Identification of Key Genes for the Precise Classification between Solenopsis invicta and S. geminata Facilitating the Quarantine Process
Abstract
1. Introduction
2. Materials and Methods
2.1. S. invicta Sample Collection and NGS Sequencing
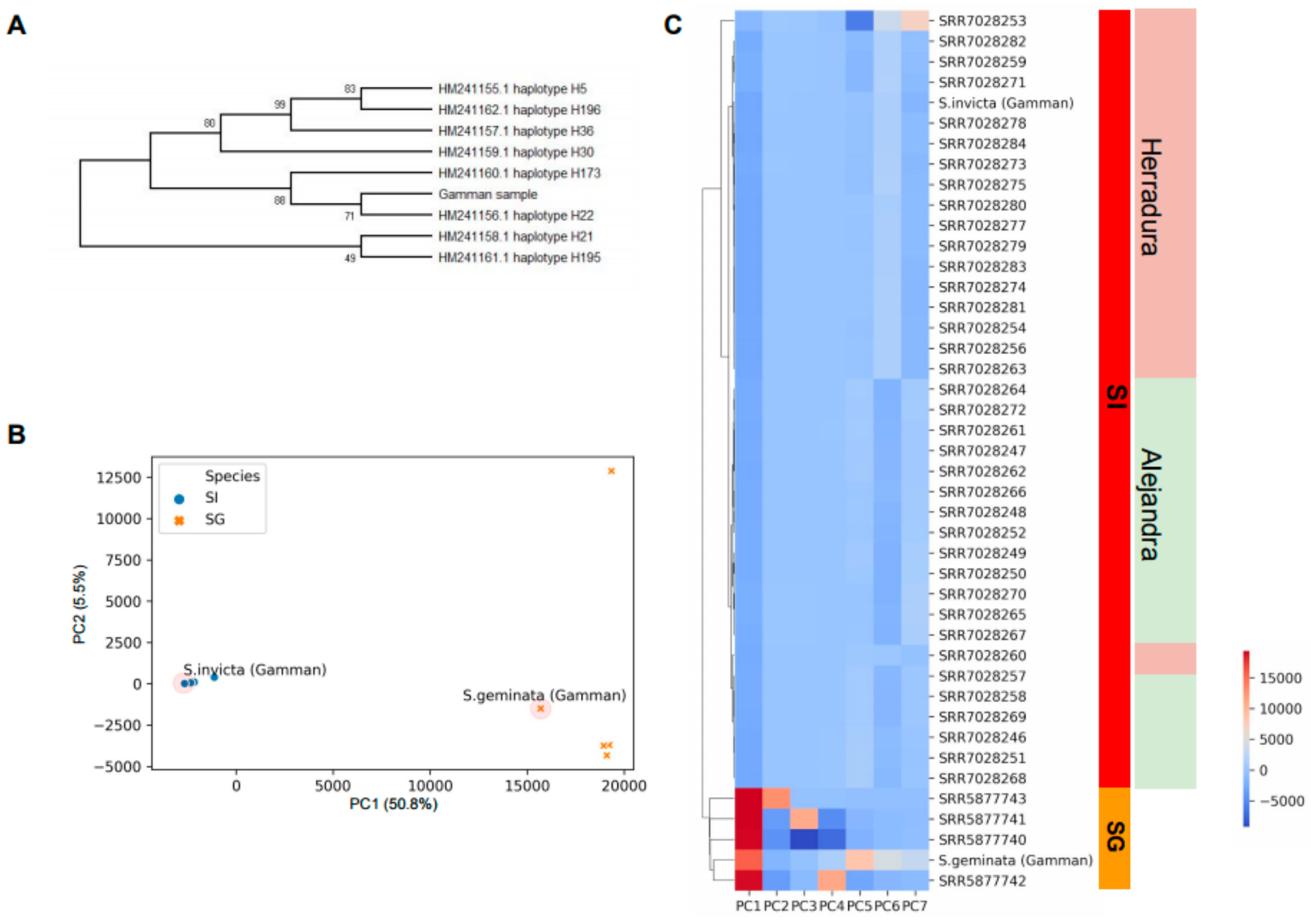
2.2. Phylogenetic Analysis and Sample Clustering
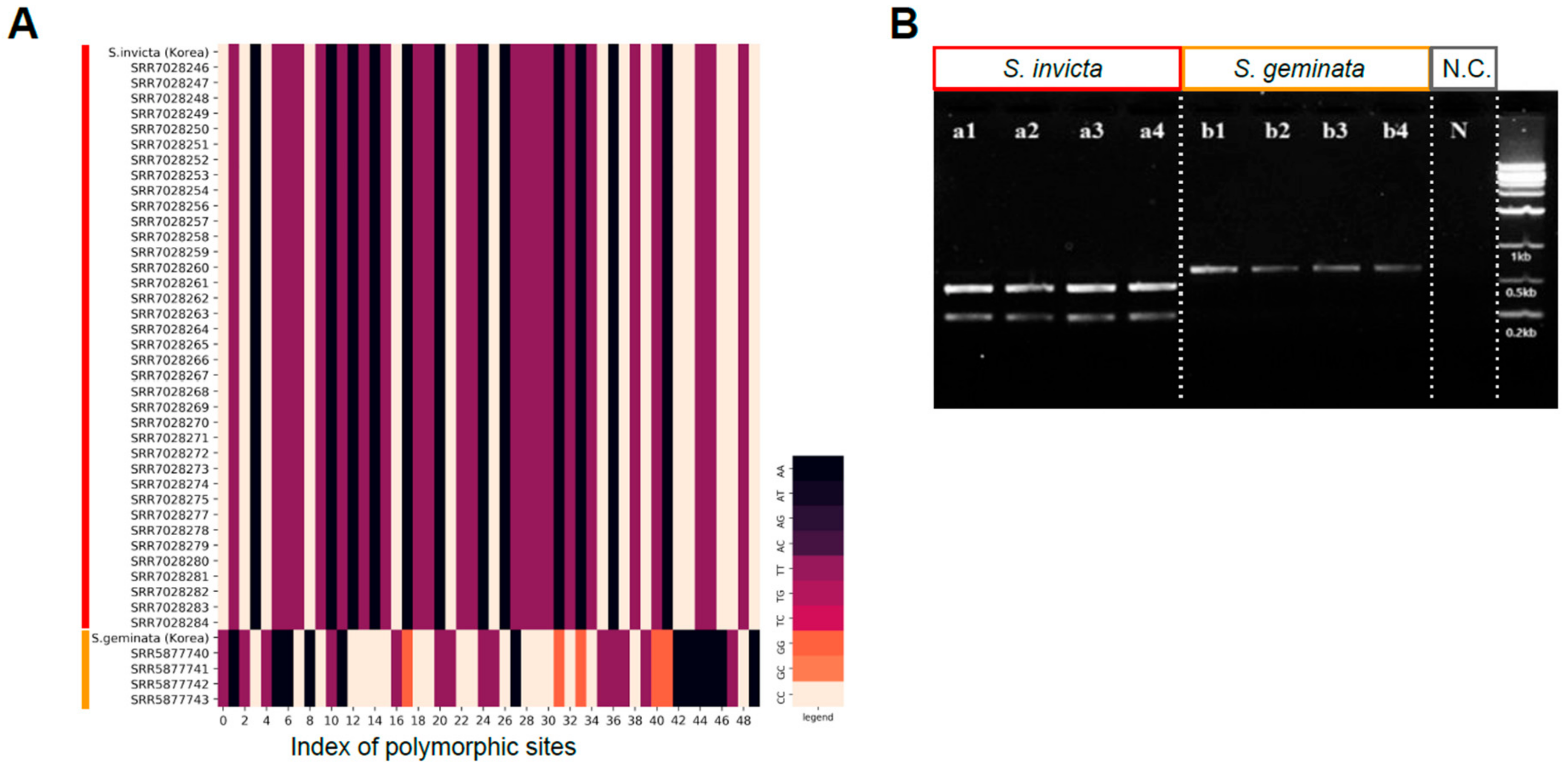
2.3. Cleaved Amplified Polymorphic Sequences (CAPS) Design and Validation Samples
3. Results
3.1. Sample Collection from Gamman Pier in Korea
3.2. Whole Genome Sequencing of Collected Fire Ant Samples
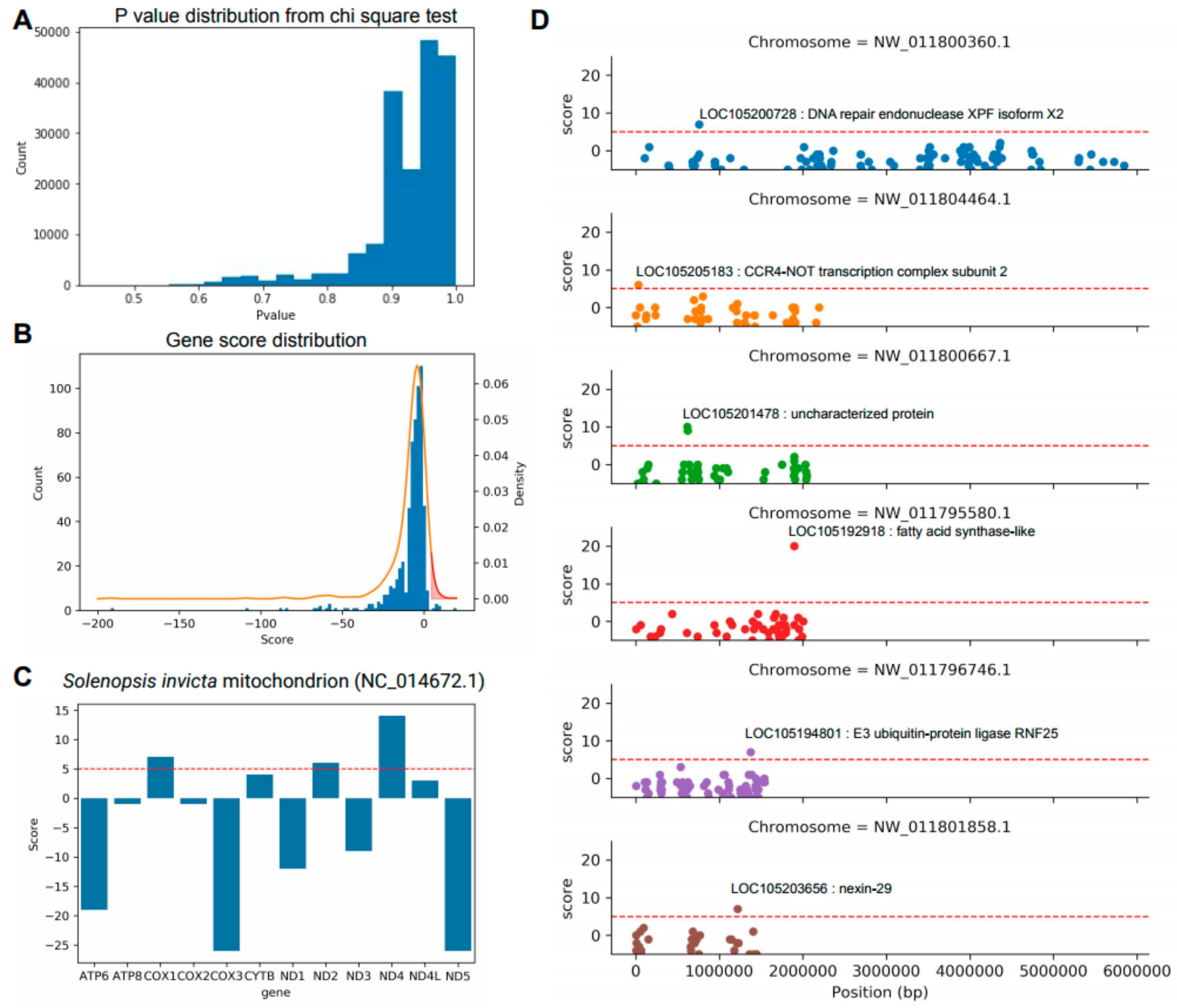
3.3. Identification of Key Genes to Distinguish between S. invicta and S. geminata
3.4. Development of CAPS Marker for Quick Classification of S. invicta and S. geminata
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lowe, S.; Browne, M.; Boudjelas, S.; De Poorter, M. 100 of the World’s Worst Invasive Alien Species: A Selection from the Global Invasive Species Database; Invasive Species Specialist Group: Auckland, New Zealand, 2000; Volume 12. [Google Scholar]
- Perrings, C.; Naeem, S.; Ahrestani, F.; Bunker, D.E.; Burkill, P.; Canziani, G.; Elmqvist, T.; Ferrati, R.; Fuhrman, J.; Jaksic, F.; et al. Conservation. Ecosystem services for 2020. Science 2010, 330, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Caldera, E.J.; Ross, K.G.; DeHeer, C.J.; Shoemaker, D.D. Putative native source of the invasive fire ant Solenopsis invicta in the USA. Biol. Invasions 2008, 10, 1457–1479. [Google Scholar] [CrossRef]
- Ascunce, M.S.; Yang, C.C.; Oakey, J.; Calcaterra, L.; Wu, W.J.; Shih, C.; Goudet, J.; Ross, K.G.; Shoemaker, D. Global Invasion History of the Fire Ant Solenopsis invicta. Science 2011, 331, 1066–1068. [Google Scholar] [CrossRef] [PubMed]
- Adams, E.S. The Fire Ants. By Walter R Tschinkel. Belknap Press. Cambridge (Massachusetts): Harvard University Press. $95.00. vii 723 p 16 pl; ill.; index. ISBN: 0–674–02207–6. 2006. Q. Rev. Biol. 2006, 81, 408–409. [Google Scholar] [CrossRef]
- Moloney, S.; Vanderwoude, C. Red Imported Fire Ants: A threat to eastern Australia’s wildlife? Ecol. Manag. Restor. 2002, 3, 167–175. [Google Scholar] [CrossRef]
- Bissmire, S. Red imported fire ants found at Whirinaki. Biosecur. Bioterror. 2006, 69, 6. [Google Scholar]
- Zhang, R.; Li, Y.; Liu, N.; Porter, S.D. An overview of the red imported fire ant (Hymenoptera: Formicidae) in mainland China. Fla. Entomol. 2007, 90, 723–732. [Google Scholar] [CrossRef]
- Lard, C.F.; Schmidt, J.; Morris, B.; Estes, L.; Ryan, C.; Bergquist, D. An economic impact of imported fire ants in the United States of America. Ph.D. Thesis, Texas A&M University, Texas Agricultural Experiment Station, College Station, TX, USA, August 2006. [Google Scholar]
- Antony, G.; Scanlan, J.C.; Francis, A.; Kloessing, K.; Nguyen, Y. Revised Benefits and Costs of Eradicating the Red Imported Fire Ant; Paper presented at the 53rd annual conference of the Australian Agricultural and Resource Economics Society: Cairns, Australia, 2009. [Google Scholar]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Peña, S.R.; Chacón-Cardosa, M.C.; Resendez-Perez, D. Identification of Fire Ants (Hymenoptera: Formicidae) from Northeastern Mexico with Morphology and Molecular Markers. Fla. Entomol. 2009, 92, 107–115. [Google Scholar] [CrossRef]
- Shokralla, S.; Gibson, J.F.; Nikbakht, H.; Janzen, D.H.; Hallwachs, W.; Hajibabaei, M. Next-generation DNA barcoding: Using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Resour. 2014, 14, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Wurm, Y.; Wang, J.; Riba-Grognuz, O.; Corona, M.; Nygaard, S.; Hunt, B.G.; Ingram, K.K.; Falquet, L.; Nipitwattanaphon, M.; Gotzek, D.; et al. The genome of the fire ant Solenopsis invicta. Proc. Natl. Acad. Sci. USA 2011, 108, 5679–5684. [Google Scholar] [CrossRef] [PubMed]
- Privman, E.; Cohen, P.; Cohanim, A.B.; Riba-Grognuz, O.; Shoemaker, D.; Keller, L. Positive selection on sociobiological traits in invasive fire ants. Mol. Ecol. 2018, 27, 3116–3130. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kemp, S.F.; deShazo, R.D.; Moffitt, J.E.; Williams, D.F.; Buhner, W.A., 2nd. Expanding habitat of the imported fire ant (Solenopsis invicta): A public health concern. J. Allergy Clin. Immunol. 2000, 105, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]

| Solenopsis invicta | S. geminata | |
|---|---|---|
| Total read bases (bp) | 25,563,097,172 | 24,794,827,340 |
| GC(%) | 36.28 | 35.66 |
| Q30(%) | 88.28 | 86.73 |
| Mapped Sites (≥1×) | 353,361,979 (99.61%) | 338,179,742 (95.33%) |
| Mapped reads | 173,616,509 | 185,123,080 |
| Mean depth (X) | 49.23 | 45.14 |
| Number of SNPs * | 1,098,989 | 5,181,818 |
| Number of insertions | 127,295 | 409,328 |
| Number of deletions | 113,590 | 364,156 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-H.; Kim, J.-S.; Cho, H.-J.; Lee, J.-H.; Jun, T.-H.; Kang, Y.J. Identification of Key Genes for the Precise Classification between Solenopsis invicta and S. geminata Facilitating the Quarantine Process. Genes 2019, 10, 812. https://doi.org/10.3390/genes10100812
Kim K-H, Kim J-S, Cho H-J, Lee J-H, Jun T-H, Kang YJ. Identification of Key Genes for the Precise Classification between Solenopsis invicta and S. geminata Facilitating the Quarantine Process. Genes. 2019; 10(10):812. https://doi.org/10.3390/genes10100812
Chicago/Turabian StyleKim, Kil-Hyun, Ji-Su Kim, Hyun-Ji Cho, Jong-Ho Lee, Tae-Hwan Jun, and Yang Jae Kang. 2019. "Identification of Key Genes for the Precise Classification between Solenopsis invicta and S. geminata Facilitating the Quarantine Process" Genes 10, no. 10: 812. https://doi.org/10.3390/genes10100812
APA StyleKim, K.-H., Kim, J.-S., Cho, H.-J., Lee, J.-H., Jun, T.-H., & Kang, Y. J. (2019). Identification of Key Genes for the Precise Classification between Solenopsis invicta and S. geminata Facilitating the Quarantine Process. Genes, 10(10), 812. https://doi.org/10.3390/genes10100812





