Evidence for Early European Neolithic Dog Dispersal: New Data on Southeastern European Subfossil Dogs from the Prehistoric and Antiquity Ages
Abstract
:1. Introduction
1.1. Ancient Dog Populations from Europe
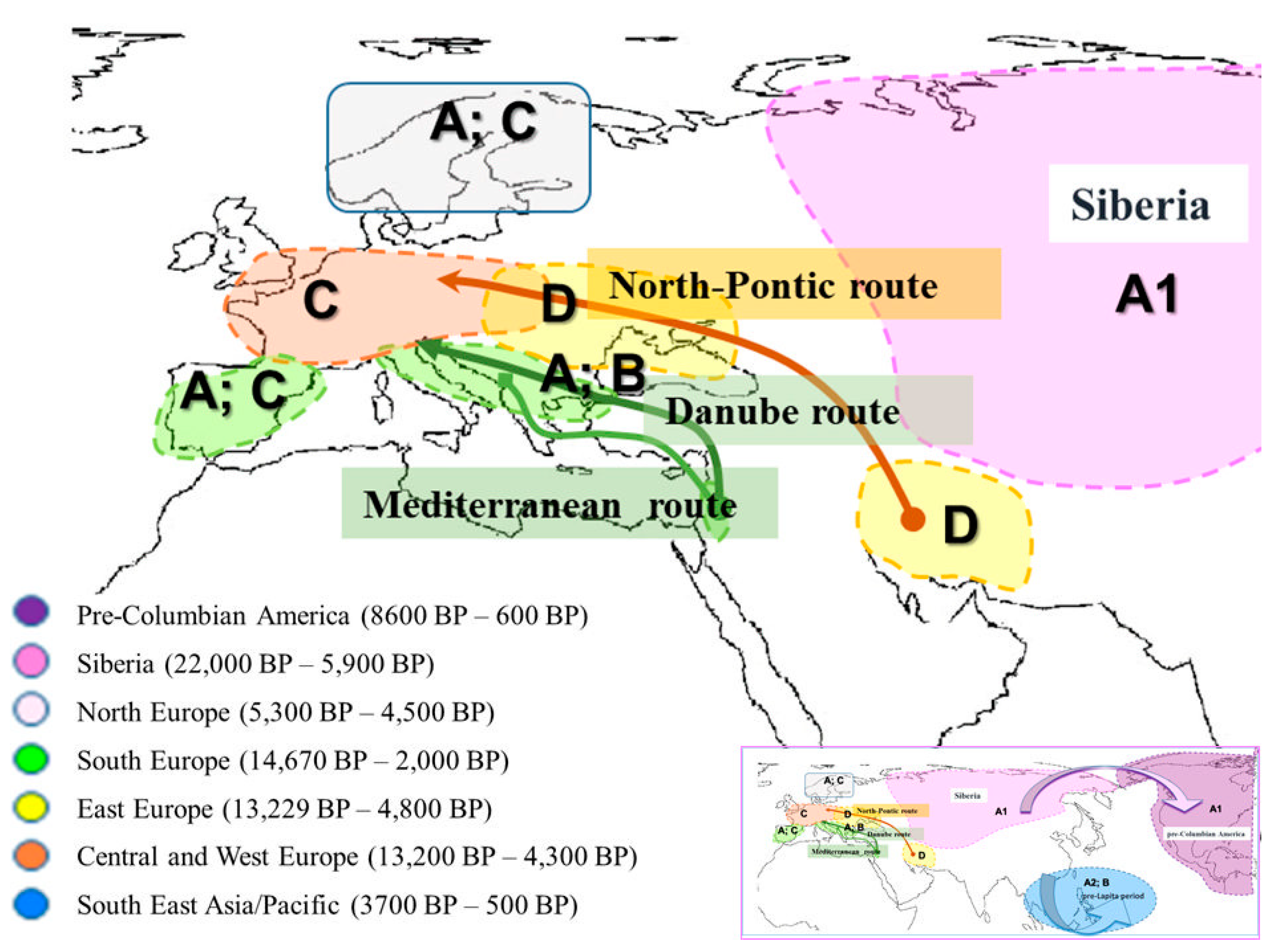
1.2. Balkan Neolithic and Chalcolithic Periods: Specifics of Human Societies and Understanding of Early European Farmer Migration Routes from the Near East
2. Materials and Methods
2.1. Archeological Sample Collection
2.2. Ancient DNA Isolation
2.3. PCR Amplification and Sequencing
2.4. Phylogenetic Reconstruction
3. Results
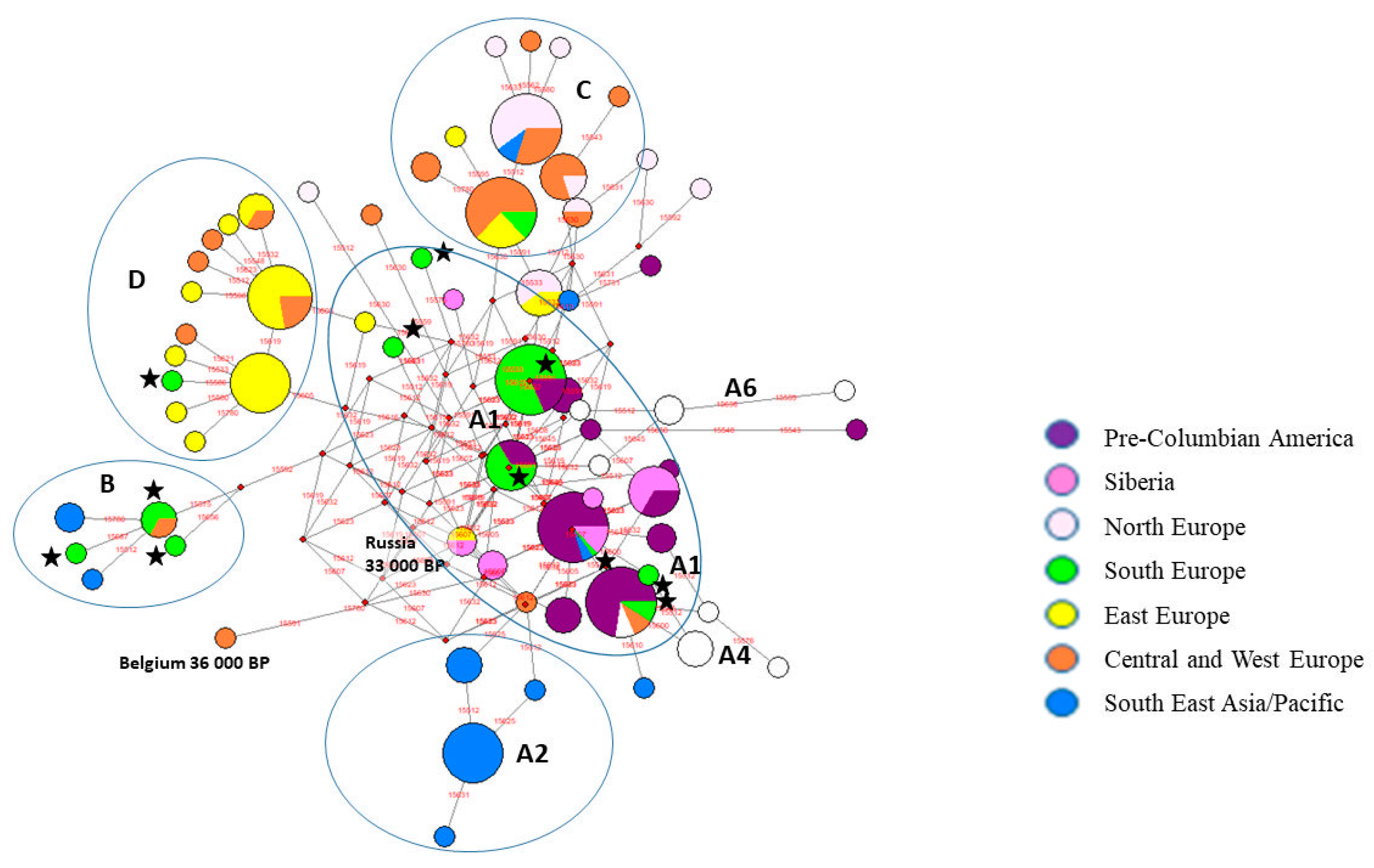
3.1. Phylogenetic Analysis and Haplogroup Classification
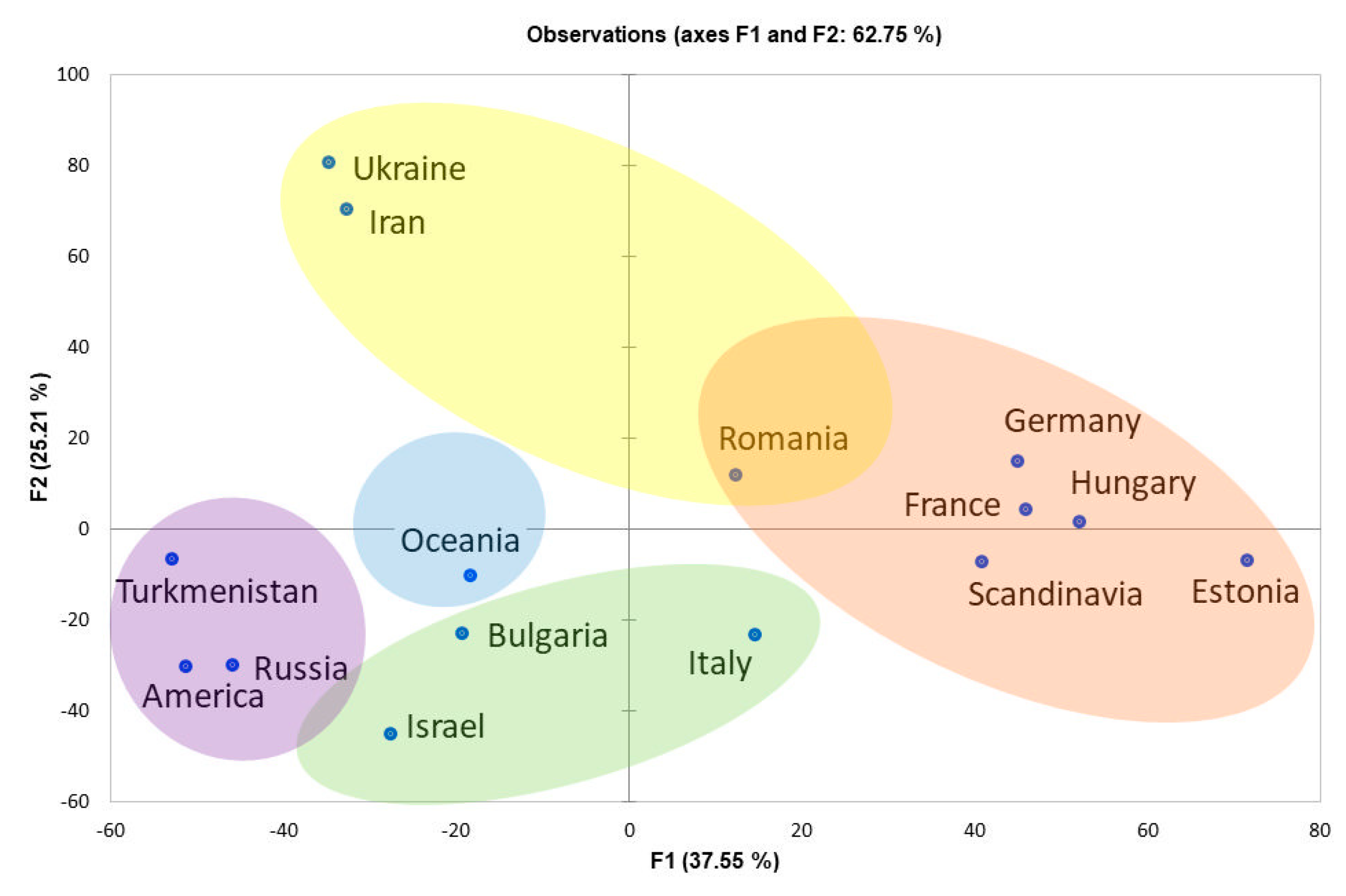
3.2. Comparative Analyses of Ancient Dog Haplotypes and Bulgarian Samples
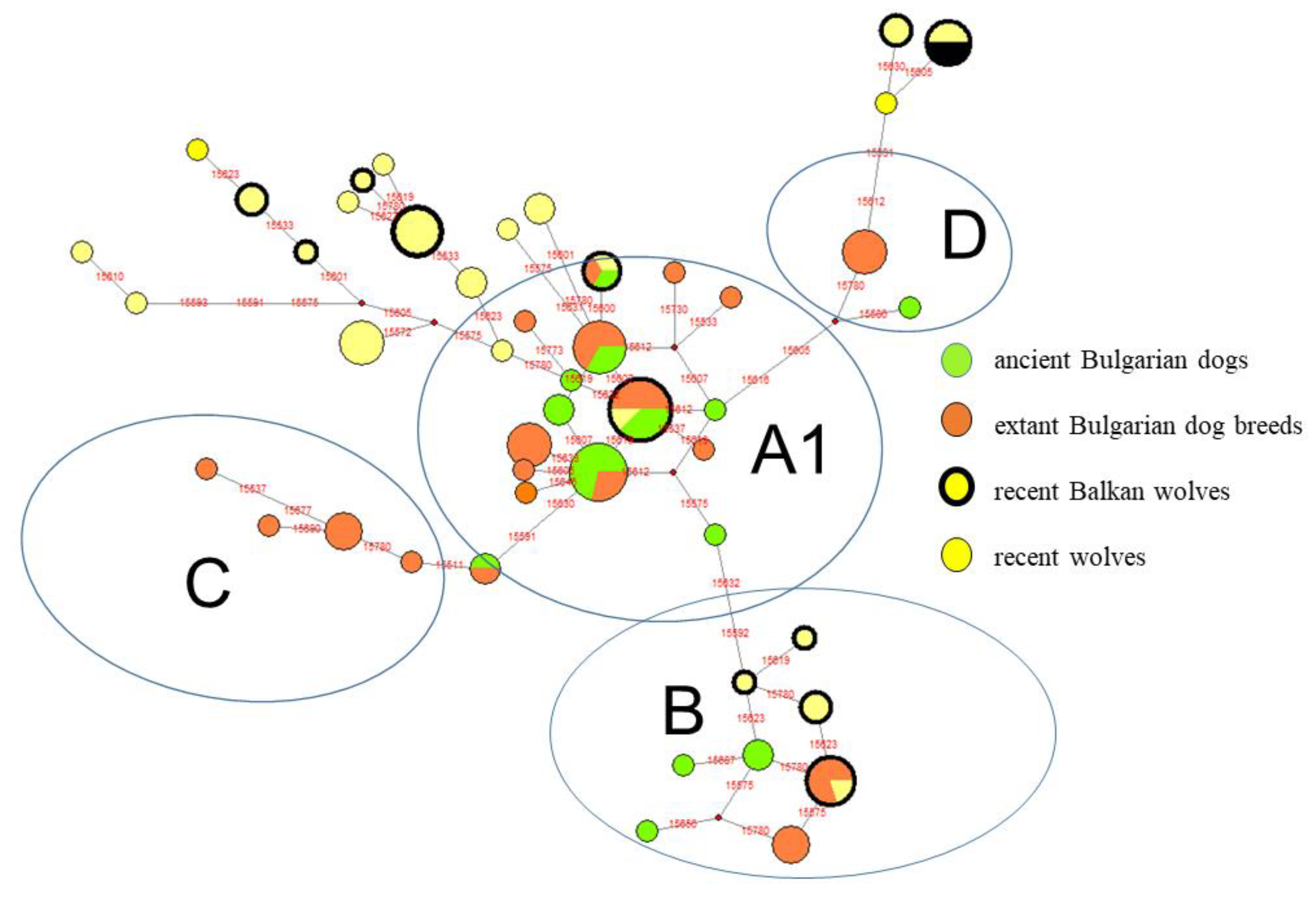
3.3. Comparative Analysis Among Ancient and Recent Balkan Dogs as well as Recent Gray Wolf Haplotype Distribution
4. Discussion
4.1. Clade A: Specific for Neolithic Southern Europe (the Mediterranean Region)
4.2. Clade B: The Possibility of a Southeastern European (Balkans) Origin of Dogs from Local Wolf Predecessors
4.3. Population Structure of Ancient Dogs from the Neolithic Period
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Sullivan, J.O. Variability in the wolf, a group hunter. In Wolf and Man; Hall, R.L., Sharp, H.S., Eds.; Cambridge Academic Press: Cambridge, UK, 1978; pp. 31–40. [Google Scholar]
- Morey, D.F.; Jeger, R. From wolf to dog: Late Pleistocene ecological dynamics, altered trophic strategies, and shifting human perceptions. Hist. Biol. 2017, 29, 895–903. [Google Scholar] [CrossRef]
- Ovodov, N.D.; Crockford, S.J.; Kuzmin, Y.V.; Higham, T.F.; Hodgins, G.W.; van der Plicht, J. A 33,000-year-old incipient dog from the Altai Mountains of Siberia: Evidence of the earliest domestication disrupted by the Last Glacial Maximum. PLoS ONE 2011, 6, e22821. [Google Scholar] [CrossRef] [PubMed]
- Germonpré, M.; Sablin, M.V.; Stevens, R.E.; Hedges, R.E.; Hofreiter, M.; Després, V.R. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: Osteometry, ancient DNA and stable isotopes. J. Archaeol. Sci. 2009, 36, 473–490. [Google Scholar] [CrossRef]
- Frantz, L.A.; Mullin, V.E.; Pionnier-Capitan, M.; Lebrasseur, O.; Ollivier, M.; Perri, A.; Tresset, A. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 2016, 352, 1228–1231. [Google Scholar] [CrossRef] [PubMed]
- Thalmann, O.; Shapiro, B.; Cui, P.; Schuenemann, V.J.; Sawyer, S.K.; Greenfield, D.L.; Napierala, H. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 2013, 342, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Boudadi-Maligne, M.; Escarguel, G. A biometric re-evaluation of recent claims for Early Upper Palaeolithic wolf domestication in Eurasia. J. Archaeol. Sci. 2014, 45, 80–89. [Google Scholar] [CrossRef]
- Savolainen, P.; Zhang, Y.P.; Luo, J.; Lundeberg, J.; Leitner, T. Genetic evidence for an East Asian origin of domestic dogs. Science 2002, 298, 1610–1613. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.F.; Kluetsch, C.; Zou, X.J.; Zhang, A.B.; Luo, L.Y.; Angleby, H.; Ardalan, A.; Ekström, C.; Sköllermo, A.; Lundeberg, J.; et al. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol. Biol. Evol. 2009, 26, 2849–2864. [Google Scholar] [CrossRef] [PubMed]
- Duleba, A.; Skonieczna, K.; Bogdanowicz, W.; Malyarchuk, B.; Grzybowski, T. Complete mitochondrial genome database and standardized classification system for Canis lupus familiaris. Forensic Sci. Int. Genet. 2015, 19, 123–129. [Google Scholar] [CrossRef]
- Ardalan, A.; Kluetsch, C.F.; Zhang, A.B.; Erdogan, M.; Uhlén, M.; Savolainen, P. Comprehensive study of mtDNA among Southwest Asian dogs contradicts independent domestication of wolf, but implies dog–wolf hybridization. Ecol. Evol. 2011, 1, 373–385. [Google Scholar] [CrossRef]
- Witt, K.E.; Judd, K.; Kitchen, A.; Grier, C.; Kohler, T.A.; Ortman, S.G.; Malhi, R.S. DNA analysis of ancient dogs of the Americas: Identifying possible founding haplotypes and reconstructing population histories. J. Hum. Evol. 2015, 79, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Greig, K.; Gosling, A.; Collins, C.J.; Boocock, J.; McDonald, K.; Addison, D.J.; Liu, F. Complex history of dog (Canis familiaris) origins and translocations in the Pacific revealed by ancient mitogenomes. Sci. Rep. 2018, 8, 9130. [Google Scholar] [CrossRef] [PubMed]
- Deguilloux, M.F.; Moquel, J.; Pemonge, M.H.; Colombeau, G. Ancient DNA supports lineage replacement in European dog gene pool: Insight into Neolithic southeast France. J. Archaeol. Sci. 2009, 36, 513–519. [Google Scholar] [CrossRef]
- Pionnier-Capitan, M. La Domestication du Chien en Eurasie: Étude de la Diversitémpassée, Approches Ostéoarchéologiques, Morphométriques et Paléogénétiques. Ph.D. Thesis, École normale supérieure (Sciences), Lyon, France, 2010. [Google Scholar]
- Ollivier, M.; Tresset, A.; Frantz, L.A.; Bréhard, S.; Bălăşescu, A.; Mashkour, M.; Bartosiewicz, L. Dogs accompanied humans during the Neolithic expansion into Europe. Biol. Lett. 2018, 14, 20180286. [Google Scholar] [CrossRef] [PubMed]
- Malmström, H.; Svensson, E.M.; Gilbert, M.T.; Willerslev, E.; Götherström, A.; Holmlund, G. More on contamination: The use of asymmetric molecular behavior to identify authentic ancient human DNA. Mol. Biol. Evol. 2007, 24, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Botigué, L.R.; Song, S.; Scheu, A.; Gopalan, S.; Pendleton, A.L.; Oetjens, M.; Bobo, D. Ancient European dog genomes reveal continuity since the Early Neolithic. Nat. Commun. 2017, 8, 16082. [Google Scholar] [CrossRef] [PubMed]
- Skoglund, P.; Ersmark, E.; Palkopoulou, E.; Dalén, L. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr. Biol. 2015, 25, 1515–1519. [Google Scholar] [CrossRef] [PubMed]
- Klütsch, C.F.; Seppälä, E.H.; Fall, T.; Uhlén, M.; Hedhammar, Å.; Lohi, H.; Savolainen, P. Regional occurrence, high frequency but low diversity of mitochondrial DNA haplogroup d1 suggests a recent dog-wolf hybridization in Scandinavia. Anim. Genet. 2011, 42, 100–103. [Google Scholar] [CrossRef]
- Adeola, A.C.; Ommeh, S.C.; Song, J.J.; Olaogun, S.C.; Sanke, O.J.; Yin, T.T.; Agwanda, B.R. A cryptic mitochondrial DNA link between North European and West African dogs. J. Genet. Genom. 2017, 44, 163–170. [Google Scholar] [CrossRef]
- Verginelli, F.; Capelli, C.; Coia, V.; Musiani, M.; Falchetti, M.; Ottini, L.; Mariani-Costantini, R. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and South-East European wolves. Mol. Biol. Evol. 2005, 22, 2541–2551. [Google Scholar] [CrossRef]
- Pires, A.E.; Detry, C.; Chikhi, L.; Rasteiro, R.; Amorim, I.R.; Simões, F.; Cardoso, J.L. The curious case of the Mesolithic Iberian dogs: An archaeogenetic study. J. Archaeol. Sci. 2019, 105, 116–129. [Google Scholar] [CrossRef]
- Vilà, C.; Savolainen, P.; Maldonado, J.E.; Amorim, I.R.; Rice, J.E.; Honeycutt, R.L.; Wayne, R.K. Multiple and ancient origins of the domestic dog. Science 1997, 276, 1687–1689. [Google Scholar] [CrossRef] [PubMed]
- Randi, E.; Lucchini, V.; Christensen, M.F.; Mucci, N.; Funk, S.M.; Dolf, G.; Loeschcke, V. Mitochondrial DNA variability in Italian and East European wolves: Detecting the consequences of small population size and hybridization. Conserv. Biol. 2000, 14, 464–473. [Google Scholar] [CrossRef]
- Koblmüller, S.; Vilà, C.; Lorente-Galdos, B.; Dabad, M.; Ramirez, O.; Marques-Bonet, T.; Leonard, J.A. Whole mitochondrial genomes illuminate ancient intercontinental dispersals of grey wolves (Canis lupus). J. Biogeogr. 2016, 43, 1728–1738. [Google Scholar] [CrossRef]
- Clark, P.U.; Dyke, A.S.; Shakun, J.D.; Carlson, A.E.; Clark, J.; Wohlfarth, B.; McCabe, A.M. The Last Glacial Maximum. Science 2009, 325, 710–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieson, I.; Alpaslan-Roodenberg, S.; Posth, C.; Szécsényi-Nagy, A.; Rohland, N.; Mallick, S.; Fernandes, D. The genomic history of southeastern Europe. Nature 2018, 555, 197–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenstra, J.; Ajmone-Marsan, P.; Beja-Pereira, A.; Bollongino, R.; Bradley, D.; Colli, L.; Ginja, C. Meta-analysis of mitochondrial DNA reveals several population bottlenecks during worldwide migrations of cattle. Diversity 2014, 6, 178–187. [Google Scholar] [CrossRef]
- Spassov, N.; Iliev, N.; Boev, Z. Animal remains from the Eneolithic site near the village of Dolnoslav, Plovdiv District, South Bulgaria. Hist. Nat. Bulg. 2001, 13, 159–179. [Google Scholar]
- Spassov, N.; Iliev, N. The animal bones from the prehistoric necropolis near Durankulak and the latest record of Equus hydruntinus Regalia (NE Bulgaria). Durankulak 2002, 2, 249–261. [Google Scholar]
- Spassov, N.; Iliev, N. CHAPTER XII. 1. Bone remains from domestic and wild animals. In Yabalkovo; Roodenberg, J., Leshtakov, K., Petrova, V., Eds.; ATE—Ars et Technica Explicatus; Sofia University “ST. KLIMENT OHRIDSKI”: Palo Alto, CA, USA, 2014; Volume 1. [Google Scholar]
- Kitchell, K.F., Jr. Animals in the Ancient World from A to Z; Routledge: London, UK, 2014. [Google Scholar]
- Bökönyi, S. Vlasac: An early site of dog domestication. In Archaeozoological Studies: Papers of the Archaeozoological Conference 1974; Clason, A., Ed.; North Holland Publisher: Groningen, Amsterdam, The Netherlands; Oxford, NY, USA, 1975; pp. 167–178. [Google Scholar]
- Dimitrijević VVuković, S. Was the dog locally domesticated in the Danube Gorges? Morphometric study of dog cranial remains from four Mesolithic–Early Neolithic archaeological sites by comparison with contemporary wolves. Int. J. Osteoarchaeol. 2015, 25, 1–30. [Google Scholar] [CrossRef]
- Pääbo, S.; Poinar, H.; Serre, D.; Jaenicke-Despres, V.; Hebler, J.; Rohland, N.; Hofreiter, M. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004, 38, 645–679. [Google Scholar] [CrossRef] [PubMed]
- Willerslev, E.; Cooper, A. Ancient DNA. Proc. Biol. Sci. 2005, 272, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.Y.; Eng, B.; Waye, J.S.; Dudar, J.C.; Saunders, S.R. Technical note: Improved DNA extraction from ancient bones using silica-based spin columns. Am. J. Phys. Anthropol. 1998, 105, 539–543. [Google Scholar] [CrossRef]
- Hristov, P.; Spassov, N.; Iliev, N.; Radoslavov, G. An independent event of Neolithic cattle domestication on the South-eastern Balkans: Evidence from prehistoric aurochs and cattle populations. Mitochondrial DNA 2017, 28, 383–391. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kim, K.S.; Lee, S.E.; Jeong, H.W.; Ha, J.H. The complete nucleotide sequence of the domestic dog (Canis familiaris) mitochondrial genome. Mol. Biol. Evol. 1998, 10, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Wang, W.Z.; Otecko, N.O.; Peng, M.S.; Zhang, Y.P. Reconciling the conflicts between mitochondrial DNA haplogroup trees of Canis lupus. Forensic Sci. Int. Genet. 2016, 23, 83–85. [Google Scholar] [CrossRef]
- Peng, M.S.; Fan, L.; Shi, N.N.; Ning, T.; Yao, Y.G.; Murphy, R.W.; Zhang, Y.P. DomeTree: A canonical toolkit for mitochondrial DNA analyses in domesticated animals. Mol. Ecol. Resour. 2015, 5, 1238–1242. [Google Scholar] [CrossRef]
- Malmstrom, H.; Stora, J.; Dalen, L.; Holmlund, G.; Gotherstrom, A. Extensive human DNA contamination in extracts from ancient dog bones and teeth. Mol. Biol. Evol. 2005, 22, 2040–2047. [Google Scholar] [CrossRef]
- Tsuda, K.; Kikkaw, Y.; Yonekawa, H.; Tanabe, Y. Extensive interbreeding occurred among multiple matriarchal ancestors during the domestication of dogs: Evidence from inter-and intraspecies polymorphisms in the D-loop region of mitochondrial DNA between dogs and wolves. Genes Genet. Syst. 1997, 72, 229–238. [Google Scholar] [CrossRef]
- Marinov, M.; Teofanova, D.; Gadjev, D.; Radoslavov, G.; Hristov, P. Mitochondrial diversity of Bulgarian native dogs suggests dual phylogenetic origin. PeerJ 2018, 6, 5060. [Google Scholar] [CrossRef]
- Achilli, A.; Olivieri, A.; Pala, M.; Metspalu, E.; Fornarino, S.; Battaglia, V.; Accetturo, M.; Kutuev, I.; Khusnutdinova, E.; Pennarun, E.; et al. Mitochondrial DNA variation of modern Tuscans supplementary orts the near eastern origin of Etruscans. Am. J. Hum. Genet. 2007, 80, 759–768. [Google Scholar] [CrossRef]
- Fregel, R.; Suárez, N.M.; Betancor, E.; González, A.M.; Cabrera, V.M.; Pestano, J. Mitochondrial DNA haplogroup phylogeny of the dog: Proposal for a cladistic nomenclature. Mitochondrion 2015, 22, 75–84. [Google Scholar] [CrossRef]
- Losey, R.J.; Garvie-Lok, S.; Leonard, J.A.; Katzenberg, M.A.; Germonpré, M.; Nomokonova, T.; Savel’ev, N.A. Burying dogs in ancient Cis-Baikal, Siberia: Temporal trends and relationships with human diet and subsistence practices. PLoS ONE 2013, 8, e63740. [Google Scholar] [CrossRef]
- Bartholdy, B.P.; Murchie, T.J.; Hacking, K.; Verwoerd, C. Dog days on the Plains: A preliminary aDNA analysis of canid bones from southern Alberta and Saskatchewan. Can. J. Archaeol. 2017, 41, 46–62. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yankova, I.; Marinov, M.; Neov, B.; Petrova, M.; Spassov, N.; Hristov, P.; Radoslavov, G. Evidence for Early European Neolithic Dog Dispersal: New Data on Southeastern European Subfossil Dogs from the Prehistoric and Antiquity Ages. Genes 2019, 10, 757. https://doi.org/10.3390/genes10100757
Yankova I, Marinov M, Neov B, Petrova M, Spassov N, Hristov P, Radoslavov G. Evidence for Early European Neolithic Dog Dispersal: New Data on Southeastern European Subfossil Dogs from the Prehistoric and Antiquity Ages. Genes. 2019; 10(10):757. https://doi.org/10.3390/genes10100757
Chicago/Turabian StyleYankova, Iskra, Miroslav Marinov, Boyko Neov, Maria Petrova, Nikolai Spassov, Peter Hristov, and Georgi Radoslavov. 2019. "Evidence for Early European Neolithic Dog Dispersal: New Data on Southeastern European Subfossil Dogs from the Prehistoric and Antiquity Ages" Genes 10, no. 10: 757. https://doi.org/10.3390/genes10100757
APA StyleYankova, I., Marinov, M., Neov, B., Petrova, M., Spassov, N., Hristov, P., & Radoslavov, G. (2019). Evidence for Early European Neolithic Dog Dispersal: New Data on Southeastern European Subfossil Dogs from the Prehistoric and Antiquity Ages. Genes, 10(10), 757. https://doi.org/10.3390/genes10100757






