Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Vectors, and Strains
2.2. De Novo Transcriptomic Assembly
2.3. Characterization and Phylogenetic Analysis
2.4. Extraction and cDNA Cloning of CtCYP82G24
2.5. Subcellular Compartmentalization of CtCYP82G24
2.6. Expression and Induction of Recombinant Protein
2.7. Vector Construction and Agrobacterium-Mediated Transformation of Arabidopsis
2.8. Metabolites Accumulation in Transgenic Plants
2.9. Quantitative RT-PCR Analysis
2.10. Statistical Analysis
3. Results
3.1. Discovery and Functional Annotation of CtCYP82G24
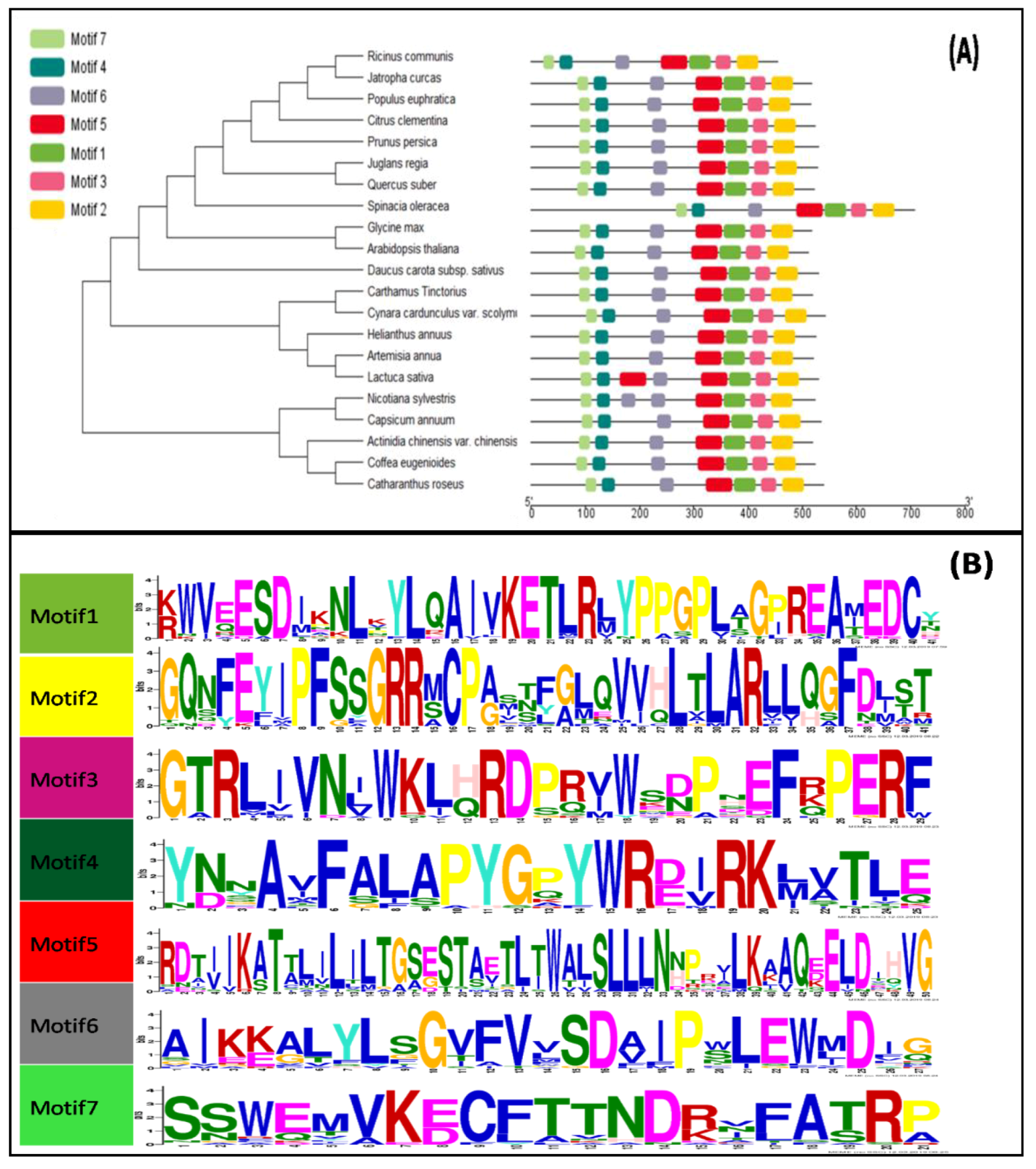
3.2. Phylogenetic and Conserved Domain Analysis
3.3. Cloning and Characterization of CtCYP82G24
3.4. Subcellular Localization of CtCYP82G24
3.5. Heterologous Expression and In Vitro Enzymatic Activity of CtCYP82G24
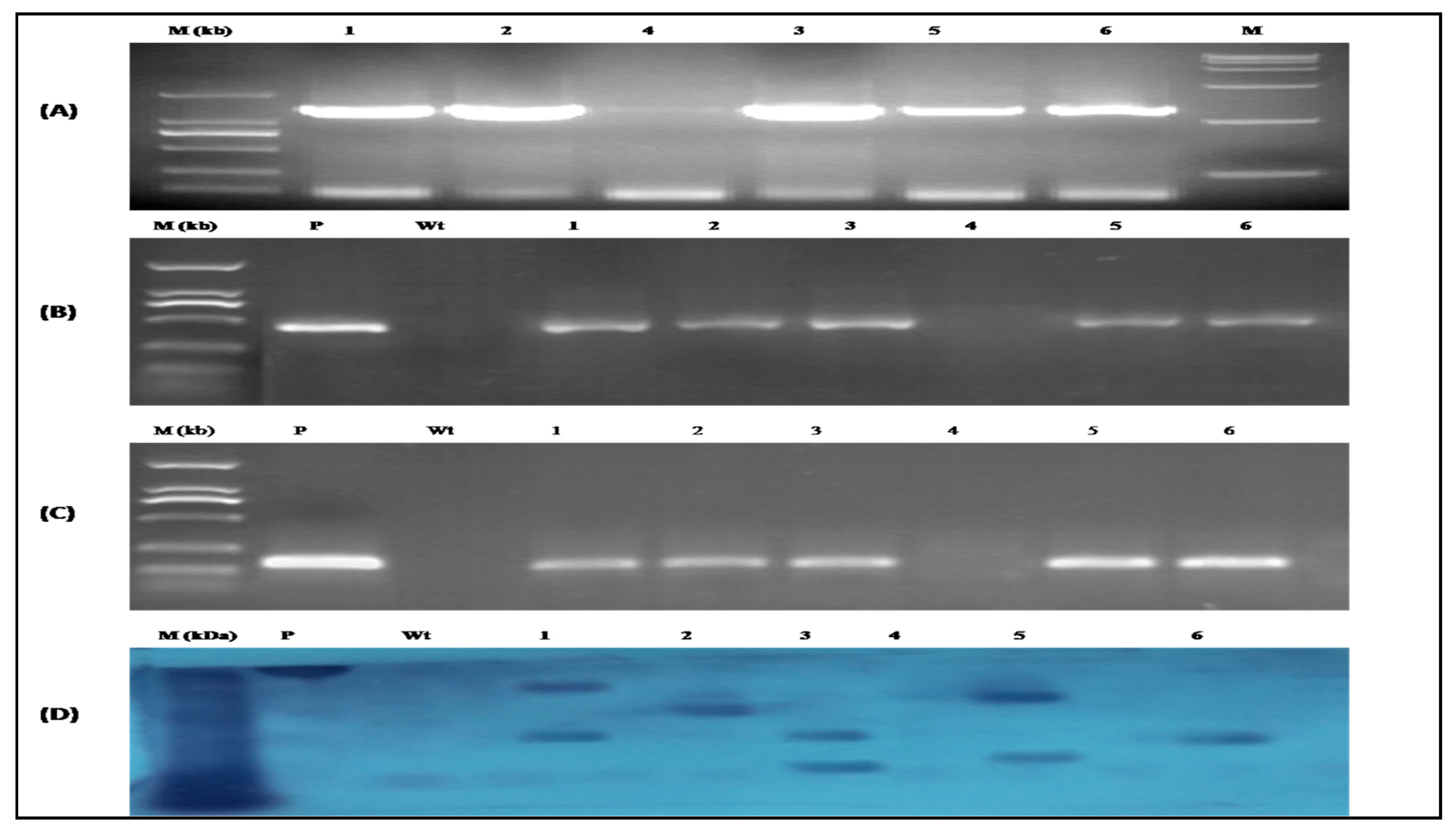
3.6. Identification of CtCYP82G24 Overexpressed Transgenic Lines of Arabidopsis
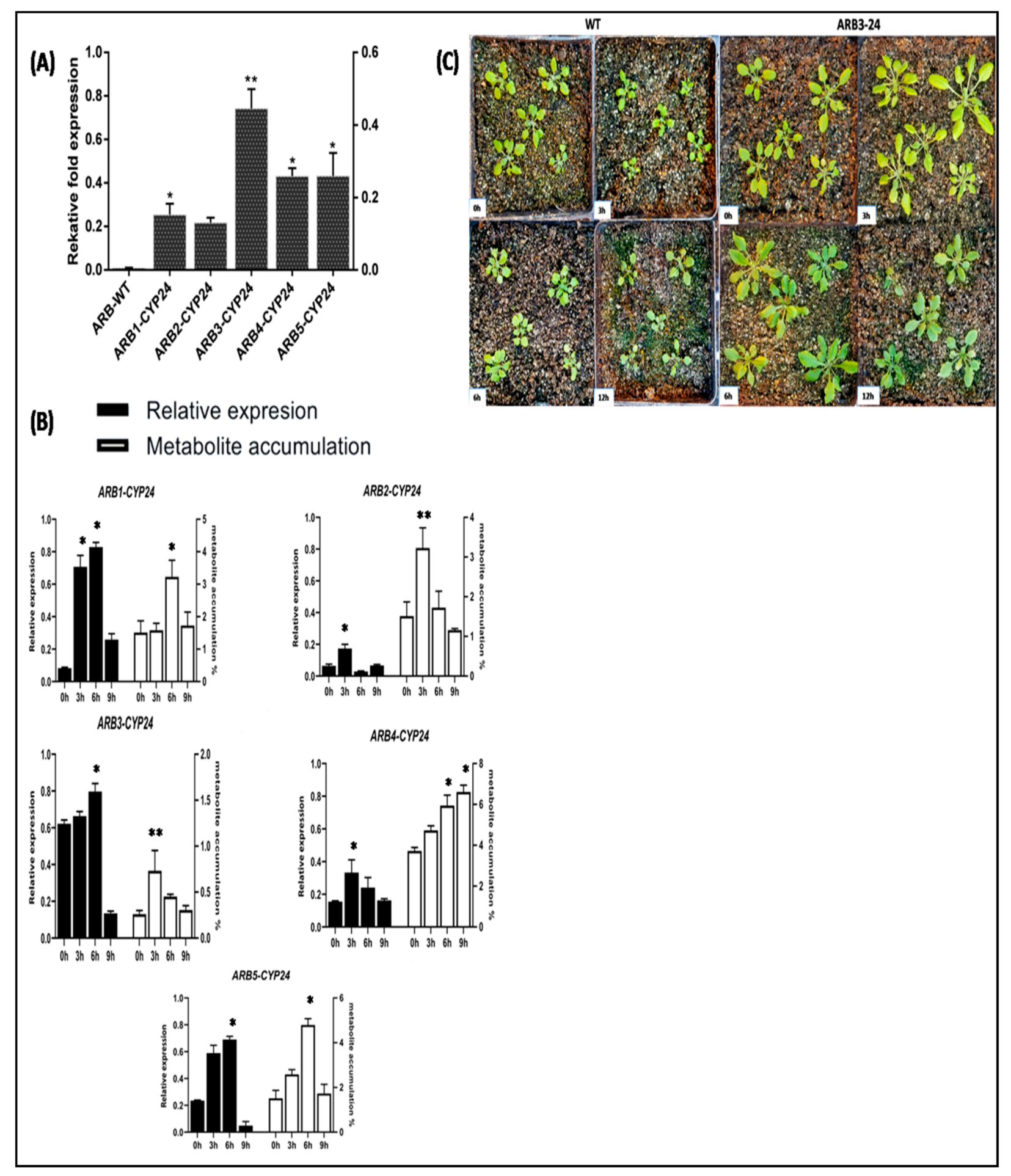
3.7. Expression Profiling of CtCYP82G24 and Metabolite Accumulation in Transgenic Plants under PEG-Induced Osmotic Stress
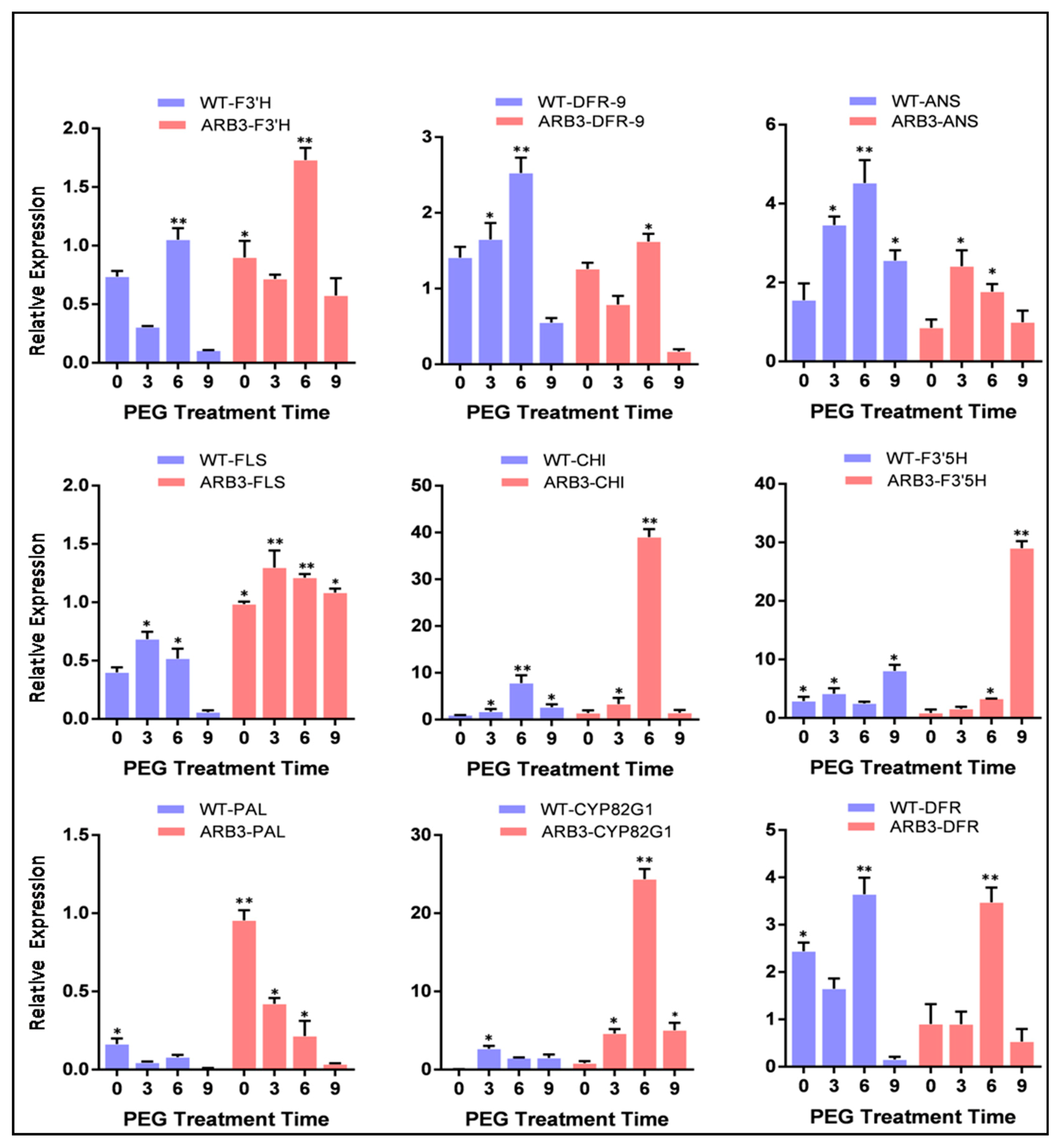
3.8. Overexpression of CtCYP82G24 Induced the Expression of Flavonoid Pathway Genes in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| GFP | Green fluorescence protein |
| KDa | Killo Dalton |
| PI | Isoelectric point |
| OD | Optical density |
| PPIase | Peptidyl-prolyl cis-trans isomerase |
| RRM | RNA recognition motif |
| ZAT | Zinc transporter family |
| MTPA2 | Metal tolerance protein |
| SDS | Sodium Dodecyl Sulfate |
References
- Winkel-Shirley, B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tang, L.; Xu, Y.; Zhou, G.; Wang, Z. Towards a better understanding of medicinal uses of Carthamus tinctorius L. in traditional Chinese medicine: A phytochemical and pharmacological review. J. Ethnopharmacol. 2014, 151, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, K.; Takahashi, T.; Sato, K.; Takeuchi, H.; Matsumoto, T.; Okuno, T. Quinochalcones and flavonoids from fresh florets in different cultivars of Carthamus tinctorius L. Biosci. Biotechnol. Biochem. 2000, 64, 1588–1599. [Google Scholar] [CrossRef] [PubMed]
- Stafford, H. Possible multienzyme complexes regulating the formation of C6-C3 phenolic compounds and lignins in higher plants. In Recent Advances in Phytochemistry; Elsevier: Warsaw, Portland, 1974; Volume 8, pp. 53–79. [Google Scholar]
- Winkel, B.S. Metabolic channeling in plants. Annu. Rev. Plant Biol. 2004, 55, 85–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Hillwig, M.L.; Wu, Y.; Peters, R.J. CYP701A8: A rice ent-kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 2012, 158, 1418–1425. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, C.A.; Chandler, P.M.; Poole, A.; Dennis, E.S.; Peacock, W.J. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc. Natl. Acad. Sci. USA 2001, 98, 2065–2070. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Ohnishi, T.; Fujioka, S.; Watanabe, B.; Mizutani, M. Rice CYP90D2 and CYP90D3 catalyze C-23 hydroxylation of brassinosteroids in vitro. Plant Physiol. Biochem. 2012, 58, 220–226. [Google Scholar] [CrossRef]
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef]
- Tanaka, Y. Flower colour and cytochromes P450. Phytochem. Rev. 2006, 5, 283–291. [Google Scholar] [CrossRef]
- Koo, A.J.; Cooke, T.F.; Howe, G.A. Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc. Natl. Acad. Sci. USA 2011, 108, 9298–9303. [Google Scholar] [CrossRef] [Green Version]
- Heitz, T.; Widemann, E.; Lugan, R.; Miesch, L.; Ullmann, P.; Désaubry, L.; Holder, E.; Grausem, B.; Kandel, S.; Miesch, M. Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J. Biol. Chem. 2012, 287, 6296–6306. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, H.; Sun, X.; Fu, F.; Zhang, X.; Wang, J.; An, J.; Ding, H. Hydroxysafflor yellow A protects rat brains against ischemia-reperfusion injury by antioxidant action. Neurosci. Lett. 2005, 386, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Yue, S.; Tang, Y.; Li, S.; Duan, J.-A. Chemical and biological properties of quinochalcone C-glycosides from the florets of Carthamus tinctorius. Molecules 2013, 18, 15220–15254. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, Z.; Ma, C.; Tian, J.; Fu, F.; Li, C.; Guo, D.; Roeder, E.; Liu, K. Neuroprotective effects of hydroxysafflor yellow A: In vivo and in vitro studies. Planta Med. 2003, 69, 429–433. [Google Scholar] [PubMed]
- Roh, J.S.; Han, J.Y.; Kim, J.H.; Hwang, J.K. Inhibitory effects of active compounds isolated from safflower (Carthamus tinctorius L.) seeds for melanogenesis. Biol. Pharm. Bull. 2004, 27, 1976–1978. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.-Y.; Gao, W.-Y. Hydroxysafflor yellow A protects neuron against hypoxia injury and suppresses inflammatory responses following focal ischemia reperfusion in rats. Arch. Pharm. Res. 2008, 31, 1010–1015. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Dong, H.; Zhao, X.; Zhou, L.; Li, X.; Liu, J.; Niu, Y. Hydroxysafflor yellow A protects against chronic carbon tetrachloride-induced liver fibrosis. Eur. J. Pharmacol. 2011, 660, 438–444. [Google Scholar] [CrossRef]
- Routaboul, J.-M.; Kerhoas, L.; Debeaujon, I.; Pourcel, L.; Caboche, M.; Einhorn, J.; Lepiniec, L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 2006, 224, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Chevreux, B.; Pfisterer, T.; Drescher, B.; Driesel, A.J.; Müller, W.E.; Wetter, T.; Suhai, S. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004, 14, 1147–1159. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Tu, L.; Deng, F.; Hu, H.; Nie, Y.; Zhang, X. A genetic and metabolic analysis revealed that cotton fiber cell development was retarded by flavonoid naringenin. Plant Physiol. 2013, 162, 86–95. [Google Scholar] [CrossRef] [PubMed]
- Saraf, A.; Sankhala, S. Simultaneous Determination of Rutin and Quercetin in Different Parts of Tecomella undulata (Seem): An Endangered Medicinal Plant. IJPR 2014, 6, 434–439. [Google Scholar]
- Seal, T. Quantitative HPLC analysis of phenolic acids, flavonoids and ascorbic acid in four different solvent extracts of two wild edible leaves, Sonchus arvensis and Oenanthe linearis of North-Eastern region in India. J. Appl. Pharm. Sci. 2016, 6, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Jailani, A.A.K.; Kumar, A.; Mandal, B.; Sivasudha, T.; Roy, A. Agroinfection of tobacco by croton yellow vein mosaic virus and designing of a replicon vector for expression of foreign gene in plant. Virus Dis. 2016, 27, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Yin, Q.; Wu, R.; Zheng, G.; Liu, J.; Dixon, R.A.; Pang, Y. Role of a chalcone isomerase-like protein in flavonoid biosynthesis in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 7165–7179. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2015, 44, D279–D285. [Google Scholar] [CrossRef]
- Yang, W.-J.; Du, Y.-T.; Zhou, Y.-B.; Chen, J.; Xu, Z.-S.; Ma, Y.-Z.; Chen, M.; Min, D.-H. Overexpression of TaCOMT Improves Melatonin Production and Enhances Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 652. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Urao, T.; Katagiri, T.; Mizoguchi, T.; Yamaguchi-Shinozaki, K.; Hayashida, N.; Shinozaki, K. Two genes that encode Ca 2+-dependent protein kinases are induced by drought and high-salt stresses in Arabidopsis thaliana. Mol. Gener. Genet. MGG 1994, 244, 331–340. [Google Scholar]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Saslowsky, D.E.; Warek, U.; Winkel, B.S. Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 2005, 280, 23735–23740. [Google Scholar] [CrossRef] [PubMed]
- Eom, S.-H.; Jin, C.-W.; Park, H.-J.; Kim, E.-H.; Chung, I.-M.; Kim, M.-J.; Yu, C.-Y.; Cho, D.-H. Far infrared ray irradiation stimulates antioxidant activity in Vitis flexuosa THUNB. Berries. Kor. J. Med. Crop Sci. 2007, 15, 319–323. [Google Scholar]
- Quattrocchio, F.; Baudry, A.; Lepiniec, L.; Grotewold, E. The regulation of flavonoid biosynthesis. In The Science of Flavonoids; Springer: New York, NY, USA, 2006; pp. 97–122. [Google Scholar]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burbulis, I.E.; Winkel-Shirley, B. Interactions among enzymes of the Arabidopsis flavonoid biosynthetic pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 12929–12934. [Google Scholar] [CrossRef] [PubMed]
- Lukačin, R.; Wellmann, F.; Britsch, L.; Martens, S.; Matern, U. Flavonol synthase from Citrus unshiu is a bifunctional dioxygenase. Phytochemistry 2003, 62, 287–292. [Google Scholar] [CrossRef]
- Ali, M.B.; Howard, S.; Chen, S.; Wang, Y.; Yu, O.; Kovacs, L.G.; Qiu, W. Berry skin development in Norton grape: Distinct patterns of transcriptional regulation and flavonoid biosynthesis. BMC Plant Biol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Zifkin, M.; Jin, A.; Ozga, J.A.; Zaharia, L.I.; Schernthaner, J.P.; Gesell, A.; Abrams, S.R.; Kennedy, J.A.; Constabel, C.P. Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 2012, 158, 200–224. [Google Scholar] [CrossRef]
- Footitt, S.; Cornah, J.E.; Pracharoenwattana, I.; Bryce, J.H.; Smith, S.M. The Arabidopsis 3-ketoacyl-CoA thiolase-2 (kat2–1) mutant exhibits increased flowering but reduced reproductive success. J. Exp. Bot. 2007, 58, 2959–2968. [Google Scholar] [CrossRef]
- Ma, D.; Sun, D.; Wang, C.; Li, Y.; Guo, T. Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol. Biochem. 2014, 80, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tang, X.; Ren, C.; Wei, B.; Wu, Y.; Wu, Q.; Pei, J. Full-length transcriptome sequences and the identification of putative genes for flavonoid biosynthesis in safflower. BMC Genom. 2018, 19, 548. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Herrera, F.; Moreno, A.A.; Tapia, R.; Reyes, F.; Araya, M.; D’Alessio, C.; Parodi, A.; Orellana, A. The UDP-glucose: Glycoprotein glucosyltransferase (UGGT), a key enzyme in ER quality control, plays a significant role in plant growth as well as biotic and abiotic stress in Arabidopsis thaliana. BMC Plant Biol. 2015, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Rabara, R.C.; Tripathi, P.; Reese, R.N.; Rushton, D.L.; Alexander, D.; Timko, M.P.; Shen, Q.J.; Rushton, P.J. Tobacco drought stress responses reveal new targets for Solanaceae crop improvement. BMC Genom. 2015, 16, 484. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Liu, J.; Xu, J.; Duan, S.; Wang, Q.; Li, G.; Jin, L. Transcriptome Profiling Reveals Effects of Drought Stress on Gene Expression in Diploid Potato Genotype P3–198. Int. J. Mol. Sci. 2019, 20, 852. [Google Scholar] [CrossRef]
- Schopfer, C.; Ebel, J. Identification of elicitor-induced cytochrome P450s of soybean (Glycine max L.) using differential display of mRNA. Mol. Gener. Genet. MGG 1998, 258, 315–322. [Google Scholar] [CrossRef]
- Kitamura, S.; Shikazono, N.; Tanaka, A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004, 37, 104–114. [Google Scholar] [CrossRef]
- Luis, L.; Foresti, O.; Denecke, J. Targeting of the plant vacuolar sorting receptor BP80 is dependent on multiple sorting signals in the cytosolic tail. Plant Cell 2006, 18, 1477–1497. [Google Scholar]
- Verweij, W.; Spelt, C.; Di Sansebastiano, G.-P.; Vermeer, J.; Reale, L.; Ferranti, F.; Koes, R.; Quattrocchio, F. An H+ P-ATPase on the tonoplast determines vacuolar pH and flower colour. Nat. Cell Biol. 2008, 10, 1456. [Google Scholar] [CrossRef]
- Abrahams, S.; Lee, E.; Walker, A.R.; Tanner, G.J.; Larkin, P.J.; Ashton, A.R. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003, 35, 624–636. [Google Scholar] [CrossRef]
- Poustka, F.; Irani, N.G.; Feller, A.; Lu, Y.; Pourcel, L.; Frame, K.; Grotewold, E. A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 2007, 145, 1323–1335. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I.R.; Young, J.C.; Armstrong, G.; Foster, N.; Bogenschutz, N.; Cordova, T.; Peer, W.A.; Hazen, S.P.; Murphy, A.S.; Harper, J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 2649–2654. [Google Scholar] [CrossRef] [PubMed]
- Feucht, W.; Dithmar, H.; Polster, J. Variation of the nuclear, subnuclear and chromosomal flavanol deposition in hemlock and rye. Int. J. Mol. Sci. 2007, 8, 635–650. [Google Scholar] [CrossRef]
- Polster, J.; Dithmar, H.; Burgemeister, R.; Friedemann, G.; Feucht, W. Flavonoids in plant nuclei: Detection by laser microdissection and pressure catapulting (LMPC), in vivo staining, and uv–visible spectroscopic titration. Physiol. Plant. 2006, 128, 163–174. [Google Scholar] [CrossRef]
- Horinouchi, S. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr. Opin. Chem. Biol. 2009, 13, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Marienhagen, J.; Bott, M. Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 2013, 163, 166–178. [Google Scholar] [CrossRef] [PubMed]
- Katsuyama, Y.; Miyahisa, I.; Funa, N.; Horinouchi, S. One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl. Microbiol. Biotechnol. 2007, 73, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Miyahisa, I.; Funa, N.; Ohnishi, Y.; Martens, S.; Moriguchi, T.; Horinouchi, S. Combinatorial biosynthesis of flavones and flavonols in Escherichia coli. Appl. Microbiol. Biotechnol. 2006, 71, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Kirchmair, J.; Williamson, M.J.; Tyzack, J.D.; Tan, L.; Bond, P.J.; Bender, A.; Glen, R.C. Computational prediction of metabolism: Sites, products, SAR, P450 enzyme dynamics, and mechanisms. J. Chem. Inf. Model. 2012, 52, 617–648. [Google Scholar] [CrossRef]
- De Bruyn Kops, C.; Stork, C.; Šícho, M.; Kochev, N.; Svozil, D.; Jeliazkova, N.; Kirchmair, J. GLORY: Generator of the Structures of Likely Cytochrome P450 Metabolites Based on Predicted Sites of Metabolism. Front. Chem. 2019, 7. [Google Scholar] [CrossRef]
- Takahama, U. Suppression of carotenoid photobleaching by kaempferol in isolated chloroplasts. Plant Cell Physiol. 1982, 23, 859–864. [Google Scholar]
- Agati, G.; Matteini, P.; Goti, A.; Tattini, M. Chloroplast-located flavonoids can scavenge singlet oxygen. New Phytol. 2007, 174, 77–89. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Normanly, J.; Tague, B.W.; Peer, W.A.; Taiz, L.; Muday, G.K. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.A.; van den Noort, R.E.; Tan, M.A.; Prinsen, E.; Lagrimini, L.M.; Thorneley, R.N. Phenol-oxidizing peroxidases contribute to the protection of plants from ultraviolet radiation stress. Plant Physiol. 2001, 126, 1012–1023. [Google Scholar] [CrossRef] [PubMed]
- Taylor, L.P.; Grotewold, E. Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 2005, 8, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Bandyopadhyay, A.; Blakeslee, J.J.; Makam, S.N.; Chen, R.J.; Masson, P.H.; Murphy, A.S. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 2004, 16, 1898–1911. [Google Scholar] [CrossRef]
- Ali, B.; Wang, B.; Ali, S.; Ghani, M.; Hayat, M.; Yang, C.; Xu, L.; Zhou, W. 5-Aminolevulinic acid ameliorates the growth, photosynthetic gas exchange capacity, and ultrastructural changes under cadmium stress in Brassica napus L. J. Plant Growth Regul. 2013, 32, 604–614. [Google Scholar] [CrossRef]
- An, Y.; Liu, L.; Chen, L.; Wang, L. ALA inhibits ABA-induced stomatal closure via reducing H2O2 and Ca2+ levels in guard cells. Front. Plant Sci. 2016, 7, 482. [Google Scholar] [CrossRef]
- Kim, T.-H.; Böhmer, M.; Hu, H.; Nishimura, N.; Schroeder, J.I. Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. [Google Scholar] [CrossRef]
- Li, D.-M.; Zhang, J.; Sun, W.-J.; Li, Q.; Dai, A.-H.; Bai, J.-G. 5-Aminolevulinic acid pretreatment mitigates drought stress of cucumber leaves through altering antioxidant enzyme activity. Sci. Horticult. 2011, 130, 820–828. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, N.; Jianyu, L.; Xu, T.; Noman, M.; Jameel, A.; Na, Y.; Yuanyuan, D.; Nan, W.; Xiaowei, L.; Fawei, W.; et al. Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis. Genes 2019, 10, 756. https://doi.org/10.3390/genes10100756
Ahmad N, Jianyu L, Xu T, Noman M, Jameel A, Na Y, Yuanyuan D, Nan W, Xiaowei L, Fawei W, et al. Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis. Genes. 2019; 10(10):756. https://doi.org/10.3390/genes10100756
Chicago/Turabian StyleAhmad, Naveed, Liu Jianyu, Tian Xu, Muhammad Noman, Aysha Jameel, Yao Na, Dong Yuanyuan, Wang Nan, Li Xiaowei, Wang Fawei, and et al. 2019. "Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis" Genes 10, no. 10: 756. https://doi.org/10.3390/genes10100756
APA StyleAhmad, N., Jianyu, L., Xu, T., Noman, M., Jameel, A., Na, Y., Yuanyuan, D., Nan, W., Xiaowei, L., Fawei, W., Xiuming, L., & Haiyan, L. (2019). Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis. Genes, 10(10), 756. https://doi.org/10.3390/genes10100756





