F.; Li, Y.W. Diethylstilbestrol arrested spermatogenesis and somatic growth in the juveniles of yellow catfish (Pelteobagrus fulvidraco), a fish with sexual dimorphic growth. Fish Physiol. Biochem. 2018, 44, 789–803. [Google Scholar] [CrossRef]
- Taranger, G.L.; Carrillo, M.; Schulz, R.W.; Fontaine, P.; Zanuy, S.; Felip, A.; Weltzien, F.A.; Dufour, S.; Karlsen, O.; Norberg, B.; et al. Control of puberty in farmed fish. Gen. Comp. Endocrinol. 2010, 165, 483–515. [Google Scholar] [CrossRef] [PubMed]
- Stamatiades, G.A.; Carroll, R.S.; Kaiser, U.B. GnRH—A Key Regulator of FSH. Endocrinology 2019, 160, 57–67. [Google Scholar] [CrossRef] [PubMed]
- McArdle, C.A.; Roberson, M.S. Chapter 10—Gonadotropes and Gonadotropin-Releasing Hormone Signaling. In Knobil and Neill’s Physiology of Reproduction, 4th ed.; Plant, T.M., Zeleznik, A.J., Eds.; Academic Press: San Diego, CA, USA, 2015; pp. 335–397. [Google Scholar] [CrossRef]
- He, L.; Jiang, H.; Cao, D.D.; Liu, L.H.; Hu, S.N.; Wang, Q. Comparative Transcriptome Analysis of the Accessory Sex Gland and Testis from the Chinese Mitten Crab (Eriocheir sinensis). PLoS ONE 2013, 8, e53915. [Google Scholar] [CrossRef] [PubMed]
- Takenaka, K.; Gotoh, Y.; Nishida, E. MAP kinase is required for the spindle assembly checkpoint but is dispensable for the normal M phase entry and exit in Xenopus egg cell cycle extracts. J. Cell Biol. 1997, 136, 1091–1097. [Google Scholar] [CrossRef]
- Gupta, G.S. Action of Phospholipases. In Proteomics of Spermatogenesis; Springer: Boston, MA, USA, 2005; pp. 539–554. [Google Scholar] [CrossRef]
- Lie, P.P.Y.; Cheng, C.Y.; Mruk, D.D. Coordinating cellular events during spermatogenesis: A biochemical model. Trends Biochem. Sci. 2009, 34, 366–373. [Google Scholar] [CrossRef]
- Sabeur, K.; Ball, B.A.; Corbin, C.J.; Conley, A. Characterization of a novel, testis-specific equine serine/threonine kinase. Mol. Reprod. Dev. 2008, 75, 867–873. [Google Scholar] [CrossRef]
- Jha, K.N.; Coleman, A.R.; Wong, L.; Salicioni, A.M.; Howcroft, E.; Johnson, G.R. Heat shock protein 90 functions to stabilize and activate the testis-specific serine/threonine kinases, a family of kinases essential for male fertility. J. Biol. Chem. 2013, 288, 16308–16320. [Google Scholar] [CrossRef]
- Shang, P.; Baarends, W.M.; Hoogerbrugge, J.; Ooms, M.P.; van Cappellen, W.A.; de Jong, A.A.W.; Dohle, G.R.; van Eenennaam, H.; Gossen, J.A.; Grootegoed, J.A. Functional transformation of the chromatoid body in mouse spermatids requires testis-specific serine/threonine kinases. J. Cell Sci. 2010, 123, 331–339. [Google Scholar] [CrossRef]
- Alekseev, O.M.; Richardson, R.T.; O’Rand, M.G. Linker histones stimulate HSPA2 ATPase activity through NASP binding and inhibit CDC2/Cyclin B1 complex formation during meiosis in the mouse. Biol. Reprod. 2009, 81, 739–748. [Google Scholar] [CrossRef]
- Hirokawa, N.; Takemura, R. Kinesin superfamily proteins and their various functions and dynamics. Exp. Cell Res. 2004, 301, 50–59. [Google Scholar] [CrossRef]
- Kardon, J.R.; Vale, R.D. Regulators of the cytoplasmic dynein motor. Nat. Rev. Mol. Cell Biol. 2009, 10, 854–865. [Google Scholar] [CrossRef] [PubMed]
- Melkov, A.; Abdu, U. Regulation of long-distance transport of mitochondria along microtubules. Cell. Mol. Life Sci. 2018, 75, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.P.; Diamant, A.G.; Urnavicius, L. How dynein and dynactin transport cargos: A structural perspective. Curr. Opin. Struct. Biol. 2016, 37, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Wen, Q.; Tang, E.I.; Lui, W.-y.; Lee, W.M.; Wong, C.K.C.; Silvestrini, B.; Cheng, C.Y. Dynein 1 supports spermatid transport and spermiation during spermatogenesis in the rat testis. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E924–E948. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.J.; Kon, T.; Knight, P.J.; Sutoh, K.; Burgess, S.A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 2013, 14, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Kikkawa, M. The role of microtubules in processive kinesin movement. Trends Cell Biol. 2008, 18, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.R.; Yang, W.X. Myosin superfamily: The multi-functional and irreplaceable factors in spermatogenesis and testicular tumors. Gene 2016, 576, 195–207. [Google Scholar] [CrossRef]
- Lie, P.P.; Mruk, D.D.; Lee, W.M.; Cheng, C.Y. Cytoskeletal dynamics and spermatogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 1581–1592. [Google Scholar] [CrossRef]
- Dunleavy, J.E.M.; O’Bryan, M.; Stanton, P.G.; O’Donnell, L. The Cytoskeleton in Spermatogenesis. Reproduction 2018, 157, R53–R72. [Google Scholar] [CrossRef]
- Venditti, M.; Minucci, S. Subcellular Localization of Prolyl Endopeptidase During the First Wave of Rat Spermatogenesis and in Rat and Human Sperm. J. Histochem. Cytochem. 2019, 67, 229–243. [Google Scholar] [CrossRef]
- Kierszenbaum, A.L.; Tres, L.L. The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch. Histol. Cytol. 2004, 67, 271–284. [Google Scholar] [CrossRef] [PubMed]
- Kierszenbaum, A.L.; Rivkin, E.; Tres, L.L. The actin-based motor myosin Va is a component of the acroplaxome, an acrosome-nuclear envelope junctional plate, and of manchette-associated vesicles. Cytogenet. Genome Res. 2003, 103, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Woolner, S.; O’Brien, L.C.; Bement, W. Myosin-10 and actin filaments are essential for mitotic spindle function. J. Cell Biol. 2008, 182, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; He, Y.; Hou, L.; Yang, W. Myosin Va participates in acrosomal formation and nuclear morphogenesis during spermatogenesis of Chinese mitten crab Eriocheir sinensis. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Isaji, M.; Lenartowska, M.; Noguchi, T.; Frank, D.J.; Miller, K.G. Myosin VI Regulates Actin Structure Specialization through Conserved Cargo-Binding Domain Sites. PLoS ONE 2011, 6, e22755. [Google Scholar] [CrossRef]
- Noguchi, T.; Lenartowska, M.; Miller, K.G. Myosin VI Stabilizes an Actin Network during Drosophila Spermatid Individualization. Mol. Biol. Cell 2006, 17, 2559–2571. [Google Scholar] [CrossRef]
- Velichkova, M.; Guttman, J.A.; Warren, C.; Eng, L.; Kline, K.; Vogl, A.W.; Hasson, T. A human homologue of Drosophila kelch associates with myosin-VIIa in specialized adhesion junctions. Cytoskeleton 2002, 51, 147–164. [Google Scholar] [CrossRef]
- Wu, Y.; Pei, Y.; Qin, Y. Developmental expression of heat shock proteins 60, 70, 90, and A2 in rabbit testis. Cell Tissue Res. 2011, 344, 355–363. [Google Scholar] [CrossRef]
- Domingos, F.F.T.; Thomé, R.G.; Martinelli, P.M.; Sato, Y.; Bazzoli, N.; Rizzo, E. Role of HSP70 in the regulation of the testicular apoptosis in a seasonal breeding teleost Prochilodus argenteus from the São Francisco river, Brazil. Microsc. Res. Tech. 2013, 76, 350–356. [Google Scholar] [CrossRef]
- Cao, W.; Huang, P.; Zhang, L.; Wu, H.Z.; Zhang, J.; Shi, F.X. Acute Heat Stress Increases HSP70 Expression in the Testis, Epididymis and Vas Deferens of Adult Male Mice. Natl. J. Androl. 2009, 15, 200. [Google Scholar]
- Mezquita, B.; Mezquita, J.; Durfort, M.; Mezquita, C. Constitutive and heat-shock induced expression of Hsp70 mRNA during chicken testicular development and regression. J. Cell. Biochem. 2001, 82, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Erata, G.O.; Kocak Toker, N.; Durlanik, O.; Kadioglu, A.; Aktan, G.; Aykac Toker, G. The role of heat shock protein 70 (Hsp 70) in male infertility: Is it a line of defense against sperm DNA fragmentation? Fertil. Steril. 2008, 90, 322–327. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Wang, Q.; Jin, X.; Wang, Y.; Chen, L.; Liu, L.; Wang, Y. Transcriptome Profiling of Testis during Sexual Maturation Stages in Eriocheir sinensis Using Illumina Sequencing. PLoS ONE 2012, 7, e33735. [Google Scholar] [CrossRef]
- Yang, F.; De La Fuente, R.; Leu, N.A.; Baumann, C.; McLaughlin, K.J.; Wang, P.J. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006, 173, 497–507. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H.; Yang, H.H.; Li, M.R.; Sun, Y.L.; Jiang, X.L.; Xie, Q.P.; Wang, T.R.; Shi, H.J.; Sun, L.N.; Zhou, L.Y.; et al. Antagonistic roles of Dmrt1 and Foxl2 in sex differentiation via estrogen production in tilapia as demonstrated by TALENs. Endocrinology 2013, 154, 4814–4825. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, G.; Shao, C.; Huang, Q.; Liu, G.; Zhang, P.; Song, W.; An, N.; Chalopin, D.; Volff, J.N.; et al. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 2014, 46, 253–260. [Google Scholar] [CrossRef]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 2009, 461, 267–271. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Lee, K.H.; Fujimoto, H.; Kadomura, K.; Yasumoto, S.; Matsuyama, M. Expression of the DMRT gene and its roles in early gonadal development of the Japanese pufferfish Takifugu Rubripes. Comp. Biochem. Physiol. Part D Genom. Proteom. 2006, 1, 59–68. [Google Scholar] [CrossRef]
- Groh, K.J.; Schonenberger, R.; Eggen, R.I.; Segner, H.; Suter, M.J. Analysis of protein expression in zebrafish during gonad differentiation by targeted proteomics. Gen. Comp. Endocrinol. 2013, 193, 210–220. [Google Scholar] [CrossRef]
- Liu, S.F.; He, S.; Liu, B.W.; Zhao, Y.; Wang, Z. Cloning and characterization of testis-specific spermatogenesis associated gene homologous to human SPATA4 in rat. Biol. Pharm. Bull. 2004, 27, 1867–1870. [Google Scholar] [CrossRef][Green Version]
- Xie, M.C.; Ai, C.; Jin, X.M.; Liu, S.F.; Tao, S.X.; Li, Z.D.; Wang, Z. Cloning and characterization of chicken SPATA4 gene and analysis of its specific expression. Mol. Cell. Biochem. 2007, 306, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Kotov, A.A.; Akulenko, N.V.; Kibanov, M.V.; Olenina, L.V. DEAD-Box RNA helicases in animal gametogenesis. Mol. Biol. 2014, 48, 16–28. [Google Scholar] [CrossRef]
- Lee, R.; Lee, W.Y.; Park, H.J.; Ha, W.T.; Woo, J.S.; Chung, H.J.; Lee, J.H.; Hong, K.; Song, H. Stage-specific expression of DDX4 and c-kit at different developmental stages of the porcine testis. Anim. Reprod. Sci. 2018, 190, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Bártfai, R.; Orbán, L. The vasa locus in zebrafish: Multiple RGG boxes from intragenic duplications. DNA Cell Biol. 2003, 22, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Tan, S.; Bao, L.; Zeng, Q.; Liu, S.; Yang, Y.; Zhong, X.; Liu, Z. DExD/H-box RNA helicase genes are differentially expressed between males and females during the critical period of male sex differentiation in channel catfish. Comp. Biochem. Physiol. Part D Genom. Proteom. 2017, 22, 109–119. [Google Scholar] [CrossRef]
- Tanaka, S.S.; Toyooka, Y.; Akasu, R.; Katohfukui, Y.; Nakahara, Y.; Suzuki, R.; Yokoyama, M.; Noce, T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000, 14, 841–853. [Google Scholar] [CrossRef]
- Rolland, A.D.; Lareyre, J.; Goupil, A.; Montfort, J.; Ricordel, M.; Esquerre, D.; Hugot, K.; Houlgatte, R.; Chalmel, F.; Gac, F.L. Expression profiling of rainbow trout testis development identifies evolutionary conserved genes involved in spermatogenesis. BMC Genom. 2009, 10, 546. [Google Scholar] [CrossRef]
- Kojima, K.; Kuramochi-Miyagawa, S.; Chuma, S.; Tanaka, T.; Nakatsuji, N.; Kimura, T.; Nakano, T. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 2009, 14, 1155–1165. [Google Scholar] [CrossRef]
- Hosokawa, M.; Shoji, M.; Kitamura, K.; Tanaka, T.; Noce, T.; Chuma, S.; Nakatsuji, N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007, 301, 38–52. [Google Scholar] [CrossRef]
- Tanaka, T.; Hosokawa, M.; Vagin, V.V.; Reuter, M.; Hayashi, E.; Mochizuki, A.L.; Kitamura, K.; Yamanaka, H.; Kondoh, G.; Okawa, K.; et al. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 10579–10584. [Google Scholar] [CrossRef]
- Bak, C.W.; Yoon, T.-K.; Choi, Y. Functions of PIWI proteins in spermatogenesis. Clin. Exp. Reprod. Med. 2011, 38, 61–67. [Google Scholar] [CrossRef] [PubMed]


| Item | Description |
|---|---|
| Investigation type | Eukaryote |
| Project name | Transcriptome for L. crocea |
| Collection date | May 2016 |
| Lat_lon | 29°86′ N, 121°56′ E |
| Geo loc name | Ningbo, China |
| Environment | Marine water |
| Biotic_relationship | Free-living |
| Trophic level | Heterotroph |
| Temp | 21 °C |
| Salinity | 24 PSU |
| Estimated size | 16.35 Gb |
| Sequencing meth | Illumina HiSeqTM 2500 |
| Mapping software | TopHat2 |
| Annotation source | Nr/Nt/Swiss-Prot/KEGG/COG/GO |
| BioProject ID of raw reads | PRJNA471154 for TES/PRJNA471574 for PMT |
| Accession number of raw reads | SRP148410 for TES/SRP148493 for PMT |
| Gene ID | Annotation | Primer | Sequence (5′→3′) |
|---|---|---|---|
| gene15399 | tex9 | F | GCTGTAGACGACTCGGCTGACTT |
| R | TGAGCATCTGAAACGCCTGATCCT | ||
| gene14348 | tex36 | F | GCAAGGAGTTGTCACACTGGCATC |
| R | TCCATCGTGGCACAGGCAGAAG | ||
| gene2626 | tex11 | F | TCGGTGAAGTCTCTGCTCTGGAAG |
| R | GGACGCCCTCTGTTGGATTCTCA | ||
| gene21265 | tcte1 | F | TGATCGGAGACAGAGGAGCCAGAG |
| R | CGGTTGAGACGCAGGTTGAGTGA | ||
| gene20395 | stpg2 | F | GCAGTCTCCAGAACCGCTCCAA |
| R | CAGTGTGTCCTCCTCGTAGCCAAA | ||
| gene3104 | spata7 | F | CTGAGGATGAGTCCAACGGCACAT |
| R | AGATTTCCCGCCTTCTGGTGAGAC | ||
| gene26146 | spata20 | F | GTTCCTGGACGACTACGCCTTCAT |
| R | TGGACGCCGACACTGAGTTAGC | ||
| gene26767 | spag5 | F | GGACATCCAGCAAGCCAATGACAG |
| R | CCTCGCCAACTCGTTCTCCATCT | ||
| gene26247 | spag17 | F | CCAGACGAGGAGGAGGACAGAGAA |
| R | TTCAGGATGGTGATGCCGAACTCA | ||
| gene16701 | pabp1l | F | AGTCCGCTAATGGAGGCTCTGTC |
| R | AGTGGTGGTCCTTGTGGTTGATGT | ||
| gene10808 | nme5h | F | TCCACGGCAGCGAGTCATTTCAT |
| R | TCAGCCAGTCAGCAAGCCAGATAC | ||
| gene21216 | meig1 | F | ACAACTCCAAGCCGAAGTCCATGA |
| R | TTGACATCACGGTCCAGGCACTC | ||
| gene13149 | lhcgrl | F | TGGCATCAAGGAGGTGGCAAGT |
| R | TGGTAGGCGGACTCTGCGATCA | ||
| gene1213 | hsp40a1 | F | AGGTCGTGGGAGTCGGAAAGGA |
| R | TGTGGACACTTGCTGGACCATACC | ||
| gene17675 | ggn | F | GCTGAAGTGCCACCTGAGTCACA |
| R | GCCGCTGTTGTATTGCTGCTCTG | ||
| XM_019257255.1 | β-actin | F | CTGTCCCTGTATGCCTCTGGTC |
| R | CTTGATGTCACGCACGATTTCC |
| Samples | TES | PMT |
|---|---|---|
| Clean reads | 26,201,689 | 28,786,700 |
| Clean bases (Gb) | 7.8 | 8.5 |
| GC percentage | 50.73% | 50.16% |
| % ≥ Q20 | 93.38% | 93.64% |
| % ≥ Q30 | 85.81% | 86.27% |
| Total mapped | 33,390,310 (63.72%) | 38,470,859 (66.82%) |
| Uniquely mapped | 30,802,638 (58.78%) | 35,380,000 (61.45%) |
| GO.ID | The Third-Level Functional Categories | All Gene Number | DEG Number | KSa |
|---|---|---|---|---|
| GO:0001539 | cilium or flagellum-dependent cell motility | 18 | 12 | 4.60E-06 |
| GO:0007018 | microtubule-based movement | 61 | 31 | 7.90E-05 |
| GO:0007283 | spermatogenesis | 20 | 11 | 0.00017 |
| GO:0003006 | developmental process involved in reproduction | 70 | 23 | 0.0035 |
| GO:0048515 | spermatid differentiation | 10 | 5 | 0.00452 |
| GO:0007126 | meiotic nuclear division | 22 | 10 | 0.00474 |
| GO:0044702 | single organism reproductive process | 89 | 30 | 0.00857 |
| GO:0071695 | anatomical structure maturation | 6 | 1 | 0.00991 |
| GO:0007286 | spermatid development | 9 | 4 | 0.01166 |
| GO:0046903 | secretion | 64 | 8 | 0.01212 |
| GO:0090068 | positive regulation of cell cycle process | 12 | 2 | 0.01577 |
| GO:0033143 | regulation of intracellular steroid hormone receptor signaling pathway | 6 | 1 | 0.01586 |
| GO:0007281 | germ cell development | 29 | 11 | 0.01627 |
| GO:0035265 | organ growth | 11 | 4 | 0.01688 |
| GO:0000910 | cytokinesis | 25 | 7 | 0.018 |
| GO:0051495 | positive regulation of cytoskeleton organization | 23 | 4 | 0.01847 |
| GO:0048729 | tissue morphogenesis | 289 | 50 | 0.01997 |
| GO:0002009 | morphogenesis of an epithelium | 236 | 38 | 0.02012 |
| GO:0051653 | spindle localization | 12 | 1 | 0.02114 |
| GO:0048608 | reproductive structure development | 38 | 9 | 0.02247 |
| GO:0061458 | reproductive system development | 38 | 9 | 0.02247 |
| GO:0007010 | cytoskeleton organization | 223 | 50 | 0.02305 |
| GO:0045010 | actin nucleation | 14 | 4 | 0.02346 |
| GO:0007548 | sex differentiation | 33 | 9 | 0.02645 |
| GO:0060070 | canonical Wnt signaling pathway | 77 | 12 | 0.03034 |
| GO:0008406 | gonad development | 28 | 8 | 0.03043 |
| GO:0060029 | convergent extension involved in organogenesis | 12 | 1 | 0.03087 |
| GO:0051493 | regulation of cytoskeleton organization | 63 | 11 | 0.03124 |
| GO:0007017 | microtubule-based process | 170 | 58 | 0.03224 |
| GO:0030029 | actin filament-based process | 132 | 30 | 0.0363 |
| GO:0046330 | positive regulation of JNK cascade | 11 | 1 | 0.04252 |
| GO:0030036 | actin cytoskeleton organization | 130 | 30 | 0.04805 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Gao, X.; Ding, J.; Liu, C.; Du, C.; Hou, C.; Zhu, J.; Lou, B. Transcriptome Sequencing Reveals the Traits of Spermatogenesis and Testicular Development in Large Yellow Croaker (Larimichthys crocea). Genes 2019, 10, 958. https://doi.org/10.3390/genes10120958
Luo S, Gao X, Ding J, Liu C, Du C, Hou C, Zhu J, Lou B. Transcriptome Sequencing Reveals the Traits of Spermatogenesis and Testicular Development in Large Yellow Croaker (Larimichthys crocea). Genes. 2019; 10(12):958. https://doi.org/10.3390/genes10120958
Chicago/Turabian StyleLuo, Shengyu, Xinming Gao, Jie Ding, Cheng Liu, Chen Du, Congcong Hou, Junquan Zhu, and Bao Lou. 2019. "Transcriptome Sequencing Reveals the Traits of Spermatogenesis and Testicular Development in Large Yellow Croaker (Larimichthys crocea)" Genes 10, no. 12: 958. https://doi.org/10.3390/genes10120958
APA StyleLuo, S., Gao, X., Ding, J., Liu, C., Du, C., Hou, C., Zhu, J., & Lou, B. (2019). Transcriptome Sequencing Reveals the Traits of Spermatogenesis and Testicular Development in Large Yellow Croaker (Larimichthys crocea). Genes, 10(12), 958. https://doi.org/10.3390/genes10120958





