miR2Diabetes: A Literature-Curated Database of microRNA Expression Patterns, in Diabetic Microvascular Complications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Database Content
2.2. Nomenclature Standardization
2.3. miRNA Dysregulation Pattern Generalization
2.4. Database and Web Application Implementation
3. Results
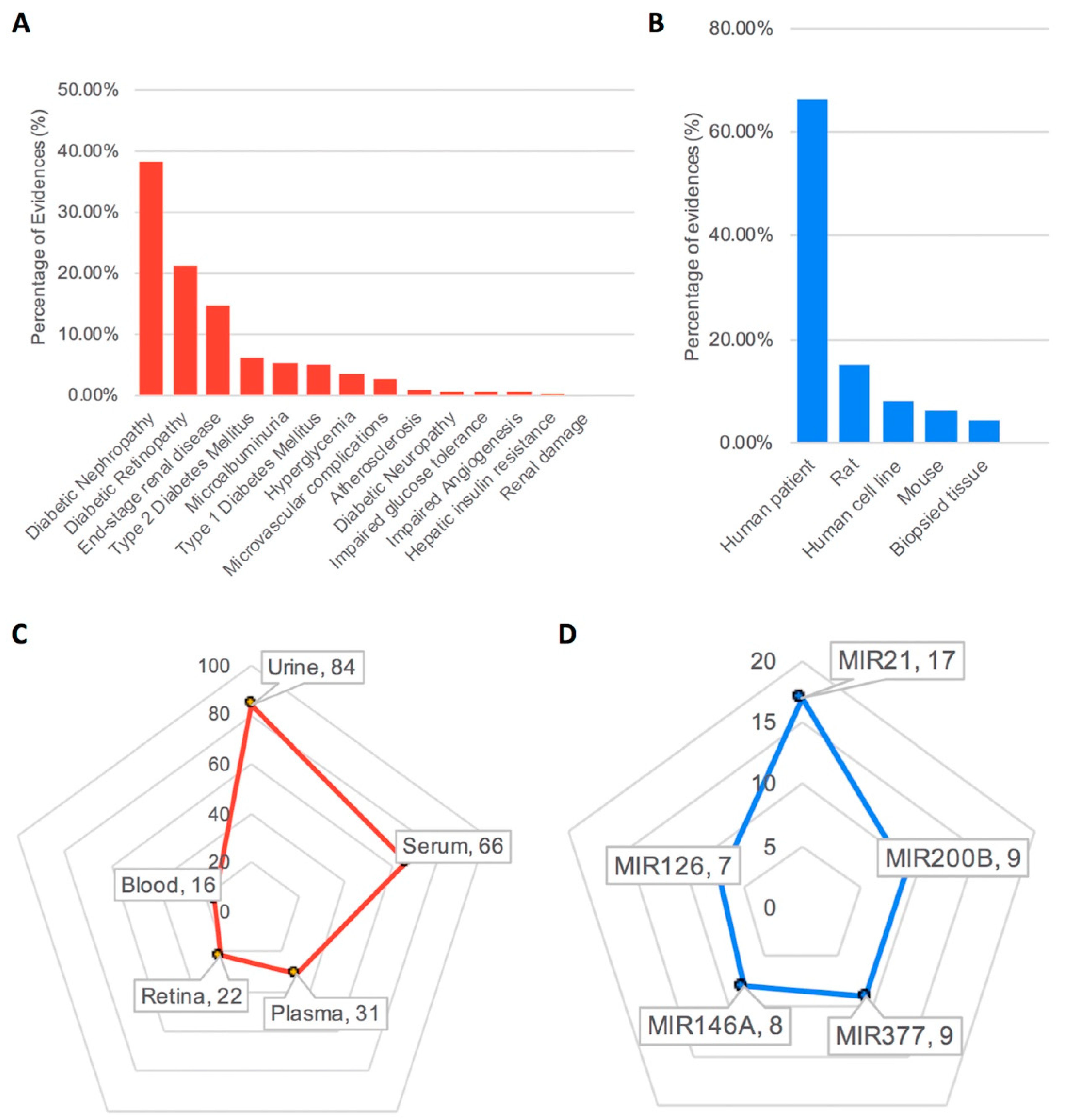
3.1. Database Content and Statistics
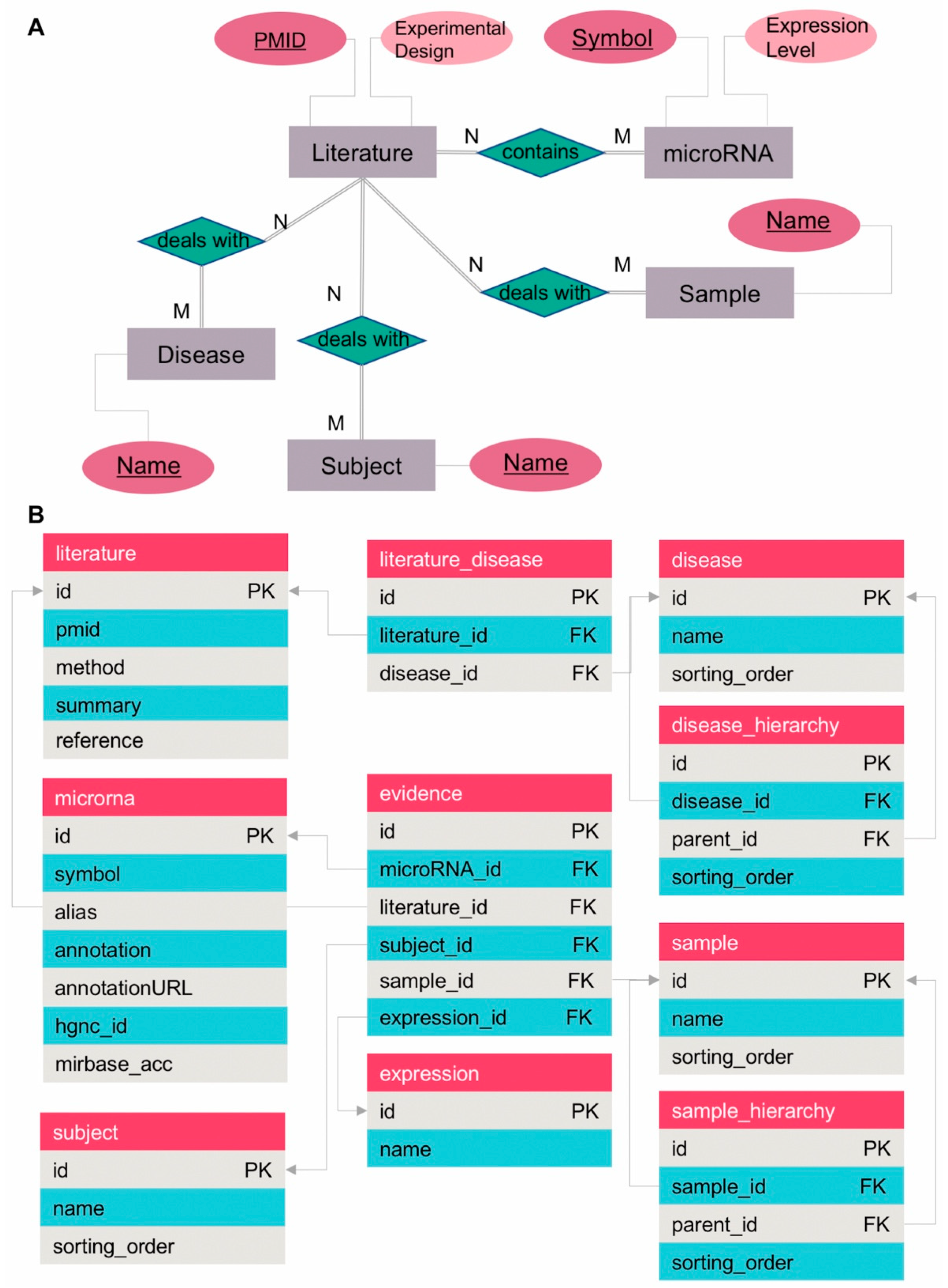
3.2. Database Model
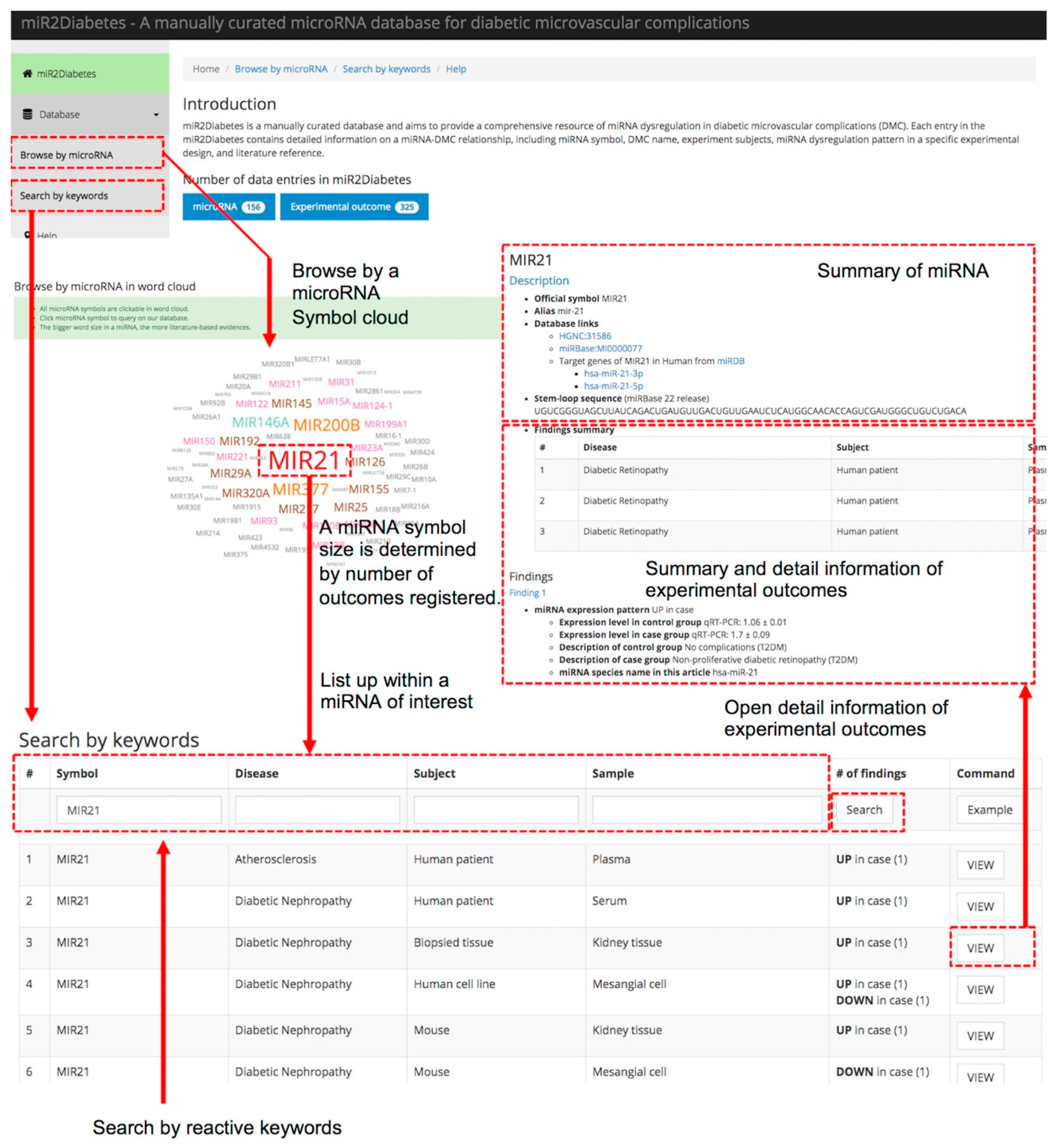
3.3. Database Access and Case Study of miR2Diabetes
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Database Availability
References
- Blair, M. Diabetes Mellitus Review. Urol. Nurs. 2016, 36, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ogurtsova, K.; da Rocha Fernandes, J.D.; Huang, Y.; Linnenkamp, U.; Guariguata, L.; Cho, N.H.; Cavan, D.; Shaw, J.E.; Makaroff, L.E. Idf Diabetes Atlas: Global Estimates for the Prevalence of Diabetes for 2015 and 2040. Diabetes Res. Clin. Pract. 2017, 128, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Zimmet, P.; Alberti, K.G.; Magliano, D.J.; Bennett, P.H. Diabetes Mellitus Statistics on Prevalence and Mortality: Facts and Fallacies. Nat. Rev. Endocrinol. 2016, 12, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, R.; Putta, S.; Kato, M. MicroRNAs and Diabetic Complications. J. Cardiovasc. Transl. Res. 2012, 5, 413–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guay, C.; Regazzi, R. Circulating MicroRNAs as Novel Biomarkers for Diabetes Mellitus. Nat. Rev. Endocrinol. 2013, 9, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Moon, S.; Lee, K.; Park, I.B.; Lee, D.H.; Nam, S. Urinary and Blood MicroRNA-126 and -770 Are Potential Noninvasive Biomarker Candidates for Diabetic Nephropathy: A Meta-Analysis. Cell. Physiol. Biochem. 2018, 46, 1331–1340. [Google Scholar] [CrossRef] [PubMed]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial Mir-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef]
- Salas-Perez, F.; Codner, E.; Valencia, E.; Pizarro, C.; Carrasco, E.; Perez-Bravo, F. MicroRNAs Mir-21a and Mir-93 Are Down Regulated in Peripheral Blood Mononuclear Cells (Pbmcs) from Patients with Type 1 Diabetes. Immunobiology 2013, 218, 733–737. [Google Scholar] [CrossRef]
- Wang, C.; Wan, S.; Yang, T.; Niu, D.; Zhang, A.; Yang, C.; Cai, J.; Wu, J.; Song, J.; Zhang, C.Y.; et al. Increased Serum Micrornas Are Closely Associated with the Presence of Microvascular Complications in Type 2 Diabetes Mellitus. Sci. Rep. 2016, 6, 20032. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Aboushahba, R.; Bekhet, M.M.; Soliman, Y. Urinary Exosomal MicroRNA Panel Unravels Novel Biomarkers for Diagnosis of Type 2 Diabetic Kidney Disease. J. Diabetes Complicat. 2016, 30, 1585–1592. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Bekhet, M.M. Clinical Verification of a Novel Urinary MicroRNA Panal: 133b, -342 and -30 as Biomarkers for Diabetic Nephropathy Identified by Bioinformatics Analysis. Biomed. Pharmacother. 2016, 83, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. Mir2disease: A Manually Curated Database for MicroRNA Deregulation in Human Disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Gu, J.; Wang, T.; Ding, Z. Oncomirdb: A Database for the Experimentally Verified Oncogenic and Tumor-Suppressive MicroRNAs. Bioinformatics 2014, 30, 2237–2238. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. Mircancer: A MicroRNA-Cancer Association Database Constructed by Text Mining on Literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Barh, D.; Kamapantula, B.; Jain, N.; Nalluri, J.; Bhattacharya, A.; Juneja, L.; Barve, N.; Tiwari, S.; Miyoshi, A.; Azevedo, V.; et al. Miregulome: A Knowledge-Base of MiRNA Regulomics and Analysis. Sci. Rep. 2015, 5, 12832. [Google Scholar] [CrossRef] [PubMed]
- White, J.A.; McAlpine, P.J.; Antonarakis, S.; Cann, H.; Eppig, J.T.; Frazer, K.; Frezal, J.; Lancet, D.; Nahmias, J.; Pearson, P.; et al. Guidelines for Human Gene Nomenclature (1997). Hugo Nomenclature Committee. Genomics 1997, 45, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Wain, H.M.; Lush, M.; Ducluzeau, F.; Povey, S. Genew: The Human Gene Nomenclature Database. Nucleic Acids Res. 2002, 30, 169–171. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. Mirbase: MicroRNA Sequences, Targets and Gene Nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Gremse, M.; Chang, A.; Schomburg, I.; Grote, A.; Scheer, M.; Ebeling, C.; Schomburg, D. The Brenda Tissue Ontology (Bto): The First All-Integrating Ontology of All Organisms for Enzyme Sources. Nucleic Acids Res. 2011, 39, D507–D513. [Google Scholar] [CrossRef]
- Schriml, L.M.; Arze, C.; Nadendla, S.; Chang, Y.W.; Mazaitis, M.; Felix, V.; Feng, G.; Kibbe, W.A. Disease Ontology: A Backbone for Disease Semantic Integration. Nucleic Acids Res. 2012, 40, D940–D946. [Google Scholar] [CrossRef]
- Wong, N.; Wang, X. Mirdb: An Online Resource for MicroRNA Target Prediction and Functional Annotations. Nucleic Acids Res. 2015, 43, D146–D152. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Moon, S.; Lee, K.; Park, I.B.; Lee, D.H.; Nam, S. miR2diabetes. Available online: http://mir2diabetes.yoonlab.or.kr (accessed on 22 September 2019).
- Chen, P.P.S. The Entity-Relationship Model—Toward a Unified View of Data. ACM Trans. Database Syst. (TODS) 1976, 1, 9–36. [Google Scholar] [CrossRef]
- Reusch, J.E.; Manson, J.E. Management of Type 2 Diabetes in 2017: Getting to Goal. JAMA 2017, 317, 1015–1016. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the Cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Wilks, C.; Cline, M.S.; Weiler, E.; Diehkans, M.; Craft, B.; Martin, C.; Murphy, D.; Pierce, H.; Black, J.; Nelson, D.; et al. The Cancer Genomics Hub (Cghub): Overcoming Cancer through the Power of Torrential Data. Database 2014, 2014, bau093. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Sen, S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016, 231, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Odell, S.G.; Lazo, G.R.; Woodhouse, M.R.; Hane, D.L.; Sen, T.Z. The Art of Curation at a Biological Database: Principles and Application. Curr. Plant. Biol. 2017, 11–12, 2–11. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, S.; Moon, S.; Lee, K.; Park, I.B.; Lee, D.H.; Nam, S. miR2Diabetes: A Literature-Curated Database of microRNA Expression Patterns, in Diabetic Microvascular Complications. Genes 2019, 10, 784. https://doi.org/10.3390/genes10100784
Park S, Moon S, Lee K, Park IB, Lee DH, Nam S. miR2Diabetes: A Literature-Curated Database of microRNA Expression Patterns, in Diabetic Microvascular Complications. Genes. 2019; 10(10):784. https://doi.org/10.3390/genes10100784
Chicago/Turabian StylePark, Sungjin, SeongRyeol Moon, Kiyoung Lee, Ie Byung Park, Dae Ho Lee, and Seungyoon Nam. 2019. "miR2Diabetes: A Literature-Curated Database of microRNA Expression Patterns, in Diabetic Microvascular Complications" Genes 10, no. 10: 784. https://doi.org/10.3390/genes10100784
APA StylePark, S., Moon, S., Lee, K., Park, I. B., Lee, D. H., & Nam, S. (2019). miR2Diabetes: A Literature-Curated Database of microRNA Expression Patterns, in Diabetic Microvascular Complications. Genes, 10(10), 784. https://doi.org/10.3390/genes10100784




