Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Research Animals
2.2. Production of Lamprey Embryos
2.3. PACT Clearing
2.4. Preparation of Metaphase Spreads
2.5. C0t DNA Isolation
2.6. Computational Prediction of Germline-Specific Repeats
2.7. Fluorescence in Situ Hybridization
3. Results and Discussion
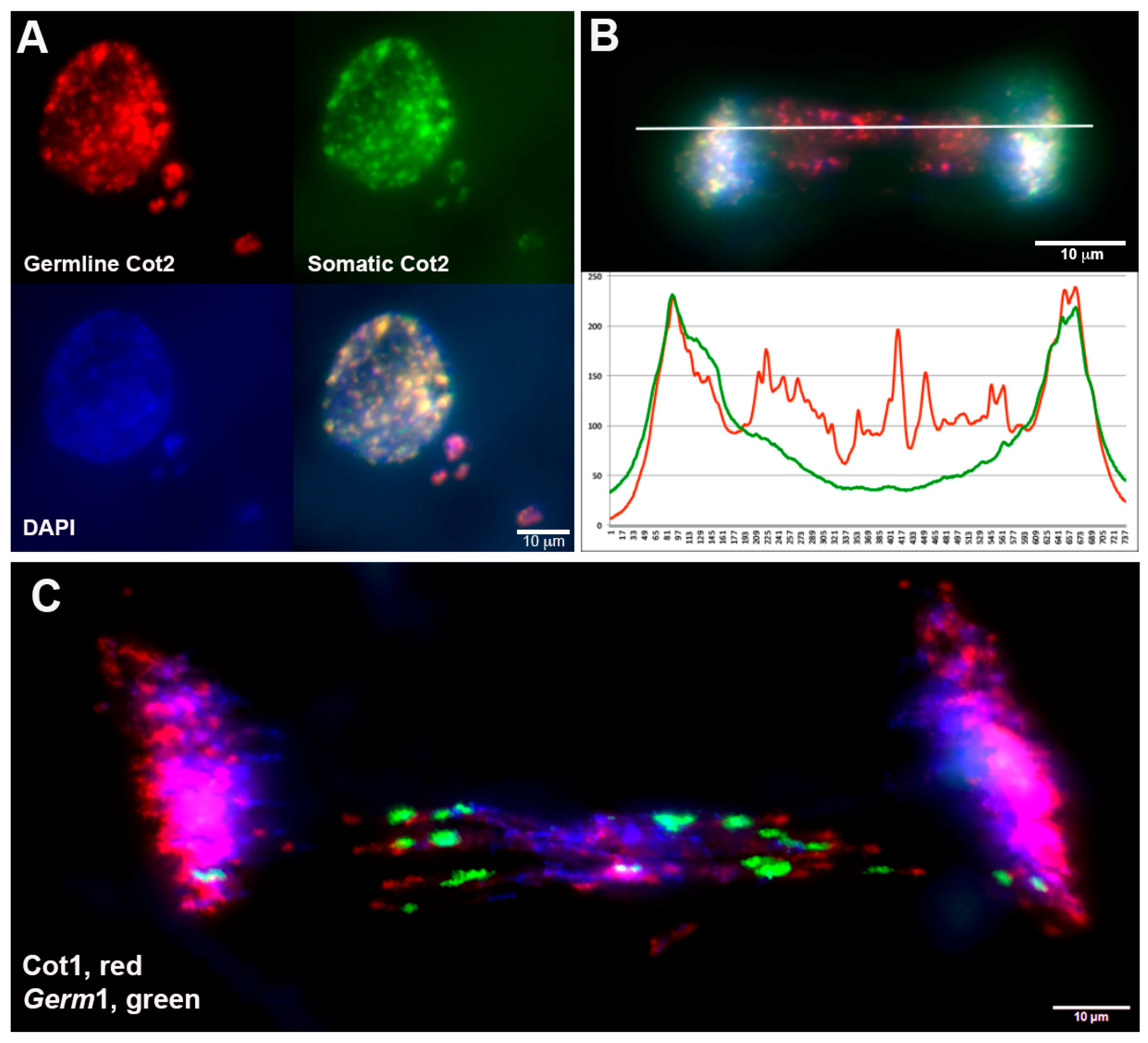
3.1. Comparative Hybridization Indicates the Presence of Germline-Specific Repeats
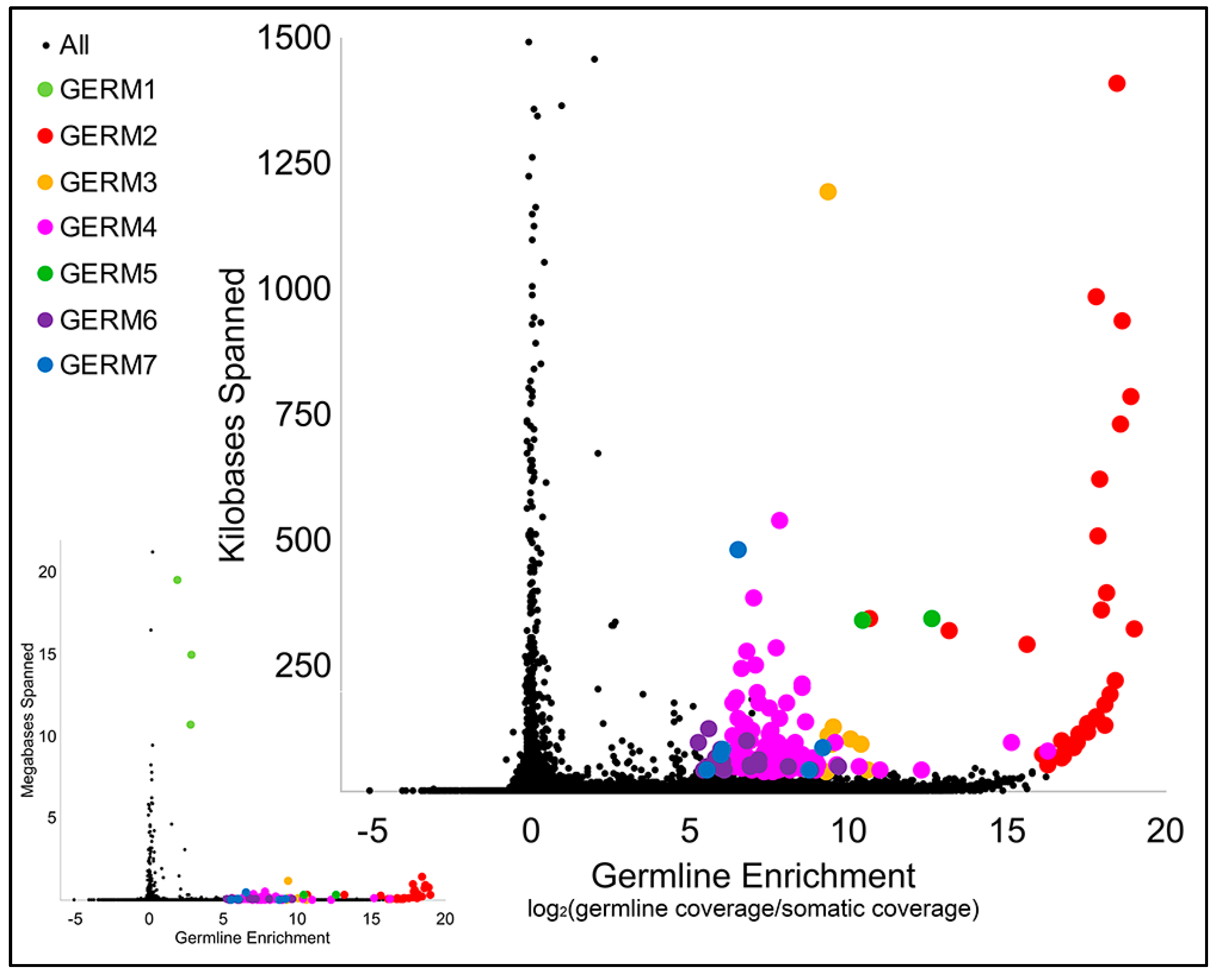
3.2. Computational Identification of Germline-Specific and Centromeric Repeats
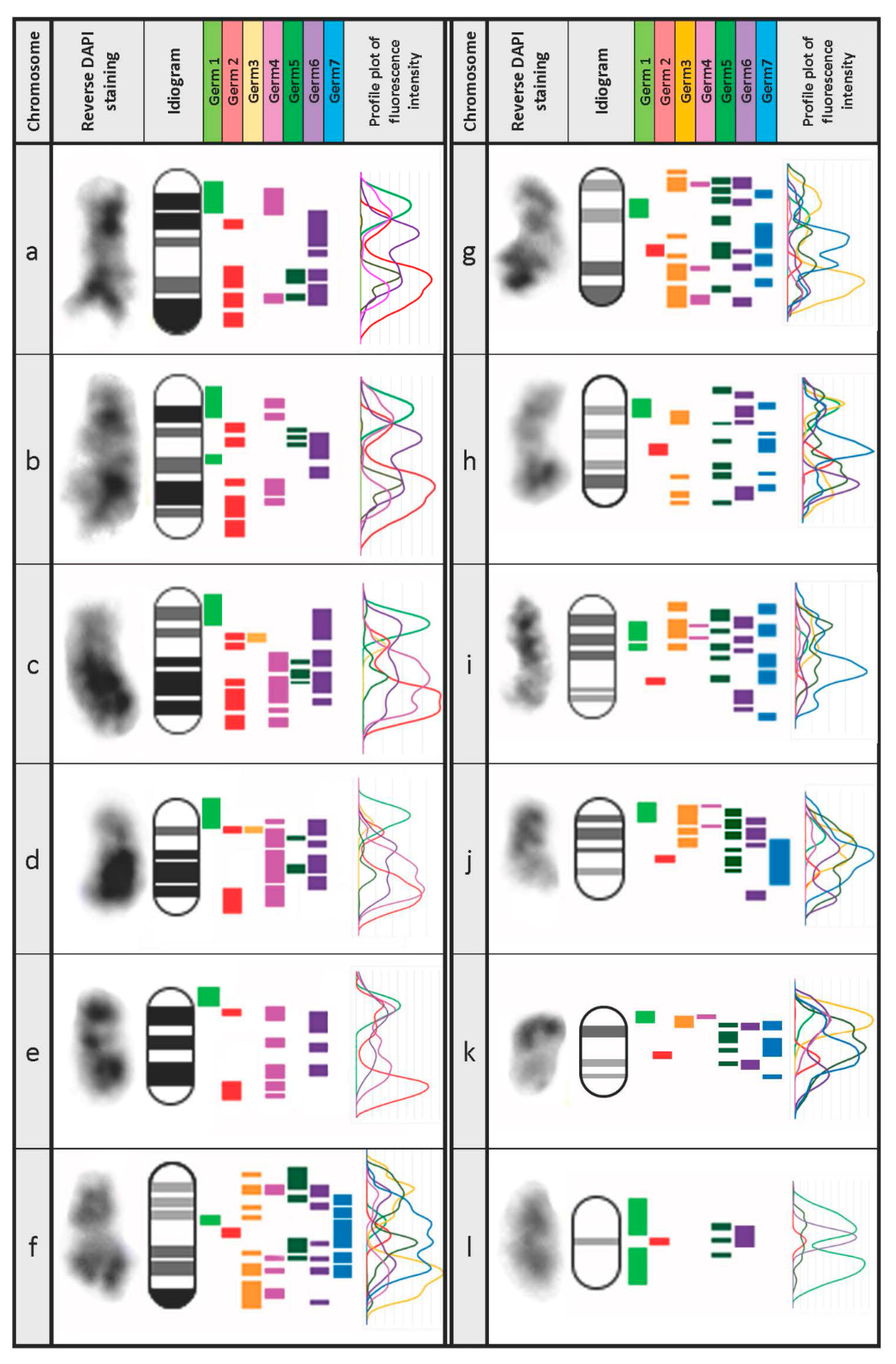
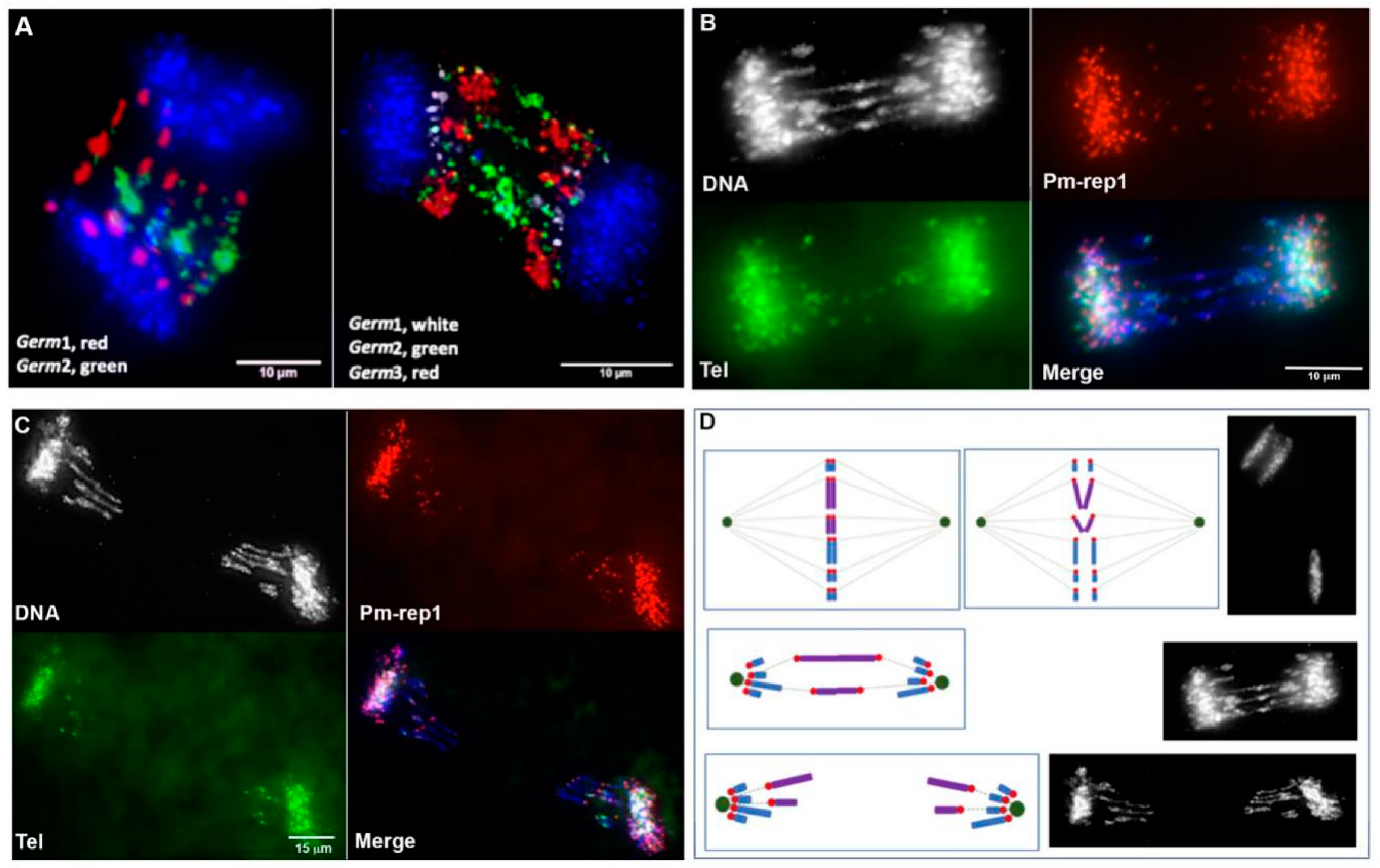
3.3. Defining the Chromosomal Localization of Germline-Restricted Repeats
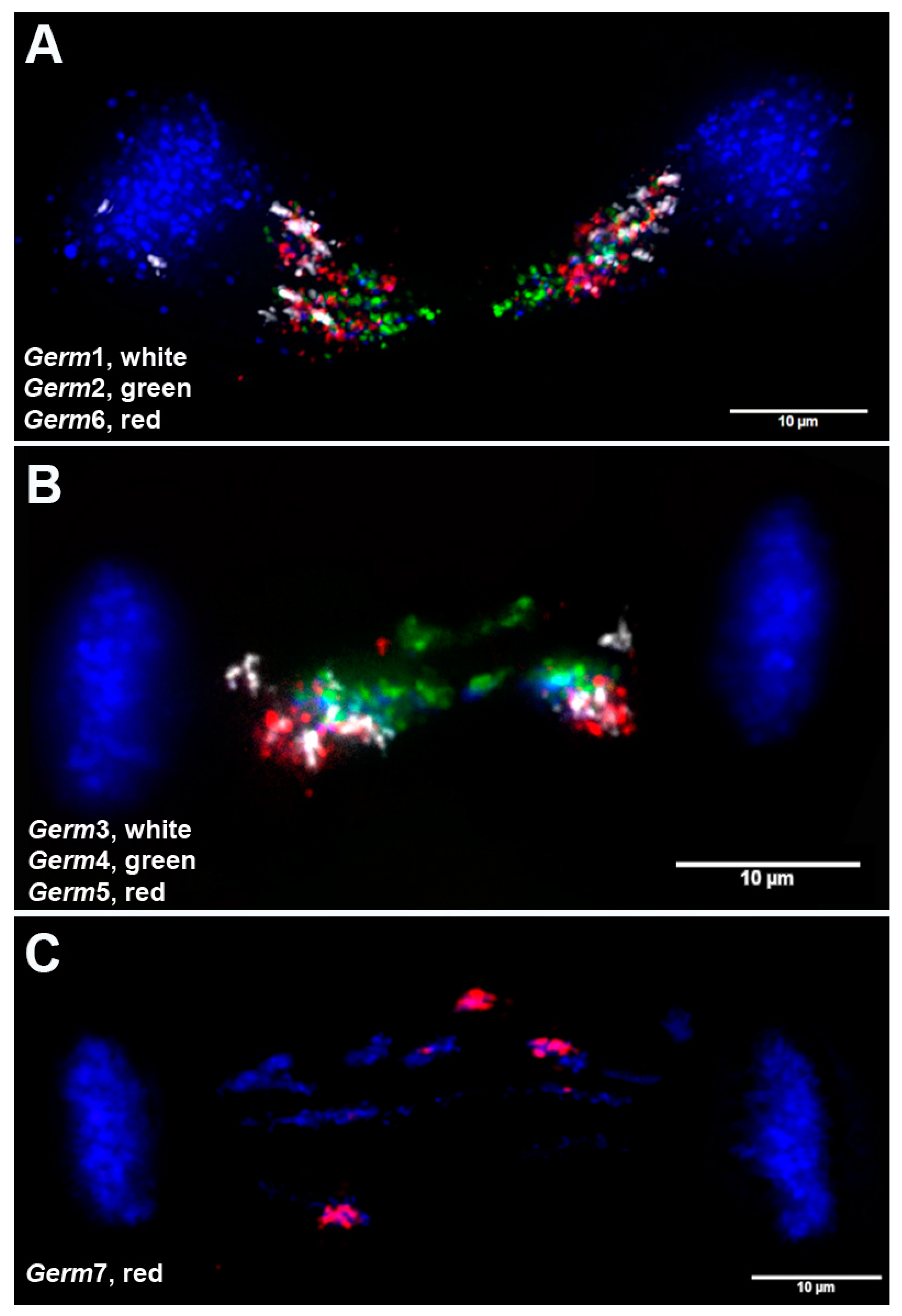
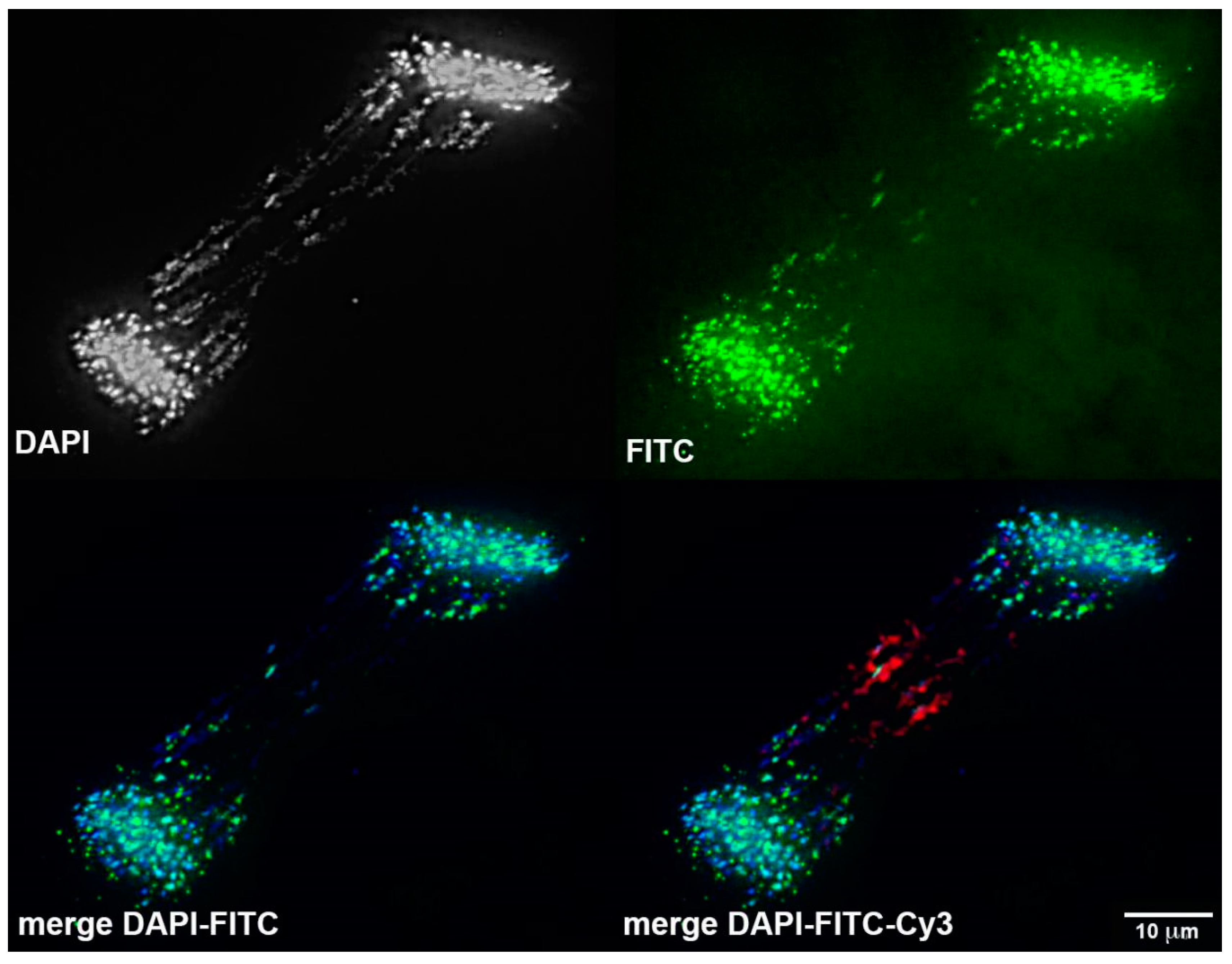
3.4. Chromosomal Localization of Germline-Restricted Repeats in Elimination Anaphases
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, J.J.; Baker, C.; Eichler, E.E.; Amemiya, C.T. Genetic consequences of programmed genome rearrangement. Curr. Biol. 2012, 22, 1524–1529. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Herdy, J.R.; Keinath, M.C.; Smith, J.J. Cellular and molecular features of developmentally programmed genome rearrangement in a vertebrate (sea lamprey: Petromyzon marinus). PLoS Genet. 2016, 12, e1006103. [Google Scholar] [CrossRef]
- Wang, J.; Davis, R.E. Programmed DNA elimination in multicellular organisms. Curr. Opin. Genet. Dev. 2014, 27, 26–34. [Google Scholar] [CrossRef]
- Bracht, J.R.; Fang, W.; Goldman, A.D.; Dolzhenko, E.; Stein, E.M.; Landweber, L.F. Genomes on the edge: Programmed genome instability in ciliates. Cell 2013, 152, 406–416. [Google Scholar] [CrossRef]
- Mochizuki, K.; Gorovsky, M.A. Small rnas in genome rearrangement in tetrahymena. Curr. Opin. Genet. Dev. 2004, 14, 181–187. [Google Scholar] [CrossRef]
- Mochizuki, K. Developmentally programmed, rna-directed genome rearrangement in tetrahymena. Dev. Growth Differ. 2012, 54, 108–119. [Google Scholar] [CrossRef]
- Boveri, T. Uber differenzierung der zellkerne wahrend der furchung des eies von ascaris megalocephala. Anat. Anz. 1887, 2, 688–693. [Google Scholar]
- Streit, A.; Wang, J.; Kang, Y.; Davis, R.E. Gene silencing and sex determination by programmed DNA elimination in parasitic nematodes. Curr. Opin. Microbiol. 2016, 32, 120–127. [Google Scholar] [CrossRef]
- Goday, C.; Esteban, M.R. Chromosome elimination in sciarid flies. Bioessays 2001, 23, 242–250. [Google Scholar] [CrossRef]
- Singh, P.B.; Belyakin, S.N. L chromosome behaviour and chromosomal imprinting in sciara coprophila. Genes 2018, 9, 440. [Google Scholar] [CrossRef]
- Grishanin, A. Chromatin diminution in copepoda (crustacea): Pattern, biological role and evolutionary aspects. Comp. Cytogenet. 2014, 8, 1–10. [Google Scholar] [CrossRef][Green Version]
- Staiber, W. Chromosome elimination in germ line-soma differentiation of acricotopus lucidus (diptera, chironomidae). Genome 2006, 49, 269–274. [Google Scholar] [CrossRef]
- Kohno, S.; Nakai, Y.; Satoh, S.; Yoshida, M.; Kobayashi, H. Chromosome elimination in the japanese hagfish, eptatretus burgeri (agnatha, cyclostomata). Cytogenet. Genome Res. 1986, 41, 209–214. [Google Scholar] [CrossRef]
- Kubota, S.; Nakai, Y.; Sato, N.; Kuroo, M.; Kohno, S. Chromosome elimination in northeast pacific hagfish, eptatretus-stoutii (cyclostomata, agnatha). J. Hered. 1994, 85, 413–415. [Google Scholar] [CrossRef]
- Nakai, Y.; Kubota, S.; Goto, Y.; Ishibashi, T.; Davison, W.; Kohno, S. Chromosome elimination in three baltic, south pacific and north-east pacific hagfish species. Chromosome Res. 1995, 3, 321–330. [Google Scholar] [CrossRef]
- Nakai, Y.; Kubota, S.; Kohno, S. Chromatin diminution and chromosome elimination in four japanese hagfish species. Cytogenet. Genome Res. 1991, 56, 196–198. [Google Scholar] [CrossRef]
- Pigozzi, M.I.; Solari, A.J. Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, taeniopygia guttata. Chromosome Res. 1998, 6, 105–113. [Google Scholar] [CrossRef]
- Torgasheva, A.A.; Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Kizilova, E.A.; Akberdina, E.A.; Pristyazhnyuk, I.E.; Shnaider, E.P.; Volodkina, V.A.; Saifitdinova, A.F.; et al. Germline-restricted chromosome (grc) is widespread among songbirds. Proc. Natl Acad. Sci. USA 2019, 116, 11845–11850. [Google Scholar] [CrossRef]
- Smith, J.J.; Antonacci, F.; Eichler, E.E.; Amemiya, C.T. Programmed loss of millions of base pairs from a vertebrate genome. Proc. Natl. Acad. Sci. USA 2009, 106, 11212–11217. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Lampman, R.T.; Hess, J.E.; Porter, L.L.; Smith, J.J. Deep ancestry of programmed genome rearrangement in lampreys. Dev. Biol. 2017, 429, 31–34. [Google Scholar] [CrossRef]
- Yao, M.C.; Chao, J.L. Rna-guided DNA deletion in tetrahymena: An rnai-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005, 39, 537–559. [Google Scholar] [CrossRef] [PubMed]
- Zufall, R.A.; Robinson, T.; Katz, L.A. Evolution of developmentally regulated genome rearrangements in eukaryotes. J. Exp. Zool. Part B 2005, 304, 448–455. [Google Scholar] [CrossRef] [PubMed]
- Kohno, S.I.; Kubota, S.; Nakai, Y. Chromatin diminution and chromosome elimination in hagfishes. In The Biology of Hagfishes; Jørgensen, J.M., Lomholt, J.P., Eds.; Chapman and Hall: London, UK, 1998; pp. 81–100. [Google Scholar]
- Del Priore, L.; Pigozzi, M.I. Histone modifications related to chromosome silencing and elimination during male meiosis in bengalese finch. Chromosoma 2014, 123, 293–302. [Google Scholar] [CrossRef]
- Muller, F.; Bernard, V.; Tobler, H. Chromatin diminution in nematodes. Bioessays 1996, 18, 133–138. [Google Scholar] [CrossRef]
- Wang, J.; Gao, S.; Mostovoy, Y.; Kang, Y.; Zagoskin, M.; Sun, Y.; Zhang, B.; White, L.K.; Easton, A.; Nutman, T.B.; et al. Comparative genome analysis of programmed DNA elimination in nematodes. Genome Res. 2017, 27, 2001–2014. [Google Scholar] [CrossRef]
- Standiford, D.M. The effects of chromatin diminution on the pattern of c-banding in the chromosomes of acanthocyclops vernalis fischer (copepoda: Crustacea). Genetica 1989, 79, 207–214. [Google Scholar] [CrossRef]
- Wyngaard, G.A.; Gregory, T.R. Temporal control of DNA replication and the adaptive value of chromatin diminution in copepods. J. Exp. Zool. 2001, 291, 310–316. [Google Scholar] [CrossRef]
- Staiber, W.; Schiffkowski, C. Structural evolution of the germ line-limited chromosomes in acricotopus. Chromosoma 2000, 109, 343–349. [Google Scholar] [CrossRef]
- Kojima, N.F.; Kojima, K.K.; Kobayakawa, S.; Higashide, N.; Hamanaka, C.; Nitta, A.; Koeda, I.; Yamaguchi, T.; Shichiri, M.; Kohno, S.; et al. Whole chromosome elimination and chromosome terminus elimination both contribute to somatic differentiation in taiwanese hagfish paramyxine sheni. Chromosome Res. 2010, 18, 383–400. [Google Scholar] [CrossRef]
- Chmielewska, M.; Dedukh, D.; Haczkiewicz, K.; Rozenblut-Koscisty, B.; Kazmierczak, M.; Kolenda, K.; Serwa, E.; Pietras-Lebioda, A.; Krasikova, A.; Ogielska, M. The programmed DNA elimination and formation of micronuclei in germ line cells of the natural hybridogenetic water frog pelophylax esculentus. Sci. Rep. 2018, 8, 7870. [Google Scholar] [CrossRef]
- Tobler, H. The differentiation of germ and somatic cell lines in nematodes. In Germ Line-Soma Differentiation: Results and Problems in Cell Differentiation; Hennig, W., Ed.; Springer: Berlin, Germany, 1986; Volume 13, pp. 1–69. [Google Scholar]
- de Saint Phalle, B.; Sullivan, W. Incomplete sister chromatid separation is the mechanism of programmed chromosome elimination during early sciara coprophila embryogenesis. Development 1996, 122, 3775–3784. [Google Scholar] [PubMed]
- Beermann, S. The diminution of heterochromatic chromosomal segments in cyclops (crustacea, copepoda). Chromosoma 1977, 60, 297–344. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.J.; Timoshevskaya, N.; Ye, C.; Holt, C.; Keinath, M.C.; Parker, H.J.; Cook, M.E.; Hess, J.E.; Narum, S.R.; Lamanna, F.; et al. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 2018, 50, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Biederman, M.K.; Nelson, M.M.; Asalone, K.C.; Pedersen, A.L.; Saldanha, C.J.; Bracht, J.R. Discovery of the first germline-restricted gene by subtractive transcriptomic analysis in the zebra finch, taeniopygia guttata. Curr. Biol. 2018, 28, 1620–1627. [Google Scholar] [CrossRef] [PubMed]
- Potter, I.C.; Rothwell, B. The mitotic chromosomes of the lamprey, petromyzon marinus L. Experientia 1970, 26, 429–430. [Google Scholar] [CrossRef] [PubMed]
- Covelo-Soto, L.; Moran, P.; Pasantes, J.J.; Perez-Garcia, C. Cytogenetic evidences of genome rearrangement and differential epigenetic chromatin modification in the sea lamprey (petromyzon marinus). Genetica 2014, 142, 545–554. [Google Scholar] [CrossRef]
- Smith, J.J.; Stuart, A.B.; Sauka-Spengler, T.; Clifton, S.W.; Amemiya, C.T. Development and analysis of a germline bac resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma 2010, 119, 381–389. [Google Scholar] [CrossRef]
- Nikitina, N.; Bronner-Fraser, M.; Sauka-Spengler, T. The sea lamprey petromyzon marinus: A model for evolutionary and developmental biology. In Emerging Model Organisms: A Laboratory Manual; CSHL Press: Cold Spring Harbor, NY, USA, 2009; Volume 1, pp. 405–429. [Google Scholar]
- Piavis, G.W. Embryological Stages in the Sea Lamprey and Effects of Temperature on Development; U. S. Fish and Wildlife Service: Washington, DC, USA, 1961; pp. 111–143.
- Harland, R.M. In situ hybridization: An improved whole-mount method for xenopus embryos. Methods Cell Biol. 1991, 36, 685–695. [Google Scholar]
- Yang, B.; Treweek, J.B.; Kulkarni, R.P.; Deverman, B.E.; Chen, C.K.; Lubeck, E.; Shah, S.; Cai, L.; Gradinaru, V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 2014, 158, 945–958. [Google Scholar] [CrossRef]
- Claussen, U.; Michel, S.; Muhlig, P.; Westermann, M.; Grummt, U.W.; Kromeyer-Hauschild, K.; Liehr, T. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet. Genome Res. 2002, 98, 136–146. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 3, p. 2344. [Google Scholar]
- Timoshevskiy, V.A.; Severson, D.W.; Debruyn, B.S.; Black, W.C.; Sharakhov, I.V.; Sharakhova, M.V. An integrated linkage, chromosome, and genome map for the yellow fever mosquito aedes aegypti. PLoS Negl. Trop. Dis. 2013, 7, e2052. [Google Scholar] [CrossRef] [PubMed]
- Marcais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Smit, A.H.R. Repeatmodeler Open-1.0. Available online: http://www.repeatmasker.org (accessed on 12 September 2019).
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Timoshevskaya, N. Difcover. Available online: https://github.com/timnat/DifCover (accessed on 12 September 2019).
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef]
- Timoshevskiy, V.A.; Sharma, A.; Sharakhov, I.V.; Sharakhova, M.V. Fluorescent in situ hybridization on mitotic chromosomes of mosquitoes. J. Vis. Exp. 2012, 67, e4215. [Google Scholar] [CrossRef]
- Solovei, I. Fluorescence in situ hybridization (fish) on tissue cryosections. Methods Mol. Biol. 2010, 659, 71–82. [Google Scholar]
- Rooney, D.E.; Czepulkowski, B.H. Human Cytogenetics: A Practical Approach, 2nd ed.; IRL Press: Oxford, UK, 1992. [Google Scholar]
- Keinath, M.C.; Timoshevskiy, V.A.; Timoshevskaya, N.Y.; Tsonis, P.A.; Voss, S.R.; Smith, J.J. Initial characterization of the large genome of the salamander ambystoma mexicanum using shotgun and laser capture chromosome sequencing. Sci. Rep. 2015, 5, 16413. [Google Scholar] [CrossRef]
- Cleland, R. The cytogenetics of oenothera. Adv. Genet. 1963, 11, 147–237. [Google Scholar]
- Syren, R.M.; Luykx, P. Permanent segmental interchange complex in the termite incisitermes schwarzi. Nature 1977, 266, 167–168. [Google Scholar] [CrossRef]
- Gazoni, T.; Gruber, S.L.; Silva, A.P.; Araujo, O.G.; Narimatsu, H.; Strussmann, C.; Haddad, C.F.; Kasahara, S. Cytogenetic analyses of eight species in the genus leptodactylus fitzinger, 1843 (amphibia, anura, leptodactylidae), including a new diploid number and a karyotype with multiple translocations. BMC Genet. 2012, 13, 109. [Google Scholar] [CrossRef] [PubMed]
- Grutzner, F.; Rens, W.; Tsend-Ayush, E.; El-Mogharbel, N.; O’Brien, P.C.; Jones, R.C.; Ferguson-Smith, M.A.; Marshall Graves, J.A. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird z and mammal x chromosomes. Nature 2004, 432, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Rens, W.; O’Brien, P.C.; Grutzner, F.; Clarke, O.; Graphodatskaya, D.; Tsend-Ayush, E.; Trifonov, V.A.; Skelton, H.; Wallis, M.C.; Johnston, S.; et al. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian z. Genome Biol. 2007, 8, R243. [Google Scholar] [CrossRef] [PubMed]
- Ashley, T. Chromosome chains and platypus sex: Kinky connections. Bioessays 2005, 27, 681–684. [Google Scholar] [CrossRef]
- Eaker, S.; Pyle, A.; Cobb, J.; Handel, M.A. Evidence for meiotic spindle checkpoint from analysis of spermatocytes from robertsonian-chromosome heterozygous mice. J. Cell Sci. 2001, 114, 2953–2965. [Google Scholar]
- Johannisson, R.; Winking, H. Synaptonemal complexes of chains and rings in mice heterozygous for multiple robertsonian translocations. Chromosome Res. 1994, 2, 137–145. [Google Scholar] [CrossRef]
- Cenci, G.; Ciapponi, L.; Gatti, M. The mechanism of telomere protection: A comparison between drosophila and humans. Chromosoma 2005, 114, 135–145. [Google Scholar] [CrossRef]
- Cenci, G.; Siriaco, G.; Raffa, G.D.; Kellum, R.; Gatti, M. The drosophila hoap protein is required for telomere capping. Nat. Cell Biol. 2003, 5, 82–84. [Google Scholar] [CrossRef]
- Iglesias, N.; Moazed, D. Silencing repetitive DNA. eLife 2017, 6, e29503. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timoshevskiy, V.A.; Timoshevskaya, N.Y.; Smith, J.J. Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes 2019, 10, 832. https://doi.org/10.3390/genes10100832
Timoshevskiy VA, Timoshevskaya NY, Smith JJ. Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes. 2019; 10(10):832. https://doi.org/10.3390/genes10100832
Chicago/Turabian StyleTimoshevskiy, Vladimir A., Nataliya Y. Timoshevskaya, and Jeramiah J. Smith. 2019. "Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus)" Genes 10, no. 10: 832. https://doi.org/10.3390/genes10100832
APA StyleTimoshevskiy, V. A., Timoshevskaya, N. Y., & Smith, J. J. (2019). Germline-Specific Repetitive Elements in Programmatically Eliminated Chromosomes of the Sea Lamprey (Petromyzon marinus). Genes, 10(10), 832. https://doi.org/10.3390/genes10100832




