A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Isolation and Evaluation
2.2. Library Preparation and Sequencing
2.3. Assembly
2.4. Curation
2.5. Genome Quality Assessment
3. Results
3.1. A Modified Protocol Allows for Library Preparation and Sequencing of Samples from as Low as 100 ng of DNA Input
3.2. De novo Assembly Using FALCON-Unzip Allows for a High-Quality Genome from a Single Anopheles coluzzii Mosquito Individual
3.3. The New Assembly Shows Improvements in Resolving Genomic Regions
4. Discussion
Availability of Data
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lewin, H.A.; Robinson, G.E.; Kress, W.J.; Baker, W.J.; Coddington, J.; Crandall, K.A.; Durbin, R.; Edwards, S.V.; Forest, F.; Gilbert, M.T.; et al. Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. USA 2018, 115, 4325–4333. [Google Scholar] [CrossRef] [PubMed]
- Leffler, E.M.; Bullaughey, K.; Matute, D.R.; Meyer, W.K.; Segurel, L.; Venkat, A.; Andolfatto, P.; Przeworski, M. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012, 10, e1001388. [Google Scholar] [CrossRef] [PubMed]
- Drosophila 12 Genomes Consortium; Clark, A.G.; Eisen, M.B.; Smith, D.R.; Bergman, C.M.; Oliver, B. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [PubMed]
- Neafsey, D.E.; Waterhouse, R.M.; Abai, M.R.; Aganezov, S.S.; Alekseyev, M.A.; Allen, J.E.; Amon, J.; Arcà, B.; Arensburger, P.; Artemov, G.; et al. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes. Science 2015, 347, 1258522. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.W.C.; Dohmen, E.; Hughes, D.S.T.; Murali, S.C.; Poelchau, M.; Glastad, K.; Anstead, C.A.; Ayoub, N.A.; Batterham, P.; Bellair, M.; et al. The Genomic Basis of Arthropod Diversity. bioRxiv 2018, 382945. [Google Scholar] [CrossRef]
- Matthews, B.J.; Dudchenko, O.; Kingan, S.B.; Koren, S.; Antoshechkin, I.; Crawford, J.E.; Glassford, W.J.; Herre, M.; Redmond, S.N.; Rose, N.H.; et al. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature 2018, 563, 501. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.-S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [PubMed]
- Holt, R.A.; Subramanian, G.M.; Halpern, A.; Sutton, G.G.; Charlab, R.; Nusskern, D.R.; Wincker, P.; Clark, A.G.; Ribeiro, J.C.; Wides, R.; et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science 2002, 298, 129–149. [Google Scholar] [CrossRef]
- Sharakhova, M.V.; Hammond, M.P.; Lobo, N.F.; Krzywinski, J.; Unger, M.F.; Hillenmeyer, M.E.; Bruggner, R.V.; Birney, E.; Collins, F.H. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007, 8, R5. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018, 19, 460. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2017, 35, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Korlach, J.; Gedman, G.; Kingan, S.B.; Chin, C.-S.; Howard, J.T.; Audet, J.-N.; Cantin, L.; Jarvis, E.D. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience 2017, 6, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 1, 7. [Google Scholar] [CrossRef] [PubMed]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Python Assembly Comparison Scripts [Internet]. Available online: https://github.com/wheaton5/assembly_comparison_scripts.
- Kukutla, P.; Lindberg, B.G.; Pei, D.; Rayl, M.; Yu, W.; Steritz, M.; Faye, I.; Xu, J. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS ONE 2014, 9, e97715. [Google Scholar] [CrossRef] [PubMed]
- Lawniczak, M.K.; Emrich, S.J.; Holloway, A.K.; Regier, A.P.; Olson, M.; White, B.; Redmond, S.; Fulton, L.; Appelbaum, E.; Godfrey, J.; et al. Widespread divergence between incipient Anopheles gambiae species revealed by whole genome sequences. Science 2010, 330, 512–514. [Google Scholar] [CrossRef]
- Ghurye, J.; Koren, S.; Small, S.T.; Redmond, S.; Howell, P.; Phillippy, A.M.; Besansky, N.J. A chromosome-scale assembly of the major African malaria vector Anopheles funestus. bioRxiv 2018, 492777. [Google Scholar] [CrossRef]
- Roach, M.J.; Schmidt, S.A.; Borneman, A.R. Purge Haplotigs: Synteny Reduction for Third-gen Diploid Genome Assemblies. bioRxiv 2018. [Google Scholar] [CrossRef]
- Sharakhova, M.V.; George, P.; Brusentsova, I.V.; Leman, S.C.; Bailey, J.A.; Smith, C.D.; Sharakhov, I.V. Genome mapping and characterization of the Anopheles gambiae heterochromatin. BMC Genom. 2010, 11, 459. [Google Scholar] [CrossRef]
- AgamP4|VectorBase. Available online: https://www.vectorbase.org/organisms/anopheles-gambiae/pest/agamp4 (accessed on 7 August 2018).
- Coetzee, M.; Hunt, R.H.; Wilkerson, R.; Torre, A.D.; Coulibaly, M.B.; Besansky, N.J. Anopheles coluzzii and Anopheles amharicus, new members of the Anopheles gambiae complex. Zootaxa 2013, 3619, 246–274. [Google Scholar] [CrossRef]
- Aboagye-Antwi, F.; Alhafez, N.; Weedall, G.D.; Brothwood, J.; Kandola, S.; Paton, D.; Fofana, A.; Olohan, L.; Betancourth, M.P.; Ekechukwu, N.E.; et al. Experimental Swap of Anopheles gambiae’s Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation. PLoS Genet. 2015, 11, e1005141. [Google Scholar] [CrossRef] [PubMed]
- Koren, S.; Rhie, A.; Walenz, B.P.; Dilthey, A.T.; Bickhart, D.M.; Kingan, S.B.; Hiendleder, S.; Williams, J.L.; Smith, T.P.; Phillippy, A.M. De novo assembly of haplotype-resolved genomes with trio binning. Nat. Biotechnol. 2018, 36, 1174. [Google Scholar] [CrossRef] [PubMed]
- Kronenberg, Z.N.; Hall, R.J.; Hiendleder, S.; Smith, T.P.; Sullivan, S.T.; Williams, J.L.; Kingan, S.B. FALCON-Phase: Integrating PacBio and Hi-C data for phased diploid genomes. bioRxiv 2018, 327064. [Google Scholar] [CrossRef]
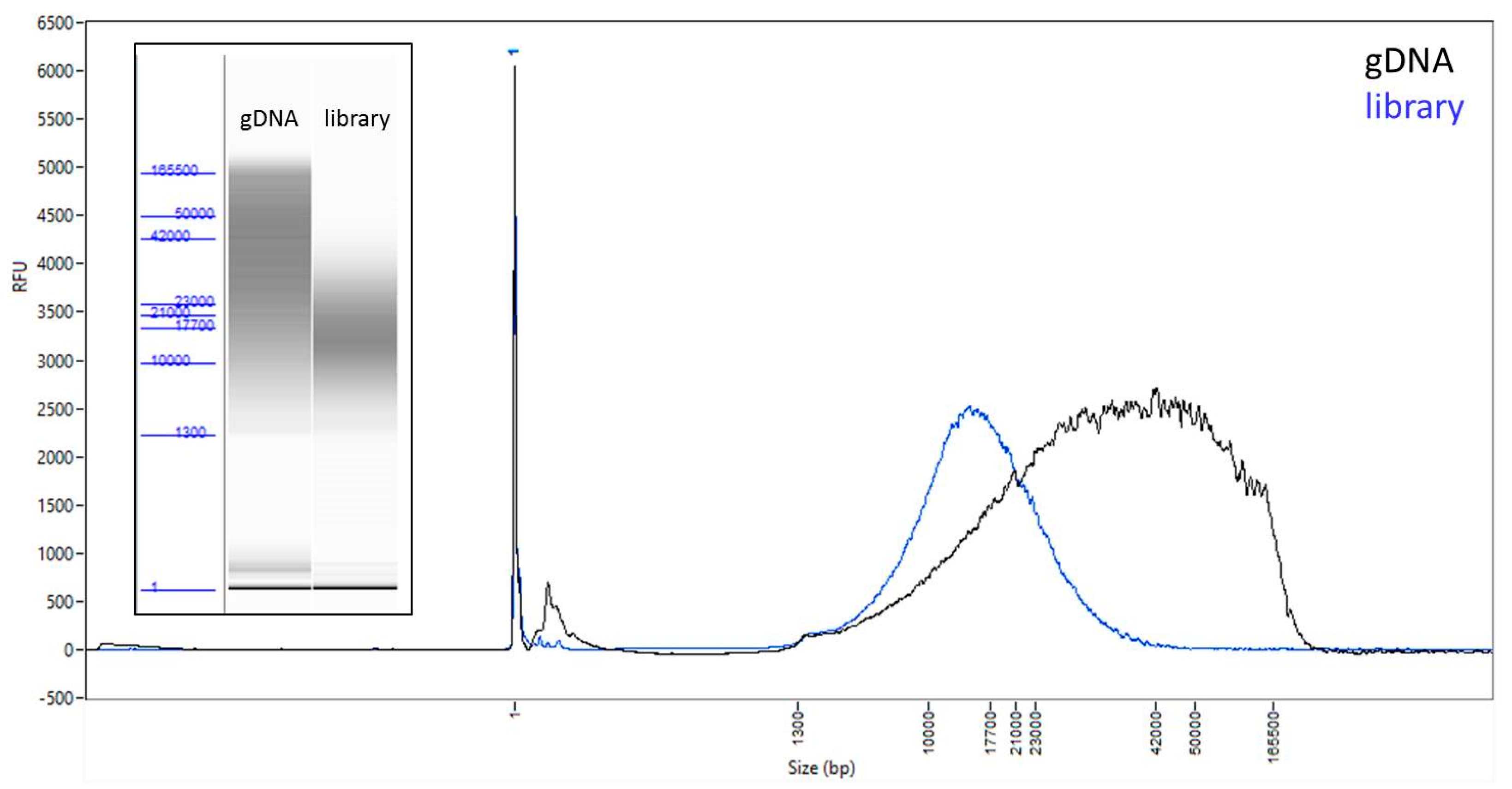
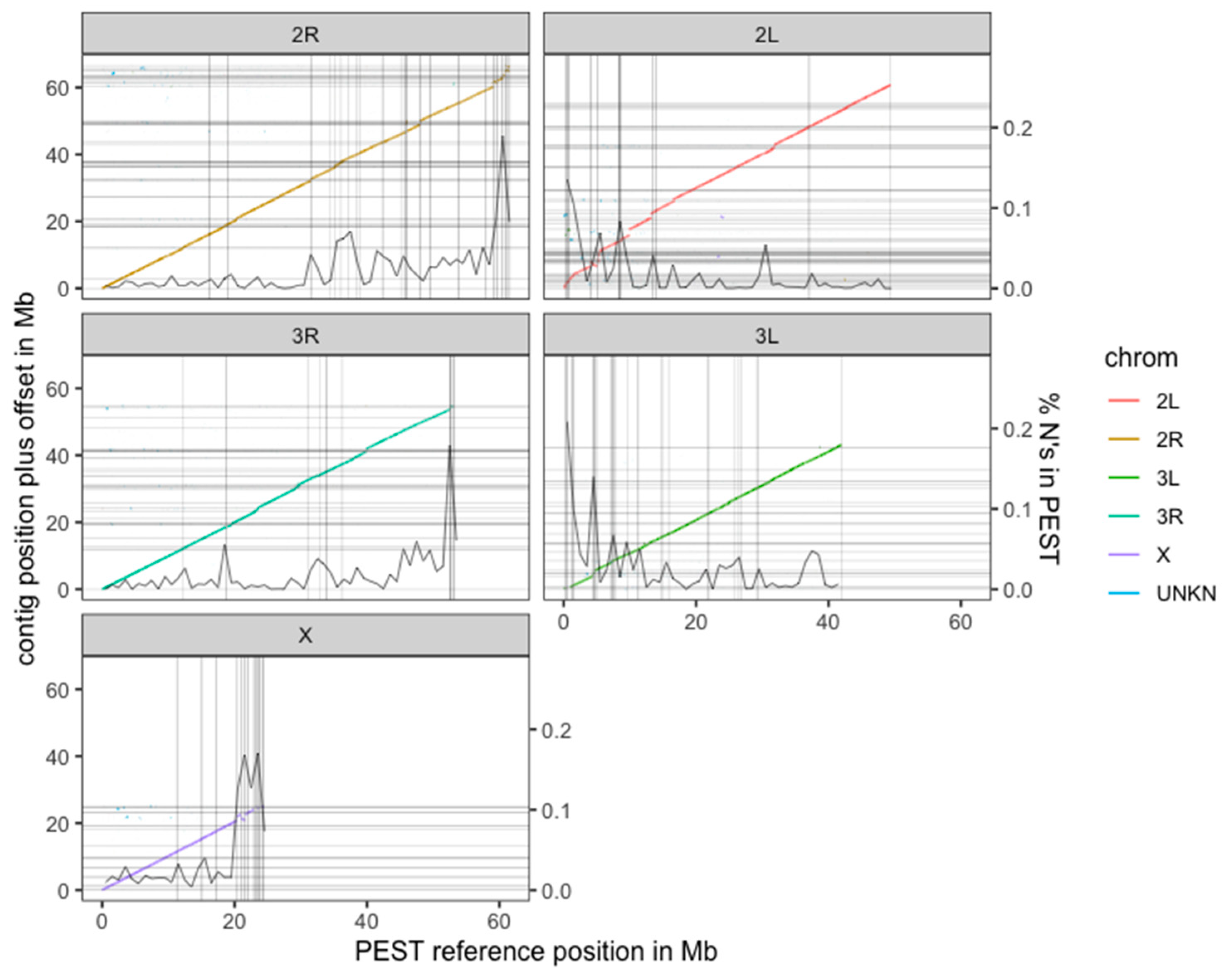
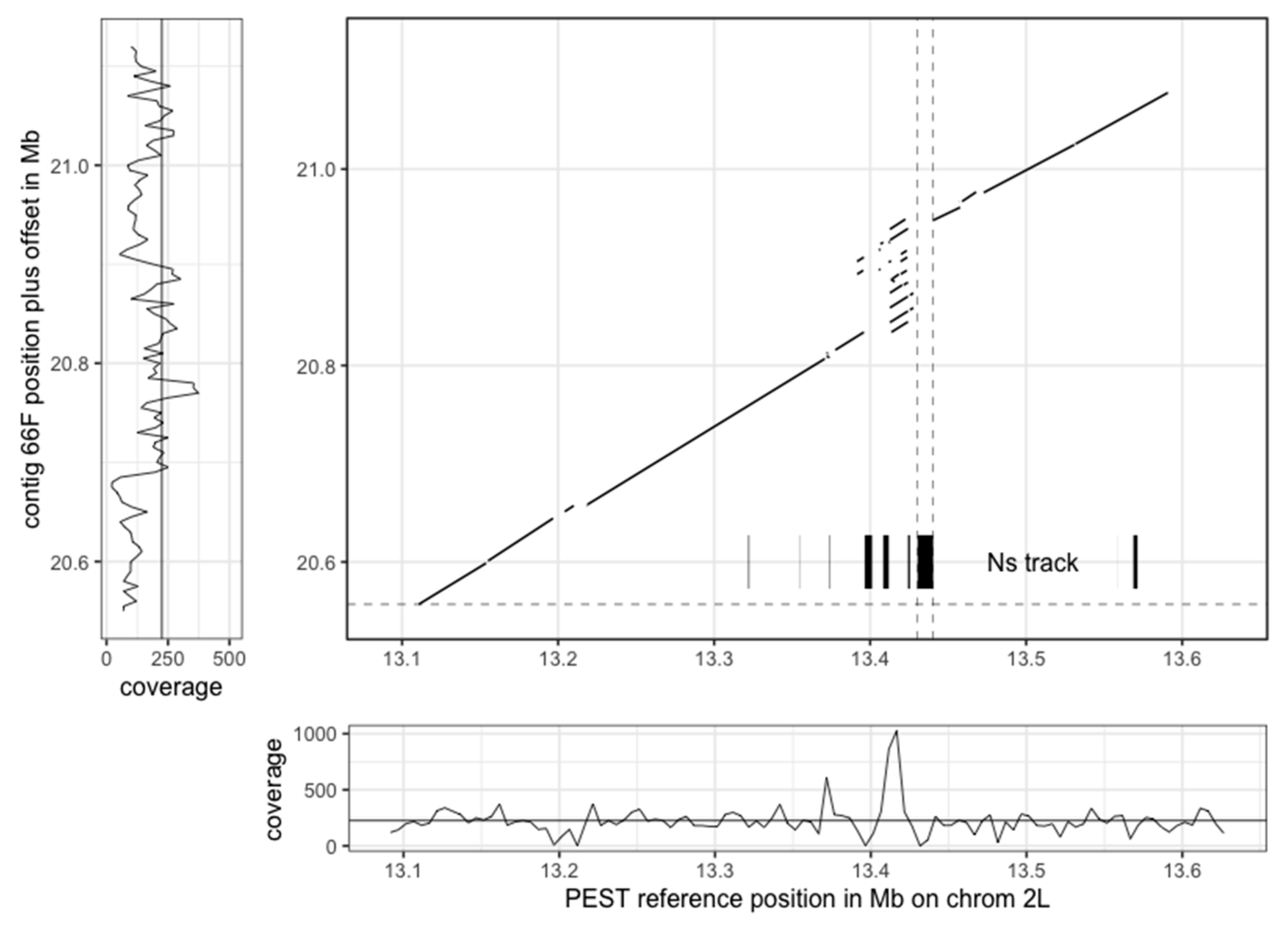
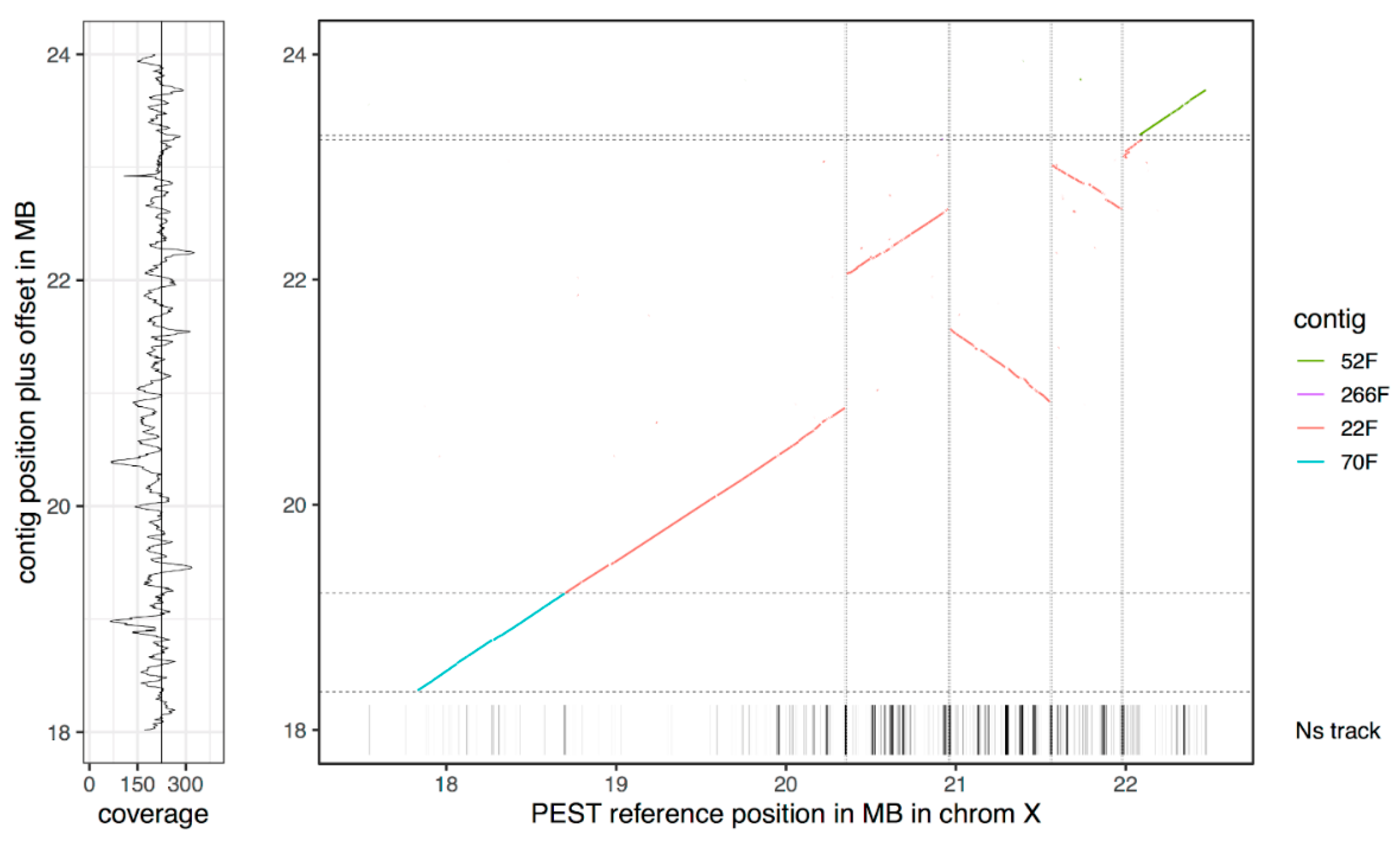
| PacBio Raw | PacBio Curated | Sanger Assembly | ||
|---|---|---|---|---|
| Primary contig assembly | Size (Mb) | 266 | 251 | 224 |
| No. contigs | 372 | 206 | 27,063 | |
| Contig N50 (Mb) | 3.52 | 3.47 | 0.025 | |
| Alternate haplotigs | Size (Mb) | 78.5 | 89.2 | unresolved |
| No. contigs | 665 | 830 | N/A | |
| Contig N50 (Mb) | 0.22 | 0.199 | N/A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kingan, S.B.; Heaton, H.; Cudini, J.; Lambert, C.C.; Baybayan, P.; Galvin, B.D.; Durbin, R.; Korlach, J.; Lawniczak, M.K.N. A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing. Genes 2019, 10, 62. https://doi.org/10.3390/genes10010062
Kingan SB, Heaton H, Cudini J, Lambert CC, Baybayan P, Galvin BD, Durbin R, Korlach J, Lawniczak MKN. A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing. Genes. 2019; 10(1):62. https://doi.org/10.3390/genes10010062
Chicago/Turabian StyleKingan, Sarah B., Haynes Heaton, Juliana Cudini, Christine C. Lambert, Primo Baybayan, Brendan D. Galvin, Richard Durbin, Jonas Korlach, and Mara K. N. Lawniczak. 2019. "A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing" Genes 10, no. 1: 62. https://doi.org/10.3390/genes10010062
APA StyleKingan, S. B., Heaton, H., Cudini, J., Lambert, C. C., Baybayan, P., Galvin, B. D., Durbin, R., Korlach, J., & Lawniczak, M. K. N. (2019). A High-Quality De novo Genome Assembly from a Single Mosquito Using PacBio Sequencing. Genes, 10(1), 62. https://doi.org/10.3390/genes10010062





