SMC5/6: Multifunctional Player in Replication
Abstract
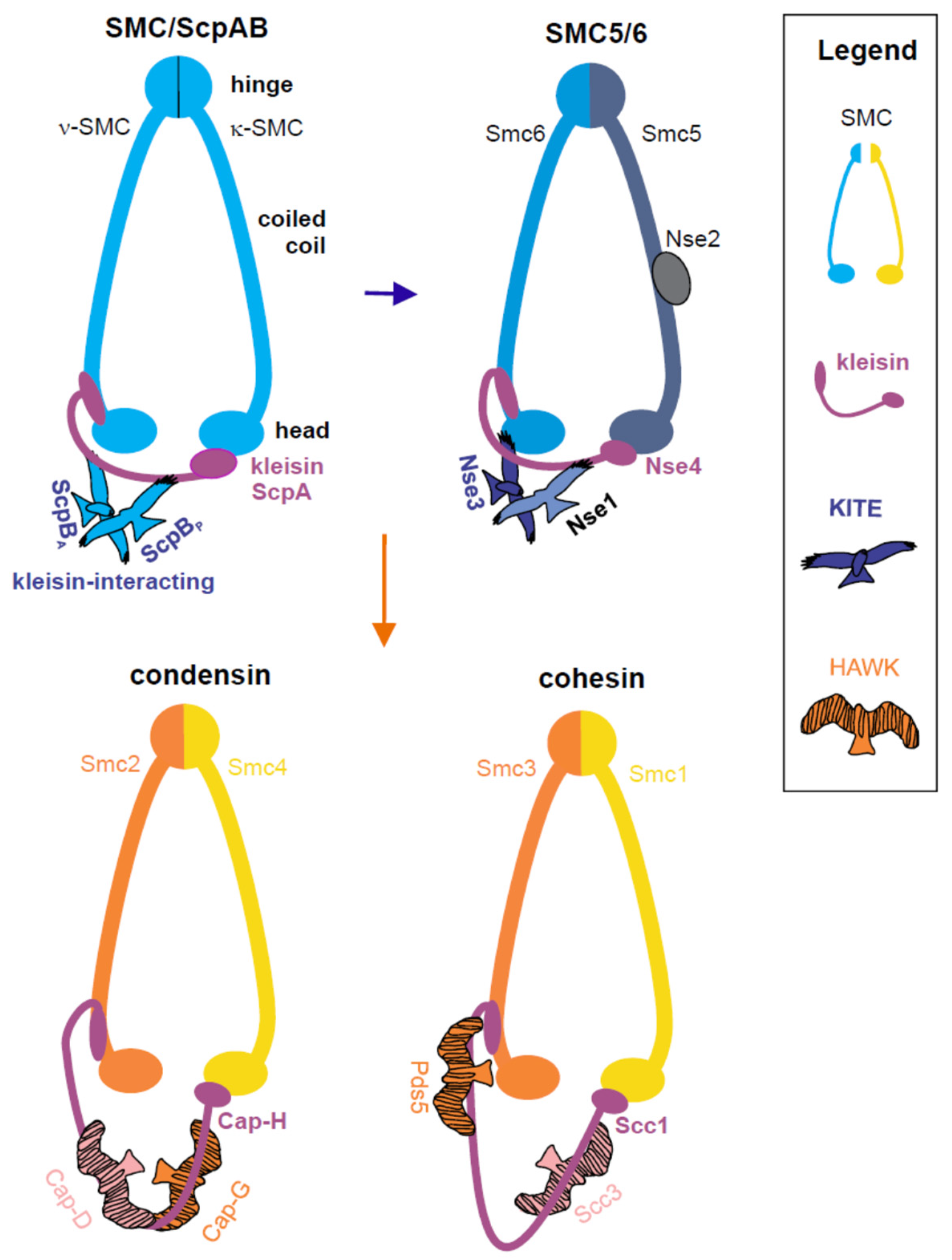
1. Introduction
2. SMC5/6 Roles in Maintenance of Genome Integrity
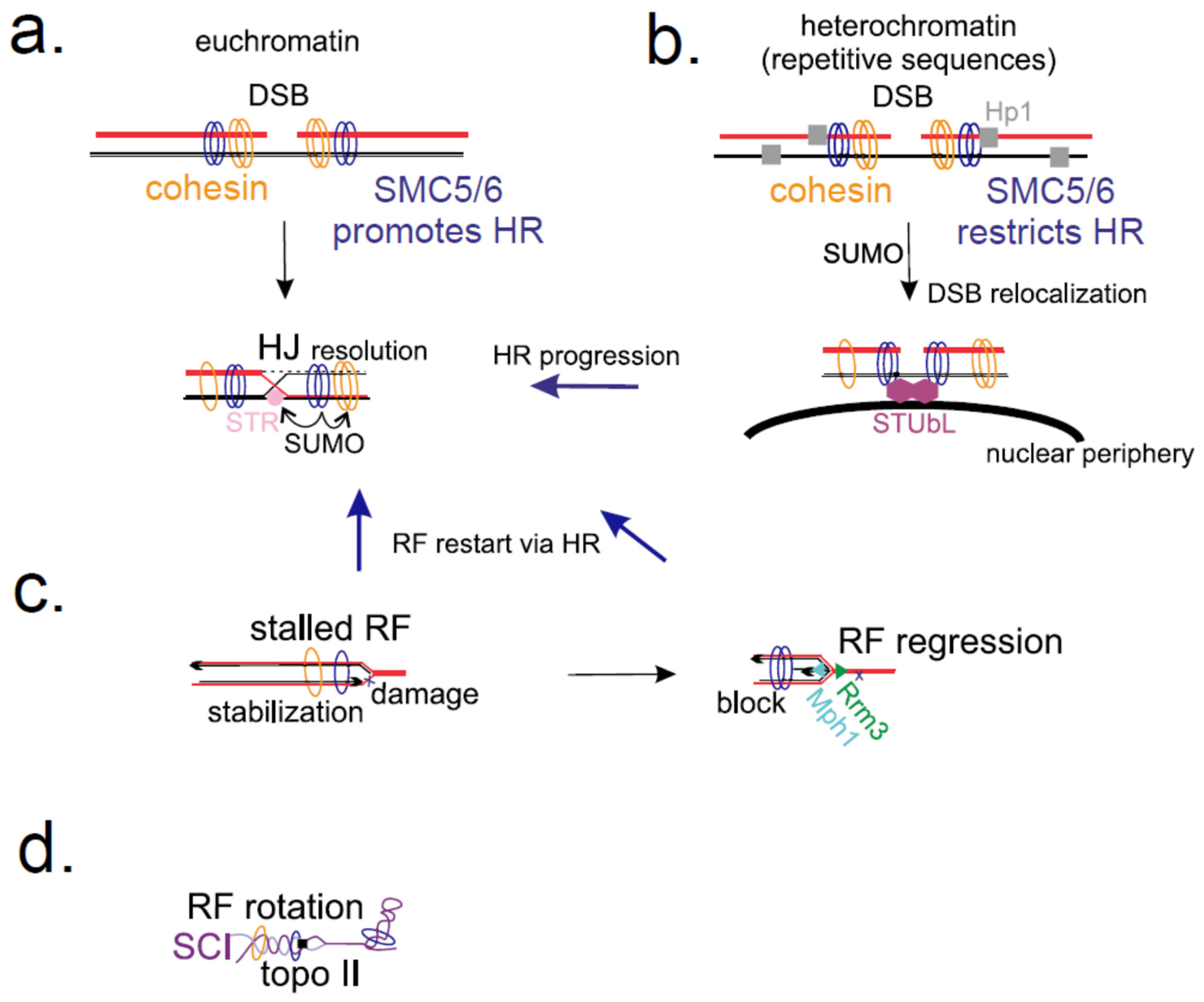
2.1. SMC5/6 in Homologous Recombination
2.2. SMC5/6 Role at Stressed Replication Forks
3. SMC5/6 Is Involved in Normal Progression of Replication
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Bürmann, F.; Gruber, S. SMC condensin: Promoting cohesion of replicon arms. Nat. Struct. Mol. Biol 2015, 22, 653–655. [Google Scholar] [CrossRef] [PubMed]
- Gruber, S. SMC complexes sweeping through the chromosome: Going with the flow and against the tide. Curr. Opin. Microbiol. 2018, 42, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Pecinka, A. Scaffolding for Repair: Understanding Molecular Functions of the SMC5/6 Complex. Genes 2018, 9, 36. [Google Scholar] [CrossRef]
- Murray, J.M.; Carr, A.M. Smc5/6: A link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 2007, 9, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, F. SMC complexes: From DNA to chromosomes. Nat. Rev. Mol. Cell Biol. 2016, 17, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef] [PubMed]
- Hassler, M.; Shaltiel, I.A.; Haering, C.H. Towards a Unified Model of SMC Complex Function. Curr. Biol. 2018, 28, R1266–R1281. [Google Scholar] [CrossRef]
- Kschonsak, M.; Haering, C.H. Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. Bioessays 2015, 37, 755–766. [Google Scholar] [CrossRef]
- Haering, C.H.; Gruber, S. SnapShot: SMC Protein Complexes Part II. Cell 2016, 164, 818. [Google Scholar] [CrossRef]
- Carter, S.D.; Sjogren, C. The SMC complexes, DNA and chromosome topology: Right or knot? Crit. Rev. Biochem. Mol. Biol. 2012, 47, 1–16. [Google Scholar] [CrossRef]
- Jeppsson, K.; Kanno, T.; Shirahige, K.; Sjögren, C. The maintenance of chromosome structure: Positioning and functioning of SMC complexes. Nat. Rev. Mol. Cell Biol. 2014, 15, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Kegel, A.; Sjögren, C. The Smc5/6 complex: More than repair? Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 179–187. [Google Scholar] [CrossRef]
- Aragon, L.; Martinez-Perez, E.; Merkenschlager, M. Condensin, cohesin and the control of chromatin states. Curr. Opin. Genet. Dev. 2013, 23, 204–211. [Google Scholar] [CrossRef][Green Version]
- De Piccoli, G.; Torres-Rosell, J.; Aragon, L. The unnamed complex: What do we know about Smc5-Smc6? Chromosome Res. 2009, 17, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Burmann, F.; Basfeld, A.; Nunez, R.V.; Diebold-Durand, M.L.; Wilhelm, L.; Gruber, S. Tuned SMC Arms Drive Chromosomal Loading of Prokaryotic Condensin. Mol. Cell 2017, 65, 861–872. [Google Scholar] [CrossRef]
- Alt, A.; Dang, H.Q.; Wells, O.S.; Polo, L.M.; Smith, M.A.; McGregor, G.A.; Welte, T.; Lehmann, A.R.; Pearl, L.H.; Murray, J.M.; et al. Specialized interfaces of Smc5/6 control hinge stability and DNA association. Nat. Commun. 2017, 8, 14011. [Google Scholar] [CrossRef] [PubMed]
- Griese, J.J.; Witte, G.; Hopfner, K.P. Structure and DNA binding activity of the mouse condensin hinge domain highlight common and diverse features of SMC proteins. Nucleic Acids Res. 2010, 38, 3454–3465. [Google Scholar] [CrossRef]
- Hirano, M.; Hirano, T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. Embo J. 2002, 21, 5733–5744. [Google Scholar] [CrossRef]
- Sergeant, J.; Taylor, E.; Palecek, J.; Fousteri, M.; Andrews, E.; Sweeney, S.; Shinagawa, H.; Watts, F.; Lehmann, A. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol. Cell. Biol. 2005, 25, 172–184. [Google Scholar] [CrossRef]
- Bürmann, F.; Shin, H.C.; Basquin, J.; Soh, Y.M.; Giménez-Oya, V.; Kim, Y.G.; Oh, B.H.; Gruber, S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol. 2013, 20, 371–379. [Google Scholar] [CrossRef]
- Gligoris, T.G.; Scheinost, J.C.; Bürmann, F.; Petela, N.; Chan, K.L.; Uluocak, P.; Beckouët, F.; Gruber, S.; Nasmyth, K.; Löwe, J. Closing the cohesin ring: Structure and function of its Smc3-kleisin interface. Science 2014, 346, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Zawadzka, K.; Zawadzki, P.; Baker, R.; Rajasekar, K.V.; Wagner, F.; Sherratt, D.J.; Arciszewska, L.K. MukB ATPases are regulated independently by the N- and C-terminal domains of MukF kleisin. Elife 2018, 7, e31522. [Google Scholar] [CrossRef] [PubMed]
- Haering, C.H.; Schoffnegger, D.; Nishino, T.; Helmhart, W.; Nasmyth, K.; Lowe, J. Structure and Stability of Cohesin’s Smc1-Kleisin Interaction. Mol. Cell 2004, 15, 951–964. [Google Scholar] [CrossRef] [PubMed]
- Palecek, J.; Vidot, S.; Feng, M.; Doherty, A.J.; Lehmann, A.R. The SMC5-6 DNA repair complex: Bridging of the SMC5-6 heads by the Kleisin, NSE4, and non-Kleisin subunits. J. Biol. Chem. 2006, 281, 36952–36959. [Google Scholar] [CrossRef]
- Haering, C.H.; Gruber, S. SnapShot: SMC Protein Complexes Part I. Cell 2016, 164, 326. [Google Scholar] [CrossRef] [PubMed]
- Cuylen, S.; Metz, J.; Haering, C.H. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 2011, 18, 894–901. [Google Scholar] [CrossRef] [PubMed]
- Haering, C.H.; Farcas, A.M.; Arumugam, P.; Metson, J.; Nasmyth, K. The cohesin ring concatenates sister DNA molecules. Nature 2008, 454, 297. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, L.; Bürmann, F.; Minnen, A.; Shin, H.C.; Toseland, C.P.; Oh, B.H.; Gruber, S. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. Elife 2015, 4, e06659. [Google Scholar] [CrossRef] [PubMed]
- Palecek, J.J.; Gruber, S. Kite Proteins: A Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure 2015, 23, 2183–2190. [Google Scholar] [CrossRef]
- Wells, J.N.; Gligoris, T.G.; Nasmyth, K.A.; Marsh, J.A. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr. Biol. 2017, 27, R17–R18. [Google Scholar] [CrossRef]
- Hudson, J.J.R.; Bednarova, K.; Kozakova, L.; Liao, C.Y.; Guerineau, M.; Colnaghi, R.; Vidot, S.; Marek, J.; Bathula, S.R.; Lehmann, A.R.; et al. Interactions between the Nse3 and Nse4 Components of the SMC5-6 Complex Identify Evolutionarily Conserved Interactions between MAGE and EID Families. PLoS ONE 2011, 6, 14. [Google Scholar] [CrossRef]
- Zabrady, K.; Adamus, M.; Vondrova, L.; Liao, C.; Skoupilova, H.; Novakova, M.; Jurcisinova, L.; Alt, A.; Oliver, A.W.; Lehmann, A.R.; et al. Chromatin association of the SMC5/6 complex is dependent on binding of its NSE3 subunit to DNA. Nucleic Acids Res. 2016, 44, 1064–1079. [Google Scholar] [CrossRef]
- Kschonsak, M.; Merkel, F.; Bisht, S.; Metz, J.; Rybin, V.; Hassler, M.; Haering, C.H. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell 2017, 171, 588–600. [Google Scholar] [CrossRef] [PubMed]
- Bisht, K.K.; Daniloski, Z.; Smith, S. SA1 binds directly to DNA through its unique AT-hook to promote sister chromatid cohesion at telomeres. J. Cell Sci. 2013, 126, 3493–3503. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J.M.; Gao, J.; Wang, J.; Yang, M.; Potts, P.R. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 2010, 39, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Kozakova, L.; Vondrova, L.; Stejskal, K.; Charalabous, P.; Kolesar, P.; Lehmann, A.R.; Uldrijan, S.; Sanderson, C.M.; Zdrahal, Z.; Palecek, J.J. The melanoma-associated antigen 1 (MAGEA1) protein stimulates the E3 ubiquitin-ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex. Cell Cycle 2015, 14, 920–930. [Google Scholar] [CrossRef]
- Andrews, E.; Palecek, J.; Sergeant, J.; Taylor, E.; Lehmann, A.; Watts, F. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 2005, 25, 185–196. [Google Scholar] [CrossRef]
- McAleenan, A.; Cordon-Preciado, V.; Clemente-Blanco, A.; Liu, I.C.; Sen, N.; Leonard, J.; Jarmuz, A.; Aragón, L. SUMOylation of the α-kleisin subunit of cohesin is required for DNA damage-induced cohesion. Curr. Biol. 2012, 22, 1564–1575. [Google Scholar] [CrossRef]
- Potts, P.R.; Yu, H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005, 25, 7021–7032. [Google Scholar] [CrossRef]
- Potts, P.R. The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair 2009, 8, 499–506. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Sunjevaric, I.; De Piccoli, G.; Sacher, M.; Eckert-Boulet, N.; Reid, R.; Jentsch, S.; Rothstein, R.; Aragón, L.; Lisby, M. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat. Cell Biol. 2007, 9, 923–931. [Google Scholar] [CrossRef]
- Varejão, N.; Ibars, E.; Lascorz, J.; Colomina, N.; Torres-Rosell, J.; Reverter, D. DNA activates the Nse2/Mms21 SUMO E3 ligase in the Smc5/6 complex. EMBO J. 2018, 37, e98306. [Google Scholar] [CrossRef]
- Wu, N.; Kong, X.; Ji, Z.; Zeng, W.; Potts, P.R.; Yokomori, K.; Yu, H. Scc1 sumoylation by Mms21 promotes sister chromatid recombination through counteracting Wapl. Genes Dev. 2012, 26, 1473–1485. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Blobel, G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. USA 2005, 102, 4777–47782. [Google Scholar] [CrossRef] [PubMed]
- Watts, F.Z.; Skilton, A.; Ho, J.C.Y.; Boyd, L.K.; Trickey, M.A.M.; Gardner, L.; Ogi, F.X.; Outwin, E.A. The role of Schizosaccharomyces pombe SUMO ligases in genome stability. Biochem. Soc. Trans. 2007, 35, 1379–1384. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Dulev, S.; Liu, X.P.; Hiller, N.J.; Zhao, X.L.; Strunnikov, A. Cooperation of Sumoylated Chromosomal Proteins in rDNA Maintenance. PLoS Genet. 2008, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Pebernard, S.; Wohlschlegel, J.; McDonald, W.H.; Yates, J.R., 3rd; Boddy, M.N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol 2006, 26, 1617–1630. [Google Scholar] [CrossRef] [PubMed]
- Räschle, M.; Smeenk, G.; Hansen, R.K.; Temu, T.; Oka, Y.; Hein, M.Y.; Nagaraj, N.; Long, D.T.; Walter, J.C.; Hofmann, K.; et al. DNA repair. Proteomics reveals dynamic assembly of repair complexes during bypass of DNA cross-links. Science 2015, 348, 1253671. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wang, W.; Marqués, J.; Mohan, R.; Saleh, A.; Durrant, W.E.; Song, J.; Dong, X. Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 2013, 52, 602–610. [Google Scholar] [CrossRef]
- Hazbun, T.R.; Malmstrom, L.; Anderson, S.; Graczyk, B.J.; Fox, B.; Riffle, M.; Sundin, B.A.; Aranda, J.D.; McDonald, W.H.; Chiu, C.H.; et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell 2003, 12, 1353–1365. [Google Scholar] [CrossRef]
- Duan, X.; Yang, Y.; Chen, Y.H.; Arenz, J.; Rangi, G.K.; Zhao, X.; Ye, H. Architecture of the Smc5/6 Complex of Saccharomyces cerevisiae Reveals a Unique Interaction between the Nse5-6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J. Biol. Chem. 2009, 284, 8507–8515. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.; Nasmyth, K. A topological interaction between cohesin rings and a circular minichromosome. Cell 2005, 122, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, D.; Nasmyth, K. A physical assay for sister chromatid cohesion in vitro. Mol. Cell 2007, 27, 300–310. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Scheinost, J.C.; Petela, N.J.; Gligoris, T.G.; Wissler, M.; Ogushi, S.; Collier, J.E.; Voulgaris, M.; Kurze, A.; Chan, K.L.; et al. The Cohesin Ring Uses Its Hinge to Organize DNA Using Non-topological as well as Topological Mechanisms. Cell 2018, 173, 1508. [Google Scholar] [CrossRef] [PubMed]
- Uhlmann, F.; Lottspeich, F.; Nasmyth, K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature 1999, 400, 37–42. [Google Scholar] [PubMed]
- Hirano, T.; Nishiyama, T.; Shirahige, K. Hot debate in hot springs: Report on the second international meeting on SMC proteins. Genes Cells 2017, 22, 934–938. [Google Scholar] [CrossRef]
- Terakawa, T.; Bisht, S.; Eeftens, J.M.; Dekker, C.; Haering, C.H.; Greene, E.C. The condensin complex is a mechanochemical motor that translocates along DNA. Science 2017, 358, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Eeftens, J.; Dekker, C. Catching DNA with hoops-biophysical approaches to clarify the mechanism of SMC proteins. Nat. Struct. Mol. Biol. 2017, 24, 1012–1020. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Hughes, A.C.; Brandao, H.B.; Walker, B.; Lierz, C.; Cochran, J.C.; Oakley, M.G.; Kruse, A.C.; Rudner, D.Z. In Vivo Evidence for ATPase-Dependent DNA Translocation by the Bacillus subtilis SMC Condensin Complex. Mol. Cell 2018, 71, 841. [Google Scholar] [CrossRef] [PubMed]
- Kamada, K.; Barilla, D. Combing Chromosomal DNA Mediated by the SMC Complex: Structure and Mechanisms. Bioessays 2018, 40, 8. [Google Scholar] [CrossRef]
- Yuen, K.C.; Gerton, J.L. Taking cohesin and condensin in context. PLoS Genet. 2018, 14, 14. [Google Scholar] [CrossRef] [PubMed]
- Fudenberg, G.; Imakaev, M.; Lu, C.; Goloborodko, A.; Abdennur, N.; Mirny, L.A. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016, 15, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Goloborodko, A.; Imakaev, M.V.; Marko, J.F.; Mirny, L. Compaction and segregation of sister chromatids via active loop extrusion. Elife 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.D.; Brandao, H.B.; Le, T.B.K.; Laub, M.T.; Rudner, D.Z. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science 2017, 355, 524–527. [Google Scholar] [CrossRef] [PubMed]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-time imaging of DNA loop extrusion by condensin. Science 2018. [CrossRef] [PubMed]
- Kanno, T.; Berta, D.G.; Sjögren, C. The Smc5/6 Complex Is an ATP-Dependent Intermolecular DNA Linker. Cell Rep. 2015, 12, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Farcas, A.M.; Uluocak, P.; Helmhart, W.; Nasmyth, K. Cohesin’s Concatenation of Sister DNAs Maintains Their Intertwining. Mol. Cell 2011, 44, 97–107. [Google Scholar] [CrossRef]
- Verver, D.E.; Hwang, G.H.; Jordan, P.W.; Hamer, G. Resolving complex chromosome structures during meiosis: Versatile deployment of Smc5/6. Chromosoma 2016, 125, 15–27. [Google Scholar] [CrossRef]
- Lehmann, A.R.; Walicka, M.; Griffiths, D.J.F.; Murray, J.M.; Watts, F.Z.; McCready, S.; Carr, A.M. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 1995, 15, 7067–7080. [Google Scholar] [CrossRef]
- McDonald, W.H.; Pavlova, Y.; Yates, J.R.; Boddy, M.N. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 2003, 278, 45460–45467. [Google Scholar] [CrossRef]
- Miyabe, I.; Morishita, T.; Hishida, T.; Yonei, S.; Shinagawa, H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell. Biol. 2006, 26, 343–353. [Google Scholar] [CrossRef] [PubMed]
- De Piccoli, G.; Cortes-Ledesma, F.; Ira, G.; Torres-Rosell, J.; Uhle, S.; Farmer, S.; Hwang, J.Y.; Machin, F.; Ceschia, A.; McAleenan, A.; et al. Smc5-Smc6 mediate DNA double-strand-break repair by promoting sister-chromatid recombination. Nat. Cell Biol. 2006, 8, 1032–1034. [Google Scholar] [CrossRef] [PubMed]
- Lindroos, H.B.; Strom, L.; Itoh, T.; Katou, Y.; Shirahige, K.; Sjogren, C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006, 22, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Potts, P.R.; Porteus, M.H.; Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006, 25, 3377–3388. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Pacher, M.; Dukowic, S.; Schubert, V.; Puchta, H.; Schubert, I. The STRUCTURAL MAINTENANCE OF CHROMOSOMES 5/6 complex promotes sister chromatid alignment and homologous recombination after DNA damage in Arabidopsis thaliana. Plant Cell 2009, 21, 2688–2699. [Google Scholar] [CrossRef] [PubMed]
- Mengiste, T.; Revenkova, E.; Bechtold, N.; Paszkowski, J. An SMC-like protein is required for efficient homologous recombination in Arabidopsis. EMBO J. 1999, 18, 4505–4512. [Google Scholar] [CrossRef]
- Stephan, A.K.; Kliszczak, M.; Dodson, H.; Cooley, C.; Morrison, C.G. Roles of vertebrate Smc5 in sister chromatid cohesion and homologous recombinational repair. Mol. Cell. Biol. 2011, 31, 1369–1381. [Google Scholar] [CrossRef]
- Strom, L.; Karlsson, C.; Lindroos, H.B.; Wedahl, S.; Katou, Y.; Shirahige, K.; Sjogren, C. Postreplicative formation of cohesion is required for repair and induced by a single DNA break. Science 2007, 317, 242–245. [Google Scholar] [CrossRef]
- Almedawar, S.; Colomina, N.; Bermúdez-López, M.; Pociño-Merino, I.; Torres-Rosell, J. A SUMO-dependent step during establishment of sister chromatid cohesion. Curr. Biol. 2012, 22, 1576–1581. [Google Scholar] [CrossRef]
- Ampatzidou, E.; Irmisch, A.; O’Connell, M.J.; Murray, J.M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 2006, 26, 9387–9401. [Google Scholar] [CrossRef]
- Torres-Rosell, J.; Machin, F.; Farmer, S.; Jarmuz, A.; Eydmann, T.; Dalgaard, J.Z.; Aragon, L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005, 7, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rosell, J.; Machin, F.; Aragón, L. Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle 2005, 4, 868–872. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pebernard, S.; Schaffer, L.; Campbell, D.; Head, S.R.; Boddy, M.N. Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 2008, 27, 3011–3023. [Google Scholar] [CrossRef] [PubMed]
- Menolfi, D.; Delamarre, A.; Lengronne, A.; Pasero, P.; Branzei, D. Essential Roles of the Smc5/6 Complex in Replication through Natural Pausing Sites and Endogenous DNA Damage Tolerance. Mol. Cell 2015, 60, 835–846. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.Y.; Smith, S.; Ceschia, A.; Torres-Rosell, J.; Aragon, L.; Myung, K. Smc5-Smc6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair 2008, 7, 1426–1436. [Google Scholar] [CrossRef] [PubMed]
- Caridi, C.P.; D’Agostino, C.; Ryu, T.; Zapotoczny, G.; Delabaere, L.; Li, X.; Khodaverdian, V.Y.; Amaral, N.; Lin, E.; Rau, A.R.; et al. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 2018, 559, 54–60. [Google Scholar] [CrossRef]
- Chiolo, I.; Minoda, A.; Colmenares, S.U.; Polyzos, A.; Costes, S.V.; Karpen, G.H. Double-Strand Breaks in Heterochromatin Move Outside of a Dynamic HP1a Domain to Complete Recombinational Repair. Cell 2011, 144, 732–744. [Google Scholar] [CrossRef]
- Amaral, N.; Ryu, T.; Li, X.; Chiolo, I. Nuclear Dynamics of Heterochromatin Repair. Trends Genet. 2017, 33, 86–100. [Google Scholar] [CrossRef]
- Ryu, T.; Spatola, B.; Delabaere, L.; Bowlin, K.; Hopp, H.; Kunitake, R.; Karpen, G.H.; Chiolo, I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biology 2015, 17, 1401–1411. [Google Scholar] [CrossRef]
- Perry, J.J.; Tainer, J.A.; Boddy, M.N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008, 33, 201–208. [Google Scholar] [CrossRef]
- Ryu, T.; Bonner, M.R.; Chiolo, I. Cervantes and Quijote protect heterochromatin from aberrant recombination and lead the way to the nuclear periphery. Nucleus 2016, 7, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Potts, P.R.; Yu, H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol 2007, 14, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rosell, J.; De Piccoli, G.; Cordon-Preciado, V.; Farmer, S.; Jamuz, A.; Machin, F.; Pasero, P.; Lisby, M.; Haber, J.E.; Aragon, L. Anaphase onset before complete DNA replication with intact checkpoint responses. Science 2007, 315, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Morikawa, H.; Morishita, T.; Kawane, S.; Iwasaki, H.; Carr, A.M.; Shinagawa, H. Rad62 protein functionally and physically associates with the smc5/smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 2004, 24, 9401–9413. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Sollier, J.; Liberi, G.; Zhao, X.; Maeda, D.; Seki, M.; Enomoto, T.; Ohta, K.; Foiani, M. Ubc9- and Mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 2006, 127, 509–522. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Driscoll, R.; Castellucci, F.; Foiani, M.; Jackson, S.P.; Branzei, D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 2009, 20, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Lopez, M.; Ceschia, A.; de Piccoli, G.; Colomina, N.; Pasero, P.; Aragon, L.; Torres-Rosell, J. The Smc5/6 complex is required for dissolution of DNA-mediated sister chromatid linkages. Nucleic Acids Res. 2010, 38, 6502–6512. [Google Scholar] [CrossRef] [PubMed]
- Bermudez-Lopez, M.; Villoria, M.T.; Esteras, M.; Jarmuz, A.; Torres-Rosell, J.; Clemente-Blanco, A.; Aragon, L. Sgs1’s roles in DNA end resection, HJ dissolution, and crossover suppression require a two-step SUMO regulation dependent on Smc5/6. Genes Dev. 2016, 30, 1339–1356. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.N.; Zhao, X.L. Replication-Associated Recombinational Repair: Lessons from Budding Yeast. Genes 2016, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Bonner, J.N.; Choi, K.Y.; Xue, X.Y.; Torres, N.P.; Szakal, B.; Wei, L.; Wan, B.B.; Arter, M.; Matos, J.; Sung, P.; et al. Smc5/6 Mediated Sumoylation of the Sgs1-Top3-Rmi1 Complex Promotes Removal of Recombination Intermediates. Cell Rep. 2016, 16, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, B.Z.; Tan, A.P.; Kolodner, R.D.; Putnam, C.D.; Zhou, H.L. SUMO E3 ligase Mms21 prevents spontaneous DNA damage induced genome rearrangements. PLoS Genet. 2018, 14, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Choi, K.; Szakal, B.; Arenz, J.; Duan, X.; Ye, H.; Branzei, D.; Zhao, X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. USA 2009, 106, 21252–21257. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Szakal, B.; Chen, Y.H.; Branzei, D.; Zhao, X. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 2010, 21, 2306–2314. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Szakal, B.; Castellucci, F.; Branzei, D.; Zhao, X.L. DNA damage checkpoint and recombinational repair differentially affect the replication stress tolerance of smc6 mutants. Mol. Biol. Cell 2013, 24, 2431–2441. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Agrawal, V.; Johnson, F.B. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 2011, 286, 5119–5125. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.Y.; Choi, K.Y.; Bonner, J.; Chiba, T.; Kwon, Y.; Xu, Y.Y.; Sanchez, H.; Wyman, C.; Niu, H.Y.; Zhao, X.L.; et al. Restriction of Replication Fork Regression Activities by a Conserved SMC Complex. Mol. Cell 2014, 56, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.Y.; Choi, K.; Bonner, J.N.; Szakal, B.; Chen, Y.H.; Papusha, A.; Saro, D.; Niu, H.Y.; Ira, G.; Branzei, D.; et al. Selective modulation of the functions of a conserved DNA motor by a histone fold complex. Genes Dev. 2015, 29, 1000–1005. [Google Scholar] [CrossRef]
- Peng, X.P.; Lim, S.; Li, S.B.; Marjavaara, L.; Chabes, A.; Zhao, X.L. Acute Smc5/6 depletion reveals its primary role in rDNA replication by restraining recombination at fork pausing sites. PLoS Genet. 2018, 14, 20. [Google Scholar] [CrossRef]
- Lafuente-Barquero, J.; Luke-Glaser, S.; Graf, M.; Silva, S.; Gómez-González, B.; Lockhart, A.; Lisby, M.; Aguilera, A.; Luke, B. The Smc5/6 complex regulates the yeast Mph1 helicase at RNA-DNA hybrid-mediated DNA damage. PLoS Genet. 2017, 13, e1007136. [Google Scholar] [CrossRef]
- Peng, J.; Feng, W. Incision of damaged DNA in the presence of an impaired Smc5/6 complex imperils genome stability. Nucleic Acids Res. 2016, 44, 10216–10229. [Google Scholar] [CrossRef]
- Irmisch, A.; Ampatzidou, E.; Mizuno, K.; O’Connell, M.J.; Murray, J.M. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009, 28, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Wani, S.; Maharshi, N.; Kothiwal, D.; Mahendrawada, L.; Kalaivani, R.; Laloraya, S. Interaction of the Saccharomyces cerevisiae RING-domain protein Nse1 with Nse3 and the Smc5/6 complex is required for chromosome replication and stability. Curr. Genet. 2018, 64, 599–617. [Google Scholar] [CrossRef] [PubMed]
- Branzei, D.; Menolfi, D. G2/M chromosome transactions essentially relying on Smc5/6. Cell Cycle 2016, 15, 611–612. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van der Crabben, S.N.; Hennus, M.P.; McGregor, G.A.; Ritter, D.I.; Nagamani, S.C.S.; Wells, O.S.; Harakalova, M.; Chinn, I.K.; Alt, A.; Vondrova, L.; et al. Destabilized SMC5/6 complex leads to chromosome breakage syndrome with severe lung disease. J. Clin. Investig. 2016, 126, 2881–2892. [Google Scholar] [CrossRef] [PubMed]
- Payne, F.; Colnaghi, R.; Rocha, N.; Seth, A.; Harris, J.; Carpenter, G.; Bottomley, W.E.; Wheeler, E.; Wong, S.; Saudek, V.; et al. Hypomorphism in human NSMCE2 linked to primordial dwarfism and insulin resistance. J. Clin. Investig. 2014, 124, 4028–4038. [Google Scholar] [CrossRef]
- Kegel, A.; Betts-Lindroos, H.; Kanno, T.; Jeppsson, K.; Ström, L.; Katou, Y.; Itoh, T.; Shirahige, K.; Sjögren, C. Chromosome length influences replication-induced topological stress. Nature 2011, 471, 392–396. [Google Scholar] [CrossRef]
- Bustard, D.E.; Menolfi, D.; Jeppsson, K.; Ball, L.G.; Dewey, S.C.; Shirahige, K.; Sjögren, C.; Branzei, D.; Cobb, J.A. During replication stress, non-SMC element 5 (NSE5) is required for Smc5/6 protein complex functionality at stalled forks. J. Biol. Chem. 2012, 287, 11374–11383. [Google Scholar] [CrossRef]
- Outwin, E.A.; Irmisch, A.; Murray, J.M.; O’Connell, M.J. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol. Cell. Biol. 2009, 29, 4363–4375. [Google Scholar] [CrossRef]
- Tapia-Alveal, C.; Outwin, E.A.; Trempolec, N.; Dziadkowiec, D.; Murray, J.M.; O’Connell, M.J. SMC complexes and topoisomerase II work together so that sister chromatids can work apart. Cell Cycle 2010, 9, 2065–2070. [Google Scholar] [CrossRef][Green Version]
- Tapia-Alveal, C.; Lin, S.J.; Yeoh, A.; Jabado, O.J.; O’Connell, M.J. H2A.Z-Dependent Regulation of Cohesin Dynamics on Chromosome Arms. Mol. Cell. Biol. 2014, 34, 2092–2104. [Google Scholar] [CrossRef]
- Jeppsson, K.; Carlborg, K.K.; Nakato, R.; Berta, D.G.; Lilienthal, I.; Kanno, T.; Lindqvist, A.; Brink, M.C.; Dantuma, N.P.; Katou, Y.; et al. The chromosomal association of the Smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet. 2014, 10, e1004680. [Google Scholar] [CrossRef] [PubMed]
- Spell, R.M.; Holm, C. Nature and distribution of chromosomal intertwinings in Saccharomyces-cerevisiae. Mol. Cell. Biol. 1994, 14, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Bermejo, R.; Doksani, Y.; Capra, T.; Katou, Y.M.; Tanaka, H.; Shirahige, K.; Foiani, M. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007, 21, 1921–1936. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.A.; Wang, J.C. Function of DNA topoisomerases as replication swivels in Saccharomyces-cerevisiae. J. Mol. Biol. 1989, 208, 257–267. [Google Scholar] [CrossRef]
- Murphy, C.M.; Xu, Y.; Li, F.; Nio, K.; Reszka-Blanco, N.; Li, X.; Wu, Y.; Yu, Y.; Xiong, Y.; Su, L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell. Rep. 2016, 16, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Decorsière, A.; Mueller, H.; van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.K.; Livingston, C.M.; Niu, C.; Fletcher, S.P.; Hantz, O.; et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016, 531, 38–389. [Google Scholar] [CrossRef] [PubMed]
- Livingston, C.M.; Ramakrishnan, D.; Strubin, M.; Fletcher, S.P.; Beran, R.K. Identifying and Characterizing Interplay between Hepatitis B Virus X Protein and Smc5/6. Viruses 2017, 9, 69. [Google Scholar] [CrossRef]
- Niu, C.; Livingston, C.M.; Li, L.; Beran, R.K.; Daffis, S.; Ramakrishnan, D.; Burdette, D.; Peiser, L.; Salas, E.; Ramos, H.; et al. The Smc5/6 Complex Restricts HBV when Localized to ND10 without Inducing an Innate Immune Response and Is Counteracted by the HBV X Protein Shortly after Infection. PLoS ONE 2017, 12, e0169648. [Google Scholar] [CrossRef]
- Xu, W.; Ma, C.; Zhang, Q.; Zhao, R.; Hu, D.; Zhang, X.; Chen, J.; Liu, F.; Wu, K.; Liu, Y.; et al. PJA1 Coordinates with the SMC5/6 Complex to Restrict DNA Viruses and Episomal Genes through Interferon-independent Manner. J. Virol. 2018. [Google Scholar] [CrossRef]
- Tsuyama, T.; Inou, K.; Seki, M.; Seki, T.; Kumata, Y.; Kobayashi, T.; Kimura, K.; Hanaoka, F.; Enomoto, T.; Tada, S. Chromatin loading of Smc5/6 is induced by DNA replication but not by DNA double-strand breaks. Biochem. Biophys. Res. Commun. 2006, 351, 935–939. [Google Scholar] [CrossRef]
- Gallego-Paez, L.M.; Tanaka, H.; Bando, M.; Takahashi, M.; Nozaki, N.; Nakato, R.; Shirahige, K.; Hirota, T. Smc5/6-mediated regulation of replication progression contributes to chromosome assembly during mitosis in human cells. Mol. Biol. Cell 2014, 25, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Pryzhkova, M.V.; Jordan, P.W. Conditional mutation of Smc5 in mouse embryonic stem cells perturbs condensin localization and mitotic progression. J. Cell Sci. 2016, 129, 1619–1634. [Google Scholar] [CrossRef] [PubMed]
- Pflumm, M.F.; Botchan, M.R. Orc mutants arrest in metaphase with abnormally condensed chromosomes. Development 2001, 128, 1697–1707. [Google Scholar] [PubMed]

© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palecek, J.J. SMC5/6: Multifunctional Player in Replication. Genes 2019, 10, 7. https://doi.org/10.3390/genes10010007
Palecek JJ. SMC5/6: Multifunctional Player in Replication. Genes. 2019; 10(1):7. https://doi.org/10.3390/genes10010007
Chicago/Turabian StylePalecek, Jan J. 2019. "SMC5/6: Multifunctional Player in Replication" Genes 10, no. 1: 7. https://doi.org/10.3390/genes10010007
APA StylePalecek, J. J. (2019). SMC5/6: Multifunctional Player in Replication. Genes, 10(1), 7. https://doi.org/10.3390/genes10010007




