Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy
Abstract
1. Introduction
2. Materials and Methods
2.1. Care and Management of Mares
2.2. RNA Isolation and Quantification
2.3. RNA Sequencing
2.4. Bioinformatic Analysis
3. Results
3.1. Sequencing Results
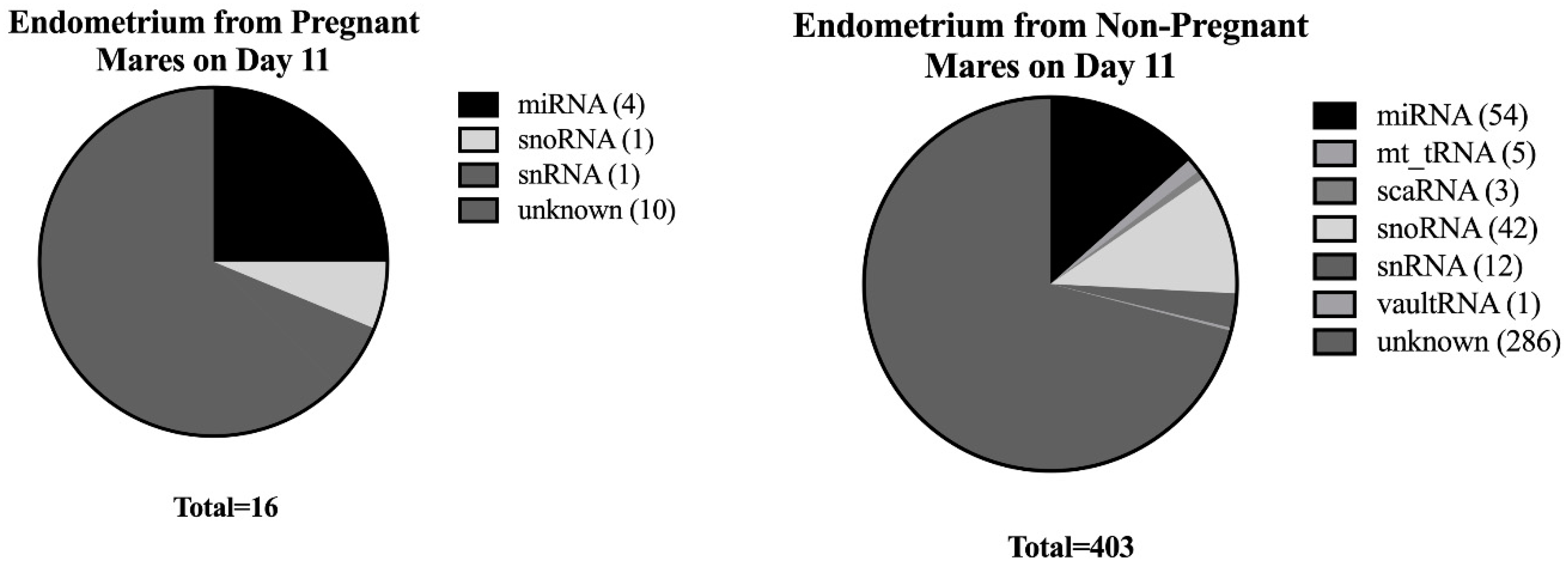
3.2. Small RNA Transcript Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McCracken, J.A.; Custer, E.E.; Lamsa, J.C. Luteolysis: A neuroendocrine-mediated event. Physiol. Rev. 1999, 79, 263–323. [Google Scholar] [CrossRef] [PubMed]
- Sharp, D.C.; Thatcher, M.J.; Salute, M.E.; Fuchs, A.R. Relationship between endometrial oxytocin receptors and oxytocin-induced prostaglandin F2 alpha release during the oestrous cycle and early pregnancy in pony mares. J. Reprod. Fertil. 1997, 109, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Rambags, B.P.; Krijtenburg, P.J.; Drie, H.F.; Lazzari, G.; Galli, C.; Pearson, P.L.; Colenbrander, B.; Stout, T.A. Numerical chromosomal abnormalities in equine embryos produced in vivo and in vitro. Mol. Reprod. Dev. 2005, 72, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Allen, W.R.; Stewart, F. Equine placentation. Reprod. Fertil. Dev. 2001, 13, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Betteridge, K.J.; Eaglesome, M.D.; Mitchell, D.; Flood, P.F.; Beriault, R. Development of horse embryos up to twenty two days after ovulation: Observations on fresh specimens. J. Anat. 1982, 135, 191–209. [Google Scholar]
- Denker, H.W. Structural dynamics and function of early embryonic coats. Cells Tissues Organs 2000, 166, 180–207. [Google Scholar] [CrossRef]
- Ginther, O.J. Mobility of the early equine conceptus. Theriogenology 1983, 19, 603–611. [Google Scholar] [CrossRef]
- McDowell, K.J.; Sharp, D.C.; Grubaugh, W.; Thatcher, W.W.; Wilcox, C.J. Restricted conceptus mobility results in failure of pregnancy maintenance in mares. Biol. Reprod. 1988, 39, 340–348. [Google Scholar] [CrossRef]
- Ginther, O.J. Internal regulation of physiological processes through local venoarterial pathways: A review. J. Anim. Sci. 1974, 39, 550–564. [Google Scholar] [CrossRef]
- Leith, G.S.; Ginther, O.J. Characterization of intrauterine mobility of the early equine conceptus. Theriogenology 1984, 22, 401–408. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Nulton, L.C.; Hess, A.M.; Bouma, G.J.; Bruemmer, J.E. The role of embryo contact and focal adhesions during maternal recognition of pregnancy. PLoS ONE 2019, 14, e0213322. [Google Scholar] [CrossRef] [PubMed]
- Klohonatz, K.M.; Coleman, S.J.; Islas-Trejo, A.D.; Medrano, J.F.; Hess, A.M.; Kalbfleisch, T.; Thomas, M.G.; Bouma, G.J.; Bruemmer, J.E. Coding RNA sequencing of equine endometrium during maternal recognition of pregnancy. Genes (Basel) 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Stout, T.A.; Allen, W.R. Role of prostaglandins in intrauterine migration of the equine conceptus. Reproduction 2001, 121, 771–775. [Google Scholar] [CrossRef]
- Baker, C.B.; Adams, M.H.; McDowell, K.J. Lack of expression of alpha or omega interferons by the horse conceptus. J. Reprod. Fertil. Suppl. 1991, 44, 439–443. [Google Scholar]
- Bazer, F.W.; Thatcher, W.W. Theory of maternal recognition of pregnancy in swine based on estrogen controlled endocrine versus exocrine secretion of prostaglandin F2alpha by the uterine endometrium. Prostaglandins 1977, 14, 397–400. [Google Scholar] [CrossRef]
- Klein, C.; Scoggin, K.E.; Ealy, A.D.; Troedsson, M.H. Transcriptional profiling of equine endometrium during the time of maternal recognition of pregnancy. Biol. Reprod. 2010, 83, 102–113. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Hess, A.M.; Hansen, T.R.; Squires, E.L.; Bouma, G.J.; Bruemmer, J.E. Equine endometrial gene expression changes during and after maternal recognition of pregnancy. J. Anim. Sci. 2015, 93, 3364–3376. [Google Scholar] [CrossRef]
- Tachibana, Y.; Nakano, Y.; Nagaoka, K.; Kikuchi, M.; Nambo, Y.; Haneda, S.; Matsui, M.; Miyake, Y.; Imakawa, K. Expression of endometrial immune-related genes possibly functioning during early pregnancy in the mare. J. Reprod. Dev. 2013, 59, 85–91. [Google Scholar] [CrossRef]
- Klohonatz, K.M.; Cameron, A.D.; Hergenreder, J.R.; da Silveira, J.C.; Belk, A.D.; Veeramachaneni, D.N.; Bouma, G.J.; Bruemmer, J.E. Circulating miRNAs as potential alternative cell signaling associated with maternal recognition of pregnancy in the mare. Biol. Reprod. 2016, 95, 124. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Kenney, R.M. Cyclic and pathologic changes of the mare endometrium as detected by biopsy, with a note on early embryonic death. J. Am. Vet. Med. Assoc. 1978, 172, 241–262. [Google Scholar]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics: Cambrige, UK, 2010. [Google Scholar]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kalbfleisch, T.S.; Rice, E.S.; DePriest, M.S., Jr.; Walenz, B.P.; Hestand, M.S.; Vermeesch, J.R.; Brendan, L.O.C.; Fiddes, I.T.; Vershinina, A.O.; Saremi, N.F.; et al. Improved reference genome for the domestic horse increases assembly contiguity and composition. Commun. Biol. 2018, 1, 197. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides accurate, fast, and bias-aware transcript expression estimates using dual-phase inference. bioRxiv 2016, 021592. [Google Scholar] [CrossRef]
- Hunt, S.E.; McLaren, W.; Gil, L.; Thormann, A.; Schuilenburg, H.; Sheppard, D.; Parton, A.; Armean, I.M.; Trevanion, S.J.; Flicek, P.; et al. Ensembl variation resources. Database (Oxford) 2018. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Kim, Y.K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Finn, K.J.; Ji, X.; Baillat, D.; Gregory, R.I.; Liebhaber, S.A.; Pasquinelli, A.E.; Shiekhattar, R. MicroRNA silencing through RISC recruitment of eIF6. Nature 2007, 447, 823–828. [Google Scholar] [CrossRef]
- Lange-Consiglio, A.; Lazzari, B.; Pizzi, F.; Stella, A.; Girani, A.; Quinte, A.; Cremonesi, F.; Capra, E. Different culture times affect microRNA cargo in equine amniotic mesenchymal cells and their microvesicles. Tissue Eng. Part. C Methods 2018, 24, 596–604. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Twenter, H.M.; Belk, A.D.; Klohonatz, K.M.; Bass, L.D.; Bouma, G.J.; Bruemmer, J.E. An investigation into miRNAs in the equine epididymis as potential regulators of spermatozoal maturation. J. Equine Vet. Sci. 2017, 48, 61–68. [Google Scholar] [CrossRef]
- Navakanitworakul, R.; Hung, W.T.; Gunewardena, S.; Davis, J.S.; Chotigeat, W.; Christenson, L.K. Characterization and small RNA content of extracellular vesicles in follicular fluid of developing bovine antral follicles. Sci. Rep. 2016, 6, 25486. [Google Scholar] [CrossRef]
- Stark, G.R.; Darnell, J.E., Jr. The JAK-STAT pathway at twenty. Immunity 2012, 36, 503–514. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Plenge, R. JAK and STAT signaling molecules in immunoregulation and immune-mediated disease. Immunity 2012, 36, 542–550. [Google Scholar] [CrossRef]
- Croker, B.A.; Kiu, H.; Nicholson, S.E. SOCS regulation of the JAK/STAT signalling pathway. Semin. Cell Dev. Biol. 2008, 19, 414–422. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- da Silveira, J.C.; Veeramachaneni, D.N.; Winger, Q.A.; Carnevale, E.M.; Bouma, G.J. Cell-secreted vesicles in equine ovarian follicular fluid contain miRNAs and proteins: A possible new form of cell communication within the ovarian follicle. Biol. Reprod. 2012, 86, 71. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-miRPath v3.0: Deciphering microRNA function with experimental support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Matera, A.G.; Terns, R.M.; Terns, M.P. Non-coding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 209–220. [Google Scholar] [CrossRef]
- Delpu, Y.; Larrieu, D.; Gayral, M.; Arvanitis, D.; Dufresne, M.; Cordelier, P.; Torrisani, J. Noncoding RNAs: Clinical and therapeutic applications. In Drug Discovery in Cancer Epigenetics; Elsevier: Amsterdam, The Netherlands, 2016; pp. 305–326. [Google Scholar]
- Weinberg, Z.; Ruzzo, W.L. Faster genome annotation of non-coding RNA families without loss of accuracy. In Proceedings of The Eighth Annual International Conference on Resaerch in Computational Molecular Biology, San Diego, CA, USA, 27–31 March 2004; pp. 243–251. [Google Scholar]
- Newman, A.J. The role of U5 snRNP in pre-mRNA splicing. EMBO J. 1997, 16, 5797–5800. [Google Scholar] [CrossRef]
- Kandels-Lewis, S.; Seraphin, B. Role of U6 snRNA in 5′splice site selection. Science 1993, 262, 2035–2039. [Google Scholar] [CrossRef]
- Kaida, D.; Berg, M.G.; Younis, I.; Kasim, M.; Singh, L.N.; Wan, L.; Dreyfuss, G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010, 468, 664–668. [Google Scholar] [CrossRef]
- Cuthbert, J.M.; Russell, S.J.; White, K.L.; Benninghoff, A.D. The maternal-to-zygotic transition in bovine in vitro-fertilized embryos is associated with marked changes in small non-coding RNAs. Biol. Reprod. 2018, 100, 331–350. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Deryusheva, S.; Gall, J.G. scaRNAs and snoRNAs: Are they limited to specific classes of substrate RNAs? RNA 2019, 25, 17–22. [Google Scholar] [CrossRef]
| Feature ID | ncRNA Name | ncRNA Description | Day 11P+ Average | Day 11NP Average | Day 11 p-Value |
|---|---|---|---|---|---|
| ENSECAT00000029757 | U109 | snoRNA | 1 | 71,765 | 2.1E-07 |
| ENSECAT00000028132 | U1 | snRNA | 0 | 21,351 | 5.4E-07 |
| ENSECAT00000032319 | eca-mir-9109 | miRNA | 9 | 15,762 | 6.3E-07 |
| ENSECAT00000055936 | eca-mir-8986b | miRNA | 448 | 0 | 1.1E-06 |
| ENSECAT00000027602 | eca-mir-323 | miRNA | 11 | 39,904 | 1.2E-06 |
| ENSECAT00000027815 | SNORA69 | snoRNA | 49 | 36,277 | 6.7E-06 |
| ENSECAT00000028354 | MIR15B | miRNA | 81 | 39,222 | 9.2E-06 |
| ENSECAT00000032694 | U6 | snRNA | 0 | 4866 | 1.5E-05 |
| ENSECAT00000027790 | MIR137 | miRNA | 2 | 2916 | 1.6E-05 |
| ENSECAT00000027318 | SNORA2B | snoRNA | 4 | 5177 | 1.7E-05 |
| ENSECAT00000028446 | SNORD65 | snoRNA | 20 | 9278 | 1.7E-05 |
| ENSECAT00000029752 | SCARNA17 | scaRNA | 16 | 5915 | 2.3E-05 |
| ENSECAT00000029923 | eca-mir-9080 | miRNA | 13 | 5718 | 4.4E-05 |
| ENSECAT00000028257 | SNORA71 | snoRNA | 22 | 8449 | 4.4E-05 |
| ENSECAT00000027964 | eca-mir-652 | miRNA | 24 | 9678 | 4.4E-05 |
| ENSECAT00000028540 | SNORA12 | snoRNA | 2 | 1924 | 4.6E-05 |
| ENSECAT00000028054 | MIR1306 | miRNA | 15 | 4741 | 5.1E-05 |
| ENSECAT00000027873 | eca-mir-582 | miRNA | 3 | 2342 | 1.0E-04 |
| ENSECAT00000028318 | SNORA36A | snoRNA | 48 | 12,262 | 1.3E-04 |
| ENSECAT00000027279 | SNORD59A | snoRNA | 246 | 38,612 | 1.8E-04 |
| KEGG Pathway | p-Value |
|---|---|
| Protein processing in endoplasmic reticulum | 1.54E-12 |
| Proteoglycans in cancer | 3.66E-12 |
| Adherens junction | 1.97E-11 |
| Viral carcinogenesis | 1.97E-11 |
| Hippo signaling pathway | 2.06E-08 |
| p53 signaling pathway | 3.44E-07 |
| Cell cycle | 3.74E-07 |
| Renal cell carcinoma | 4.62E-07 |
| Pancreatic cancer | 1.06E-06 |
| Ubiquitin mediated proteolysis | 1.54E-06 |
| Neurotrophin signaling pathway | 1.54E-06 |
| TGF-beta signaling pathway | 2.25E-06 |
| Prostate cancer | 2.25E-06 |
| Epstein–Barr virus infection | 2.25E-06 |
| Pathways in cancer | 2.25E-06 |
| Glioma | 2.77E-06 |
| mRNA surveillance pathway | 4.95E-06 |
| Chronic myeloid leukemia | 5.02E-06 |
| Hepatitis B | 5.02E-06 |
| Endocytosis | 6.27E-06 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klohonatz, K.M.; Coleman, S.J.; Cameron, A.D.; Hess, A.M.; Reed, K.J.; Canovas, A.; Medrano, J.F.; Islas-Trejo, A.D.; Kalbfleisch, T.; Bouma, G.J.; et al. Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy. Genes 2019, 10, 821. https://doi.org/10.3390/genes10100821
Klohonatz KM, Coleman SJ, Cameron AD, Hess AM, Reed KJ, Canovas A, Medrano JF, Islas-Trejo AD, Kalbfleisch T, Bouma GJ, et al. Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy. Genes. 2019; 10(10):821. https://doi.org/10.3390/genes10100821
Chicago/Turabian StyleKlohonatz, Kristin M., Stephen J. Coleman, Ashley D. Cameron, Ann M. Hess, Kailee J. Reed, Angela Canovas, Juan F. Medrano, Alma D. Islas-Trejo, Ted Kalbfleisch, Gerrit J. Bouma, and et al. 2019. "Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy" Genes 10, no. 10: 821. https://doi.org/10.3390/genes10100821
APA StyleKlohonatz, K. M., Coleman, S. J., Cameron, A. D., Hess, A. M., Reed, K. J., Canovas, A., Medrano, J. F., Islas-Trejo, A. D., Kalbfleisch, T., Bouma, G. J., & Bruemmer, J. E. (2019). Non-Coding RNA Sequencing of Equine Endometrium During Maternal Recognition of Pregnancy. Genes, 10(10), 821. https://doi.org/10.3390/genes10100821






