Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Culture Conditions
2.2. Determination of the Minimal Inhibitory Concentration and Generation of Pt-Resistant Mutants
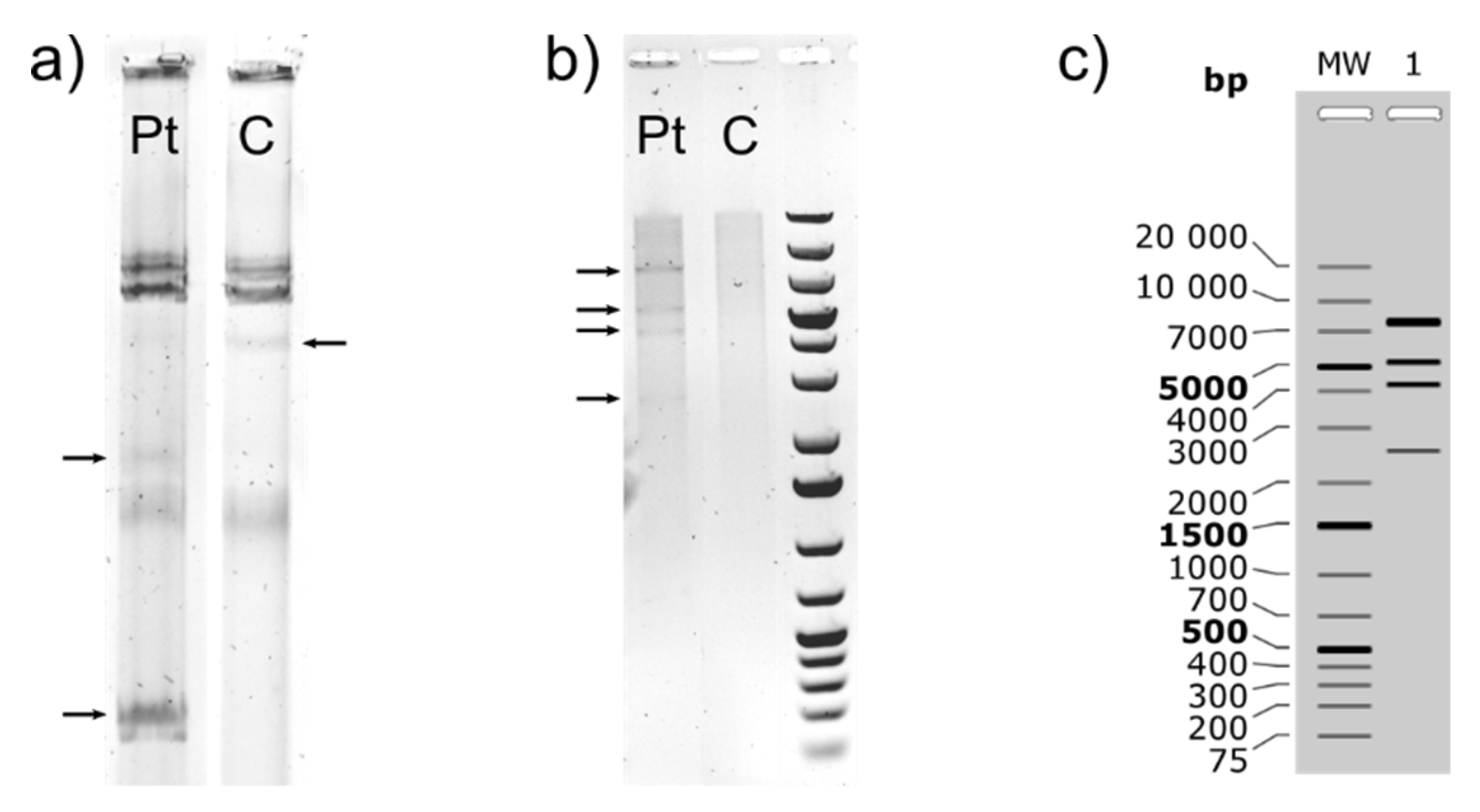
2.3. Plasmid Isolation and Restriction Digestion
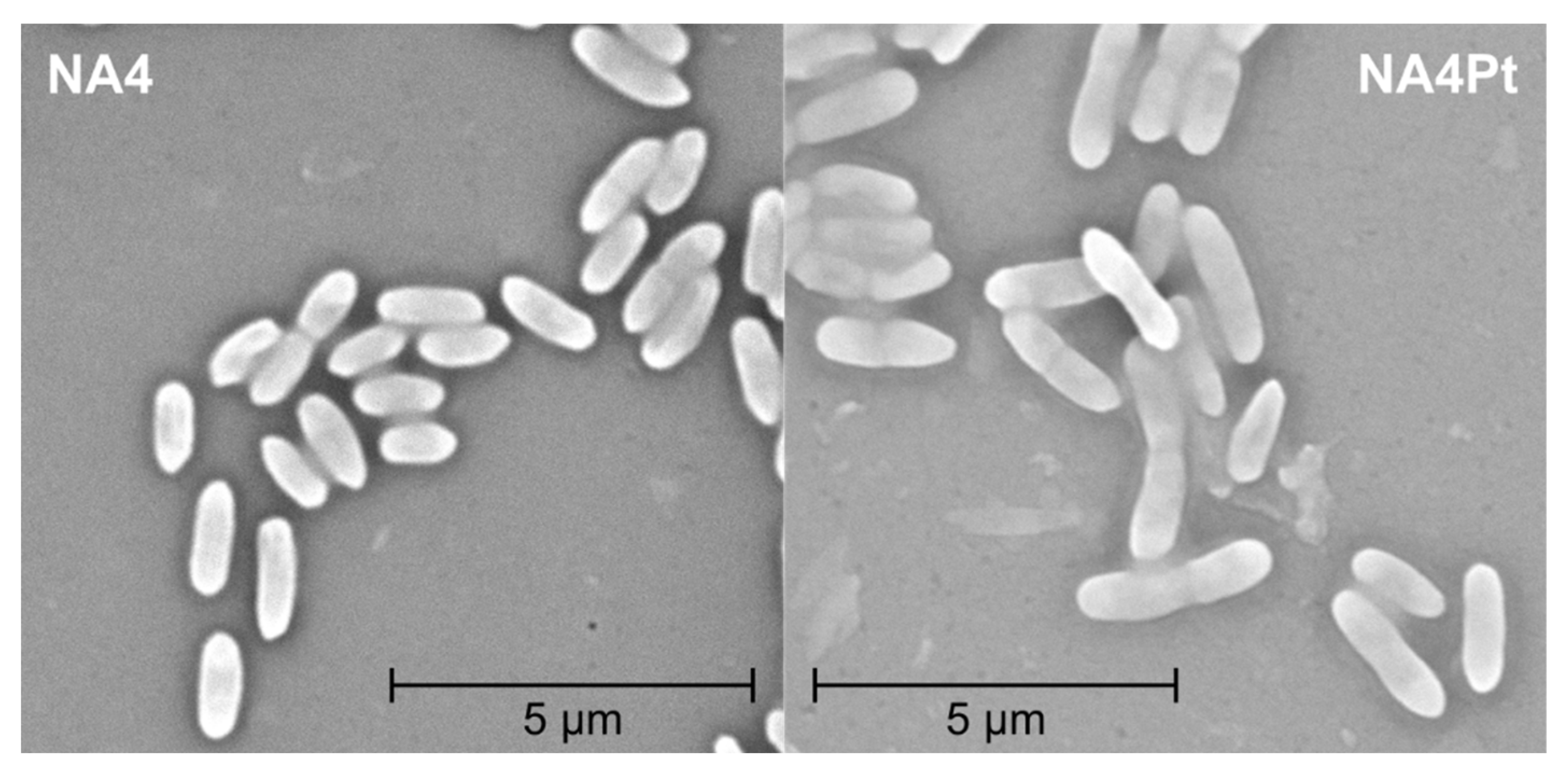
2.4. Motility, Scanning Electron Microscopy (SEM), and Flow Cytometry
2.5. Genome Sequencing
2.6. Genome Assembly
2.7. Transcriptomic Analysis Using RNA-Seq
2.8. RNA-Seq Data Analysis
2.9. Functional Analysis
3. Results and Discussion
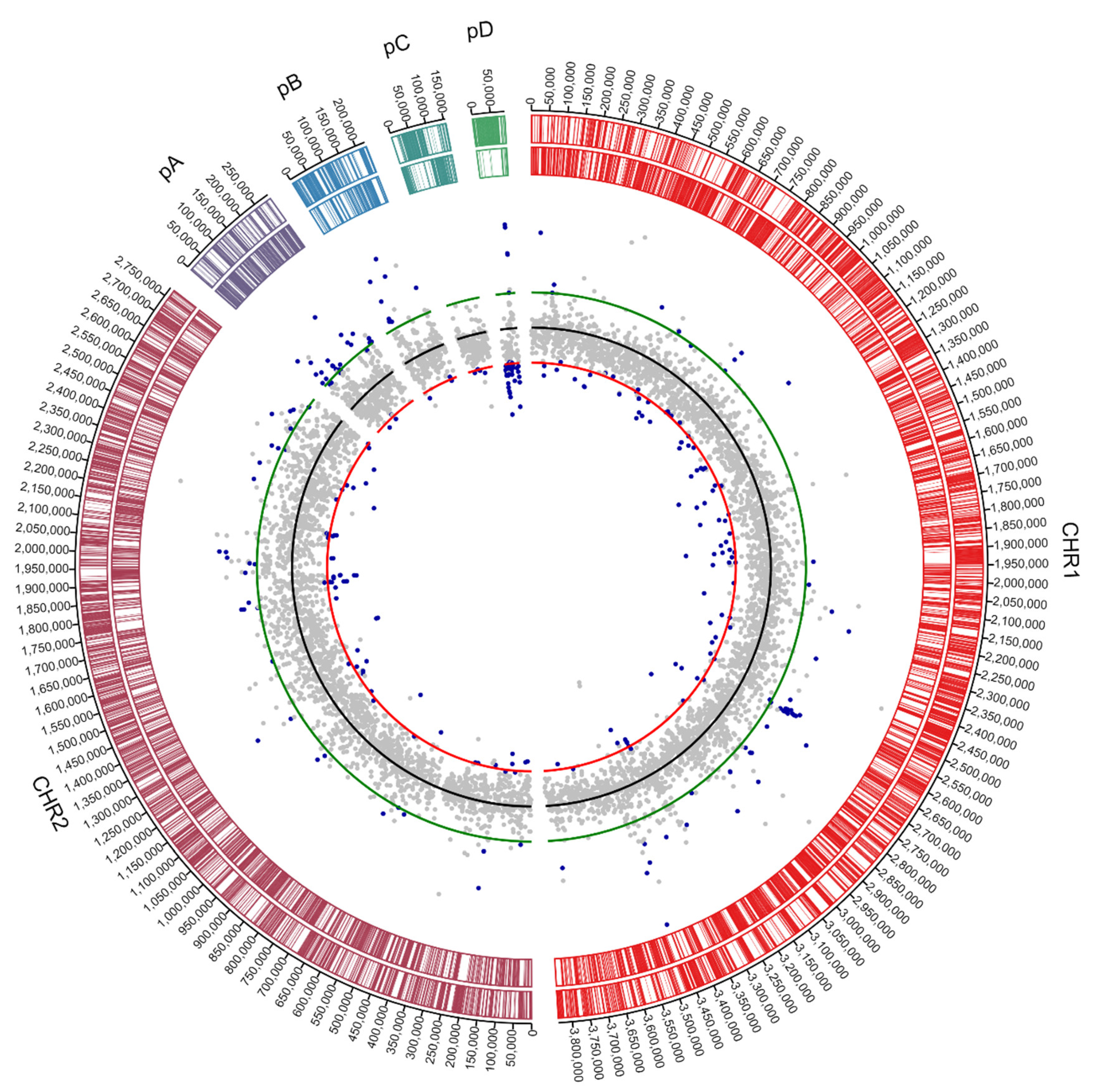
3.1. Genome Analysis of C. metallidurans NA4
3.1.1. Characterization of (Mega)Plasmids
3.1.2. Insertion Sequence Elements Distribution
3.2. Analysis of C. metallidurans NA4Pt
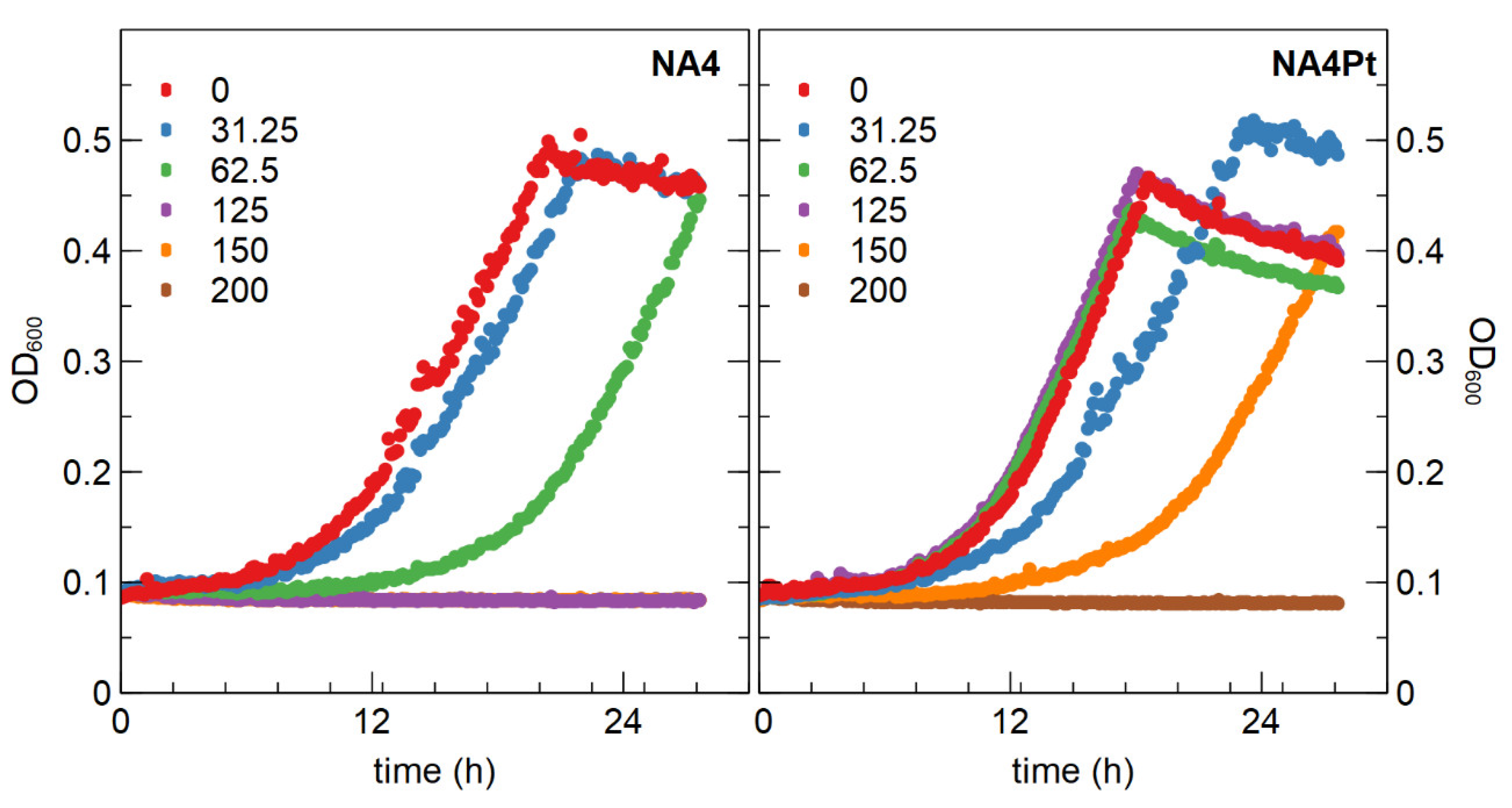
3.2.1. Determination of Minimal Inhibitory Concentration and Generation of a Pt4+ Resistant Mutant
3.2.2. Sequence Analysis of C. metallidurans NA4Pt
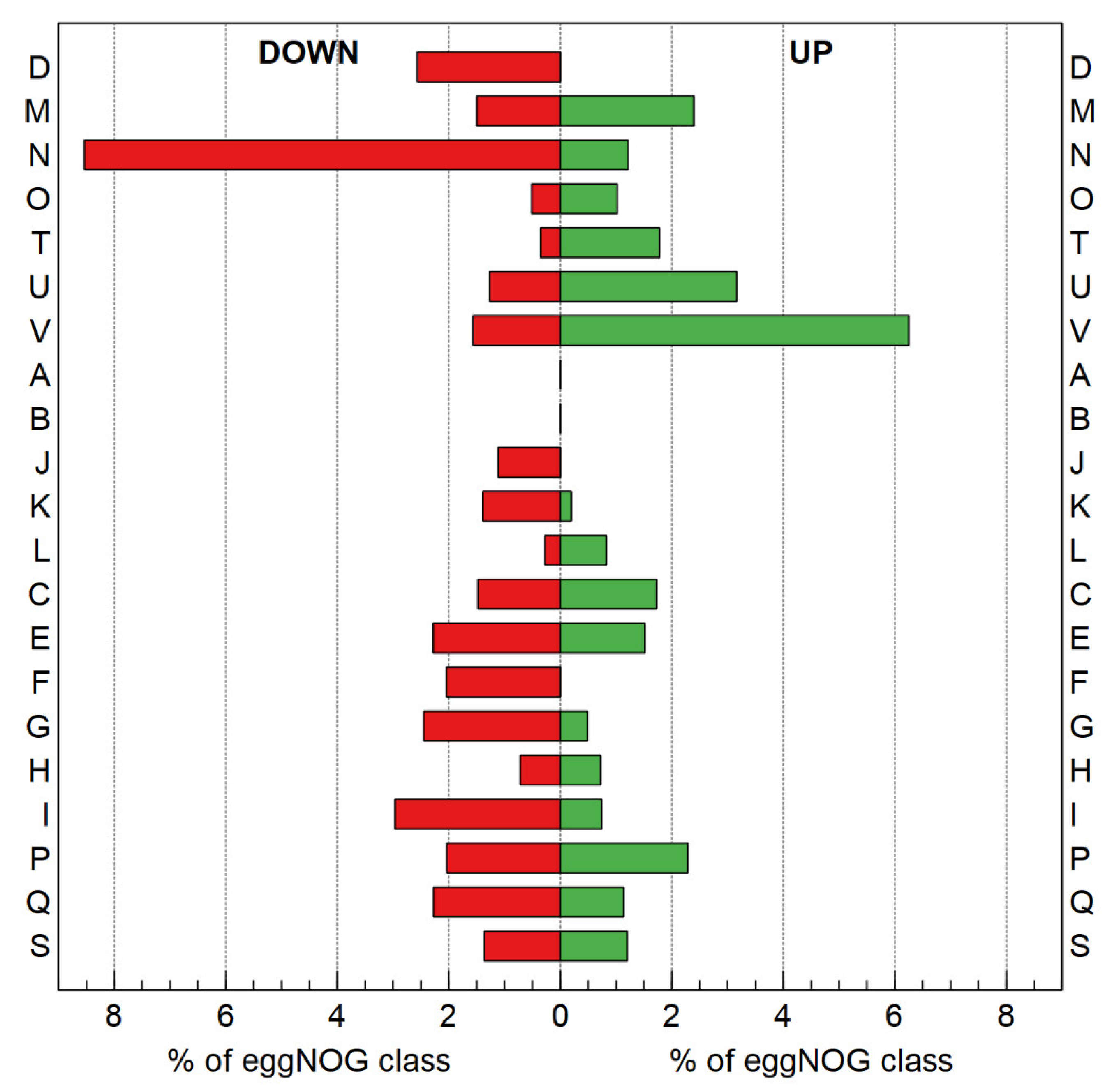
3.2.3. Transcriptome Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rauch, S.; Morrison, G.M. Environmental relevance of the platinum-group elements. Elements 2008, 4, 259–263. [Google Scholar] [CrossRef]
- Wedepohl, K.H. The Composition of the Continental-Crust. Geochim. Cosmochim. Acta 1995, 59, 1217–1232. [Google Scholar] [CrossRef]
- Ravindra, K.; Bencs, L.; Van Grieken, R. Platinum group elements in the environment and their health risk. Sci. Total Environ. 2004. [Google Scholar] [CrossRef]
- Florea, A.M.; Busselberg, D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers 2011, 3, 1351–1371. [Google Scholar] [CrossRef] [PubMed]
- Maes, S.; Props, R.; Fitts, J.P.; Smet, R.D.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Vanhaecke, F.; Boon, N.; Hennebel, T. Platinum Recovery from Synthetic Extreme Environments by Halophilic Bacteria. Environ. Sci. Technol. 2016, 50, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Hennebel, T.; Boon, N.; Maes, S.; Lenz, M. Biotechnologies for critical raw material recovery from primary and secondary sources: R&D priorities and future perspectives. New Biotechnol. 2015, 32, 121–127. [Google Scholar] [CrossRef]
- Reith, F.; Zammit, C.M.; Shar, S.S.; Etschmann, B.; Bottrill, R.; Southam, G.; Ta, C.; Kilburn, M.; Oberthür, T.; Ball, A.S.; et al. Biological role in the transformation of platinum-group mineral grains. Nat. Geosci. 2016, 9, 294–298. [Google Scholar] [CrossRef]
- Zereini, F.; Wiseman, C.L.S. Platinum Metals in the Environment; Springer-Verlag: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Maes, S.; Claus, M.; Verbeken, K.; Wallaert, E.; De Smet, R.; Vanhaecke, F.; Boon, N.; Hennebel, T. Platinum recovery from industrial process streams by halophilic bacteria: Influence of salt species and platinum speciation. Water Res. 2016, 105, 436–443. [Google Scholar] [CrossRef]
- de Vargas, I.; Macaskie, L.E.; Guibal, E. Biosorption of palladium and platinum by sulfate-reducing bacteria. J. Chem. Technol. Biotechnol. 2004, 79, 49–56. [Google Scholar] [CrossRef]
- Maes, S.; Props, R.; Fitts, J.P.; De Smet, R.; Vanhaecke, F.; Boon, N.; Hennebel, T. Biological Recovery of Platinum Complexes from Diluted Aqueous Streams by Axenic Cultures. PLoS ONE 2017. [Google Scholar] [CrossRef]
- Campbell, G.; MacLean, L.; Reith, F.; Brewe, D.; Gordon, R.A.; Southam, G. Immobilisation of Platinum by Cupriavidus metallidurans. Minerals 2018, 8, 10. [Google Scholar] [CrossRef]
- Konishi, Y.; Ohno, K.; Saitoh, N.; Nomura, T.; Nagamine, S.; Hishida, H.; Takahashi, Y.; Uruga, T. Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae. J. Biotechnol. 2007, 128, 648–653. [Google Scholar] [CrossRef] [PubMed]
- Yong, P.; Rowson, N.A.; Farr, J.P.G.; Harris, I.R.; Macaskie, L.E. Bioaccumulation of palladium by Desulfovibrio desulfuricans. J. Chem. Technol. Biotechnol. 2002, 77, 593–601. [Google Scholar] [CrossRef]
- Rapp-Giles, B.J.; Casalot, L.; English, R.S.; Ringbauer, J.A.; Dolla, A.; Wall, J.D. Cytochrome c3 Mutants of Desulfovibrio desulfuricans. Appl. Environ. Microbiol. 2000, 66, 671–677. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Elias, D.A.; Suflita, J.M.; McInerney, M.J.; Krumholz, L.R. Periplasmic cytochrome c3 of Desulfovibrio vulgaris is directly involved in H2-mediated metal but not sulfate reduction. Appl. Environ. Microbiol. 2004, 70, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Capeness, M.J.; Edmundson, M.C.; Horsfall, L.E. Nickel and platinum group metal nanoparticle production by Desulfovibrio alaskensis G20. New Biotechnol. 2015, 32, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Gauthier, D.; Sobjerg, L.S.; Jensen, K.M.; Lindhardt, A.T.; Bunge, M.; Finster, K.; Meyer, R.L.; Skrydstrup, T. Environmentally benign recovery and reactivation of palladium from industrial waste by using gram-negative bacteria. ChemSusChem 2010, 3, 1036–1039. [Google Scholar] [CrossRef]
- Rosenberg, B.; Renshaw, E.; Vancamp, L.; Hartwick, J.; Drobnik, J. Platinum-induced filamentous growth in Escherichia coli. J. Bacteriol. 1967, 93, 716–721. [Google Scholar]
- Moore, R.L.; Brubaker, R.R. Effect of cis-platinum(II)diamminodichloride on cell division of Hyphomicrobium and Caulobacter. J. Bacteriol. 1976, 125, 317–323. [Google Scholar]
- Beck, D.J.; Brubaker, R.R. Effect of cis-platinum(II)diamminodichloride on wild type and deoxyribonucleic acid repair deficient mutants of Escherichia coli. J. Bacteriol. 1973, 116, 1247–1252. [Google Scholar]
- Drobnik, J.; Urbankova, M.; Krekulova, A. The effect of cis-dichlorodiammineplatinum(II) on Escherichia coli B. The role of fil, exr and hcr markers. Mutat. Res. 1973, 17, 13–20. [Google Scholar] [CrossRef]
- Reslova, S. The induction of lysogenic strains of Escherichia coli by cis-dichlorodiammineplatinum (II). Chem. Biol. Interact. 1971, 4, 66–70. [Google Scholar] [CrossRef]
- Beck, D.J.; Fisch, J.E. Mutagenicity of platinum coordination complexes in Salmonella typhimurium. Mutat. Res. 1980, 77, 45–54. [Google Scholar] [CrossRef]
- Maboeta, M.S.; Claassens, S.; Van Rensburg, L.; Van Rensburg, P.J.J. The effects of platinum mining on the environment from a soil microbial perspective. Water Air Soil Pollut. 2006, 175, 149–161. [Google Scholar] [CrossRef]
- Janssen, P.J.; Van Houdt, R.; Moors, H.; Monsieurs, P.; Morin, N.; Michaux, A.; Benotmane, M.A.; Leys, N.; Vallaeys, T.; Lapidus, A.; et al. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 2010, 5, e10433. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Van Houdt, R. Metal Response in Cupriavidus metallidurans, Volume I: FROM Habitats to Genes and Proteins; Springer International Publishing: Basel, Switzerland, 2015; p. 89. [Google Scholar]
- Monsieurs, P.; Moors, H.; Van Houdt, R.; Janssen, P.J.; Janssen, A.; Coninx, I.; Mergeay, M.; Leys, N. Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network. Biometals 2011, 24, 1133–1151. [Google Scholar] [CrossRef]
- Monsieurs, P.; Mijnendonckx, K.; Provoost, A.; Venkateswaran, K.; Ott, C.M.; Leys, N.; Van Houdt, R. Genome Sequences of Cupriavidus metallidurans Strains NA1, NA4, and NE12, Isolated from Space Equipment. Genome Announc. 2014. [Google Scholar] [CrossRef]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; Van Gijsegem, F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Randall, C.P.; Oyama, L.B.; Bostock, J.M.; Chopra, I.; O’Neill, A.J. The silver cation (Ag+): Antistaphylococcal activity, mode of action and resistance studies. J. Antimicrob. Chemother. 2013, 68, 131–138. [Google Scholar] [CrossRef]
- Andrup, L.; Barfod, K.K.; Jensen, G.B.; Smidt, L. Detection of large plasmids from the Bacillus cereus group. Plasmid 2008, 59, 139–143. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef] [PubMed]
- Deatherage, D.E.; Barrick, J.E. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 2014, 1151, 165–188. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Le Fevre, F.; Longin, C.; Mornico, D.; Roche, D.; et al. MicroScope—An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef]
- Varani, A.M.; Siguier, P.; Gourbeyre, E.; Charneau, V.; Chandler, M. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 2011, 12, R30. [Google Scholar] [CrossRef]
- Robertson, J.; Nash, J.H.E. MOB-suite: Software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb. Genom. 2018. [Google Scholar] [CrossRef] [PubMed]
- Ankenbrand, M.J.; Hohlfeld, S.; Hackl, T.; Forster, F. AliTV-interactive visualization of whole genome comparisons. PEERJ Comput. Sci. 2017. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Mijnendonckx, K.; Provoost, A.; Ott, C.M.; Venkateswaran, K.; Mahillon, J.; Leys, N.; Van Houdt, R. Characterization of the survival ability of Cupriavidus metallidurans and Ralstonia pickettii from space-related environments. Microb. Ecol. 2013, 65, 347–360. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Monchy, S.; Janssen, P.; Houdt, R.V.; Leys, N. Megaplasmids in Cupriavidus Genus and Metal Resistance. In Microbial Megaplasmids; Schwartz, E., Ed.; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Van Houdt, R.; Monsieurs, P.; Mijnendonckx, K.; Provoost, A.; Janssen, A.; Mergeay, M.; Leys, N. Variation in genomic islands contribute to genome plasticity in Cupriavidus metallidurans. BMC Genom. 2012, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Van Houdt, R.; Monchy, S.; Leys, N.; Mergeay, M. New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria. Antonie Van Leeuwenhoek 2009, 96, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Henne, K.L.; Nakatsu, C.H.; Thompson, D.K.; Konopka, A.E. High-level chromate resistance in Arthrobacter sp. strain FB24 requires previously uncharacterized accessory genes. BMC Microbiol. 2009, 9, 199. [Google Scholar] [CrossRef]
- Henne, K.L.; Turse, J.E.; Nicora, C.D.; Lipton, M.S.; Tollaksen, S.L.; Lindberg, C.; Babnigg, G.; Giometti, C.S.; Nakatsu, C.H.; Thompson, D.K.; et al. Global proteomic analysis of the chromate response in Arthrobacter sp. strain FB24. J. Proteome Res. 2009, 8, 1704–1716. [Google Scholar] [CrossRef]
- Van Houdt, R.; Provoost, A.; Van Assche, A.; Leys, N.; Lievens, B.; Mijnendonckx, K.; Monsieurs, P. Cupriavidus metallidurans Strains with Different Mobilomes and from Distinct Environments Have Comparable Phenomes. Genes 2018, 9, 507. [Google Scholar] [CrossRef]
- Shintani, M.; Sanchez, Z.K.; Kimbara, K. Genomics of microbial plasmids: Classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 2015, 6, 242. [Google Scholar] [CrossRef]
- Ramsay, J.P.; Kwong, S.M.; Murphy, R.J.; Yui Eto, K.; Price, K.J.; Nguyen, Q.T.; O’Brien, F.G.; Grubb, W.B.; Coombs, G.W.; Firth, N. An updated view of plasmid conjugation and mobilization in Staphylococcus. Mob. Genet. Elements 2016, 6, e1208317. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Bertini, A.; Villa, L.; Falbo, V.; Hopkins, K.L.; Threlfall, E.J. Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 2005, 63, 219–228. [Google Scholar] [CrossRef]
- Garcillan-Barcia, M.P.; Francia, M.V.; de la Cruz, F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 2009, 33, 657–687. [Google Scholar] [CrossRef] [PubMed]
- Ilangovan, A.; Connery, S.; Waksman, G. Structural biology of the Gram-negative bacterial conjugation systems. Trends Microbiol. 2015, 23, 301–310. [Google Scholar] [CrossRef]
- Orlek, A.; Stoesser, N.; Anjum, M.F.; Doumith, M.; Ellington, M.J.; Peto, T.; Crook, D.; Woodford, N.; Walker, A.S.; Phan, H.; et al. Plasmid Classification in an Era of Whole-Genome Sequencing: Application in Studies of Antibiotic Resistance Epidemiology. Front. Microbiol. 2017, 8, 182. [Google Scholar] [CrossRef] [PubMed]
- Vandecraen, J.; Chandler, M.; Aertsen, A.; Van Houdt, R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit. Rev. Microbiol. 2017, 43, 709–730. [Google Scholar] [CrossRef]
- Siguier, P.; Gourbeyre, E.; Chandler, M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014, 38, 865–891. [Google Scholar] [CrossRef]
- Ricker, N.; Qian, H.; Fulthorpe, R.R. The limitations of draft assemblies for understanding prokaryotic adaptation and evolution. Genomics 2012, 100, 167–175. [Google Scholar] [CrossRef]
- Siguier, P.; Perochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef]
- Vaidya, M.Y.; McBain, A.J.; Butler, J.A.; Banks, C.E.; Whitehead, K.A. Antimicrobial Efficacy and Synergy of Metal Ions against Enterococcus faecium, Klebsiella pneumoniae and Acinetobacter baumannii in Planktonic and Biofilm Phenotypes. Sci. Rep. 2017, 7, 5911. [Google Scholar] [CrossRef]
- Chattoraj, D.K.; Snyder, K.M.; Abeles, A.L. P1 Plasmid Replication—Multiple Functions of Repa Protein at the Origin. Proc. Natl. Acad. Sci. USA 1985, 82, 2588–2592. [Google Scholar] [CrossRef] [PubMed]
- Dı́az-López, T.; Lages-Gonzalo, M.; Serrano-López, A.; Alfonso, C.; Rivas, G.; Dı́az-Orejas, R.; Giraldo, R. Structural changes in RepA, a plasmid replication initiator, upon binding to origin DNA. J. Biol. Chem. 2003, 278, 18606–18616. [Google Scholar] [CrossRef] [PubMed]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Morton, E.R.; Merritt, P.M.; Bever, J.D.; Fuqua, C. Large Deletions in the pAtC58 Megaplasmid of Agrobacterium tumefaciens Can Confer Reduced Carriage Cost and Increased Expression of Virulence Genes. Genome Biol. Evol. 2013, 5, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Konig, C.; Eulberg, D.; Groning, J.; Lakner, S.; Seibert, V.; Kaschabek, S.R.; Schlomann, M. A linear megaplasmid, p1CP, carrying the genes for chlorocatechol catabolism of Rhodococcus opacus 1CP. Microbiology 2004, 150, 3075–3087. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.C.; Marx, C.J. Repeated, selection-driven genome reduction of accessory genes in experimental populations. PLoS Genet. 2012, 8, e1002651. [Google Scholar] [CrossRef]
- Koskiniemi, S.; Sun, S.; Berg, O.G.; Andersson, D.I. Selection-driven gene loss in bacteria. PLoS Genet. 2012, 8, e1002787. [Google Scholar] [CrossRef]
- Fernandez-Lopez, R.; Del Campo, I.; Revilla, C.; Cuevas, A.; de la Cruz, F. Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet. 2014, 10, e1004171. [Google Scholar] [CrossRef]
- Zahrl, D.; Wagner, M.; Bischof, K.; Koraimann, G. Expression and assembly of a functional type IV secretion system elicit extracytoplasmic and cytoplasmic stress responses in Escherichia coli. J. Bacteriol. 2006, 188, 6611–6621. [Google Scholar] [CrossRef]
- Porse, A.; Schonning, K.; Munck, C.; Sommer, M.O. Survival and Evolution of a Large Multidrug Resistance Plasmid in New Clinical Bacterial Hosts. Mol. Biol. Evol. 2016, 33, 2860–2873. [Google Scholar] [CrossRef]
- Romanchuk, A.; Jones, C.D.; Karkare, K.; Moore, A.; Smith, B.A.; Jones, C.; Dougherty, K.; Baltrus, D.A. Bigger is not always better: Transmission and fitness burden of approximately 1MB Pseudomonas syringae megaplasmid pMPPla107. Plasmid 2014, 73, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 2002, 108, 583–586. [Google Scholar] [CrossRef]
- Dufresne, A.; Garczarek, L.; Partensky, F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005, 6, R14. [Google Scholar] [CrossRef] [PubMed]
- Lenski, R.E. What is adaptation by natural selection? Perspectives of an experimental microbiologist. PLoS Genet. 2017, 13, e1006668. [Google Scholar] [CrossRef] [PubMed]
- LaCroix, R.A.; Sandberg, T.E.; O’Brien, E.J.; Utrilla, J.; Ebrahim, A.; Guzman, G.I.; Szubin, R.; Palsson, B.O.; Feist, A.M. Use of Adaptive Laboratory Evolution To Discover Key Mutations Enabling Rapid Growth of Escherichia coli K-12 MG1655 on Glucose Minimal Medium. Appl. Environ. Microbiol. 2015, 81, 17–30. [Google Scholar] [CrossRef] [PubMed]
- McCloskey, D.; Xu, S.; Sandberg, T.E.; Brunk, E.; Hefner, Y.; Szubin, R.; Feist, A.M.; Palsson, B.O. Growth Adaptation of gnd and sdhCB Escherichia coli Deletion Strains Diverges From a Similar Initial Perturbation of the Transcriptome. Front. Microbiol. 2018, 9, 1793. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Pedersen, M.; LaCroix, R.A.; Ebrahim, A.; Bonde, M.; Herrgard, M.J.; Palsson, B.O.; Sommer, M.; Feist, A.M. Evolution of Escherichia coli to 42 degrees C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol. Biol. Evol. 2014, 31, 2647–2662. [Google Scholar] [CrossRef]
- Powell, S.; Szklarczyk, D.; Trachana, K.; Roth, A.; Kuhn, M.; Muller, J.; Arnold, R.; Rattei, T.; Letunic, I.; Doerks, T.; et al. eggNOG v3.0: Orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 2012, 40, D284–D289. [Google Scholar] [CrossRef]
- Franke, S.; Grass, G.; Rensing, C.; Nies, D.H. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 2003, 185, 3804–3812. [Google Scholar] [CrossRef]
- Mergeay, M.; Monchy, S.; Vallaeys, T.; Auquier, V.; Benotmane, A.; Bertin, P.; Taghavi, S.; Dunn, J.; van der Lelie, D.; Wattiez, R. Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: Towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 2003, 27, 385–410. [Google Scholar] [CrossRef]
- Liesegang, H.; Lemke, K.; Siddiqui, R.A.; Schlegel, H.G. Characterization of the inducible nickel and cobalt resistance determinant cnr from pMOL28 of Alcaligenes eutrophus CH34. J. Bacteriol. 1993, 175, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.P.; Yoon, J.M.; Cho, M.H.; Lee, S.W. Prokaryotic 2-component systems and the OmpR/PhoB superfamily. Can. J. Microbiol. 2015, 61, 799–810. [Google Scholar] [CrossRef] [PubMed]
- Szurmant, H.; Ordal, G.W. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 2004, 68, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Kato, T.; Hiraoka, K.D.; Miyata, T.; Minamino, T.; Chevance, F.F.; Hughes, K.T.; Namba, K. Identical folds used for distinct mechanical functions of the bacterial flagellar rod and hook. Nat. Commun. 2017, 8, 14276. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.J.; Hughes, K.T. Rod-to-hook transition for extracellular flagellum assembly is catalyzed by the L-ring-dependent rod scaffold removal. J. Bacteriol. 2014, 196, 2387–2395. [Google Scholar] [CrossRef]
- Alm, R.A.; Hallinan, J.P.; Watson, A.A.; Mattick, J.S. Fimbrial biogenesis genes of Pseudomonas aeruginosa: PILW and PILX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol. Microbiol. 1996, 22, 161–173. [Google Scholar]
- Scheurwater, E.; Reid, C.W.; Clarke, A.J. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 2008, 40, 586–591. [Google Scholar] [CrossRef]
- Monchy, S.; Benotmane, M.A.; Janssen, P.; Vallaeys, T.; Taghavi, S.; van der Lelie, D.; Mergeay, M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. J. Bacteriol. 2007, 189, 7417–7425. [Google Scholar] [CrossRef]
- Bertini, I.; Cavallaro, G.; Rosato, A. Cytochrome c: Occurrence and functions. Chem. Rev. 2006, 106, 90–115. [Google Scholar] [CrossRef]
- Cianciotto, N.P.; White, R.C. Expanding Role of Type II Secretion in Bacterial Pathogenesis and Beyond. Infect. Immun. 2017. [Google Scholar] [CrossRef]
- Xu, H.; Denny, T.P. Native and Foreign Proteins Secreted by the Cupriavidus metallidurans Type II System and an Alternative Mechanism. J. Microbiol. Biotechnol. 2017, 27, 791–807. [Google Scholar] [CrossRef] [PubMed]
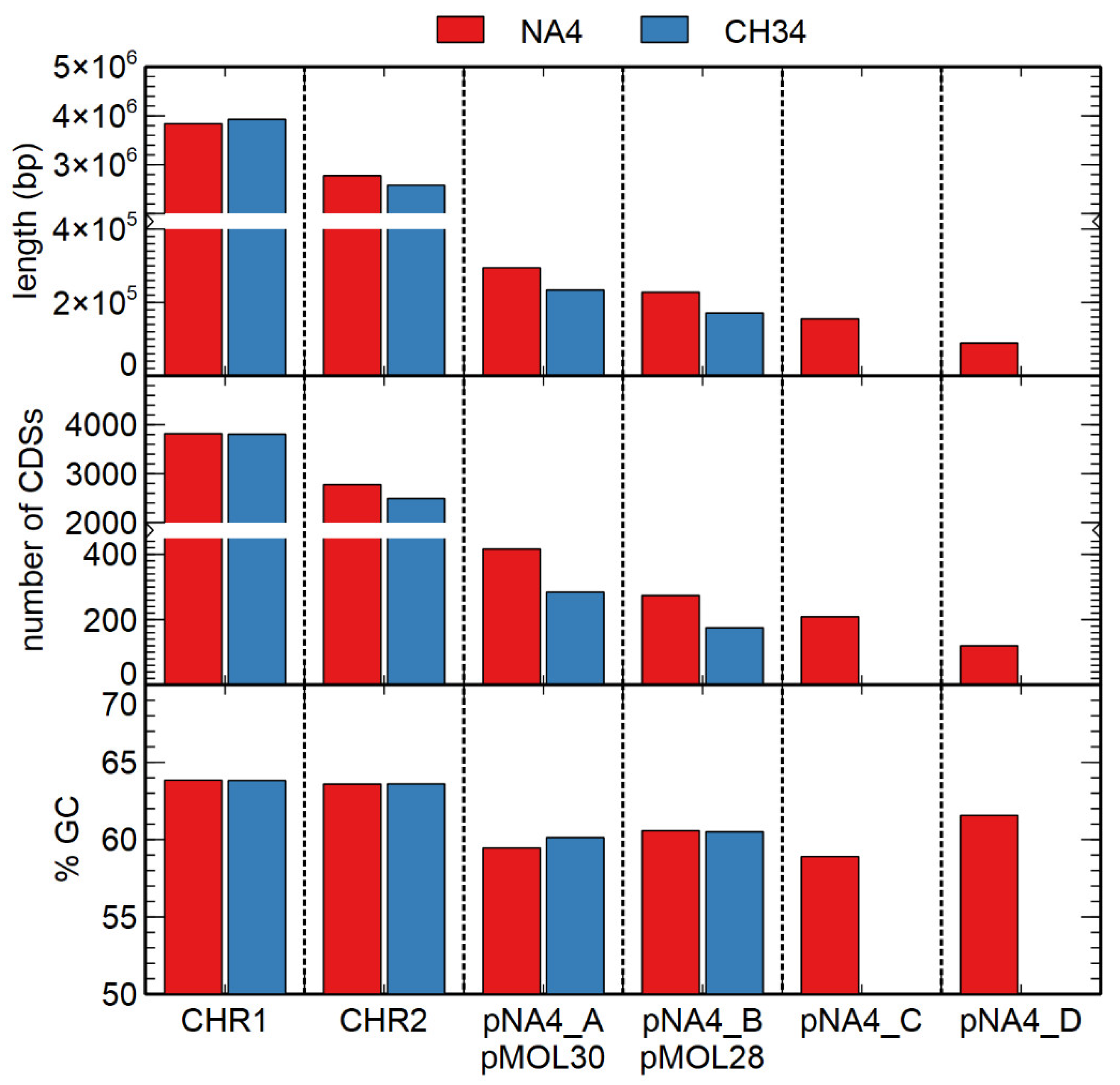
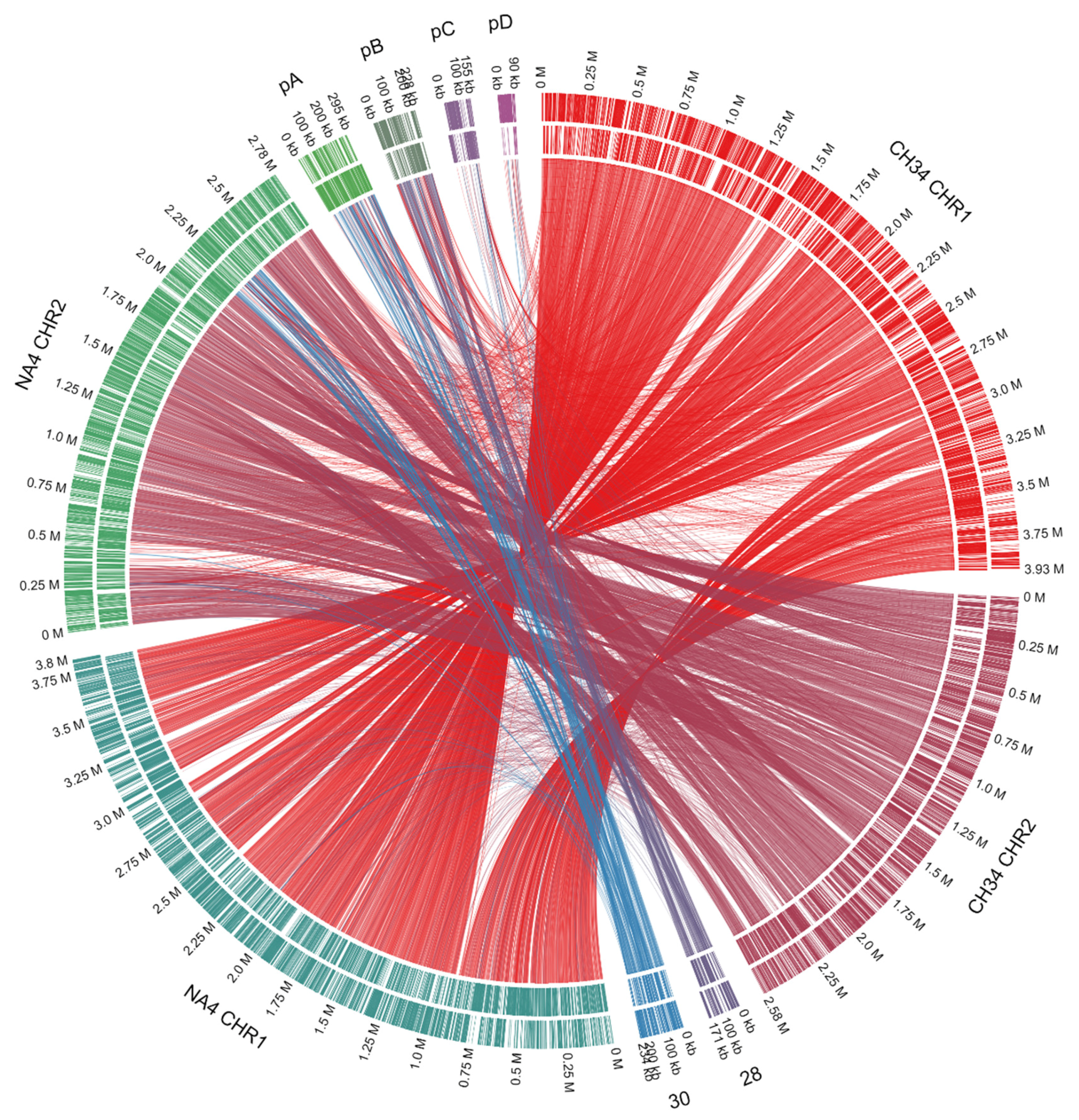
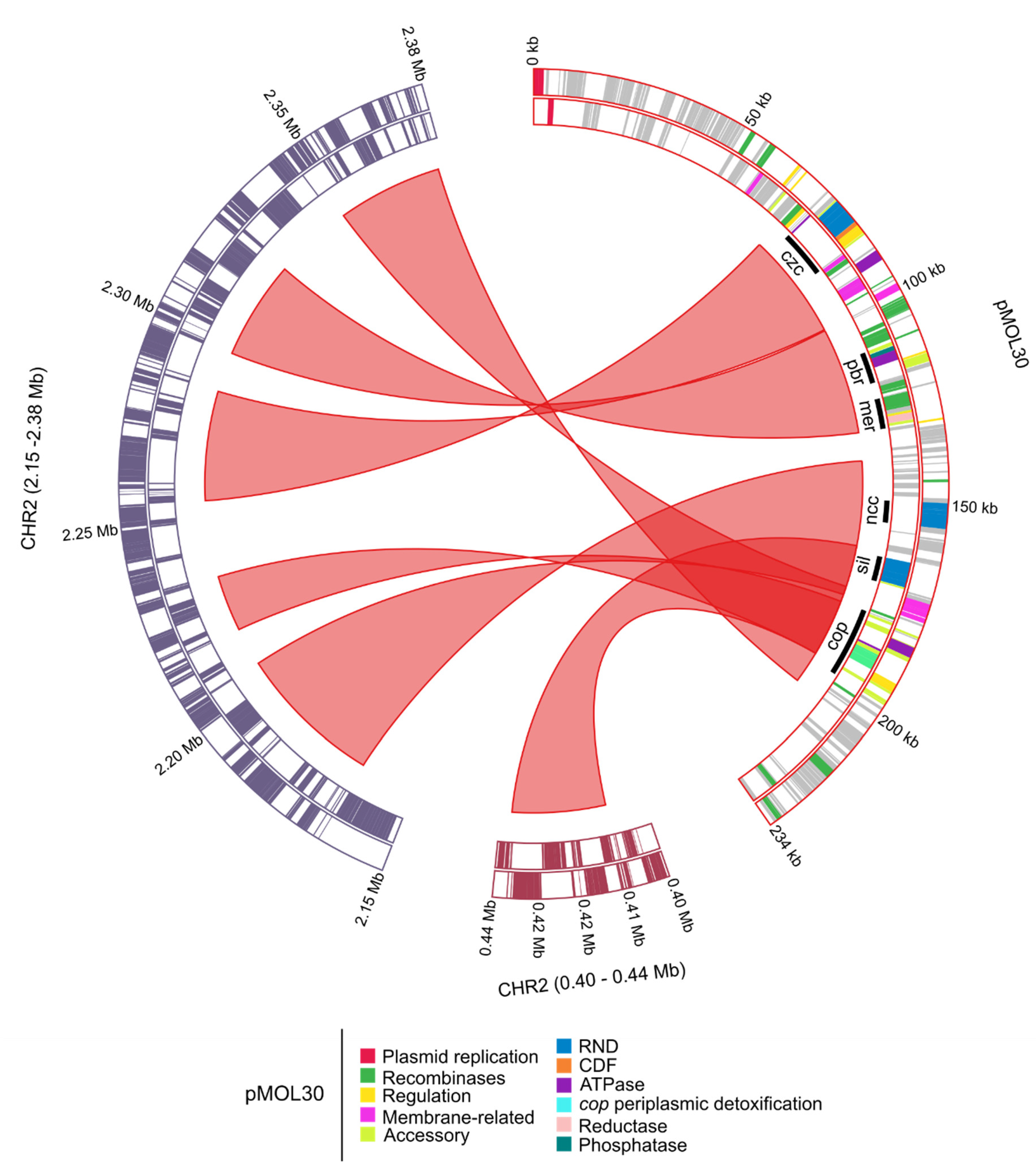
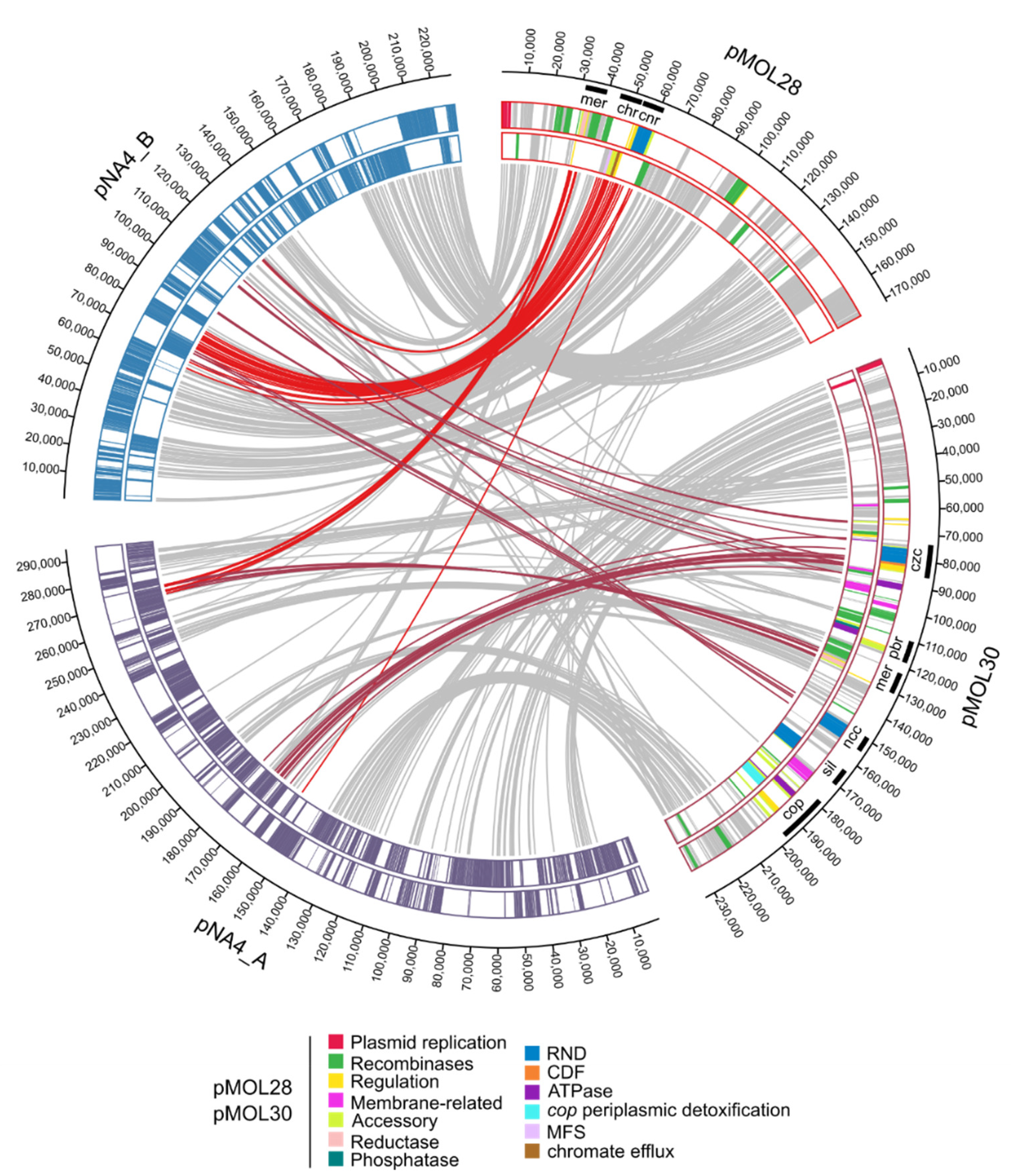
| Replicon | Length (bp) | % GC 1 | # CDSs 2 |
|---|---|---|---|
| Chromosome | 3,838,195 | 63.84 | 3818 |
| Chromid | 2,776,395 | 63.59 | 2774 |
| Plasmid pNA4_A | 294,575 | 59.45 | 416 |
| Plasmid pNA4_B | 227,796 | 60.57 | 274 |
| Plasmid pNA4_C | 155,041 | 58.90 | 209 |
| Plasmid pNA4_D | 89,606 | 61.56 | 120 |
| Replicon | Rep Type | Relaxase Type 1 | MPF Type 2 | Predicted Mobility | MASH Nearest Neighbor 3 |
|---|---|---|---|---|---|
| pMOL28 | 591 | MOBH | MPFT | Conjugative | - |
| pMOL30 | 332 | (MOBP) | MPFF | Conjugative | - |
| pNA4_A | 332 | - | MPFF | Non-mobilizable | pMOL30 |
| pNA4_B | 591 | MOBH | MPFT | Conjugative | pMOL28 |
| pNA4_C | - | - | - | Non-mobilizable | pHS87a (Pseudomonas aeruginosa) |
| pNA4_D | 1864 | MOBF | MPFF | Conjugative | pACP3.3 (Acidovorax sp. P3) |
| Element | Family | Size (%) 1 | CHR1 | CHR2 | pNA4_A | pNA4_B | pNA4_C | pNA4_D |
|---|---|---|---|---|---|---|---|---|
| IS1071 | Tn3 | 3204 (99.9%) | 1 | |||||
| 2991 (93.4%) | 2 | |||||||
| ISRme4 | IS21 | 2469 (100%) | 4 | 3 | ||||
| ISRme9 | IS21 | 2674 (94.8%) | 1 | |||||
| ISRme10 | IS30 | 1063 (100%) | 1 | |||||
| ISRme3 | IS3 | 1288 (100%) | 1 | 2 | 2 | |||
| ISPst3 | IS21 | 2605 (97.8%) | 1 | 2 | ||||
| ISPa45 | IS4 | 1637 (100%) | 1 |
| Pt4+ (µM) | Pd2+ (µM) | Ag+ (µM) | Zn2+ (mM) | Ni2+ (mM) | Cu2+ (mM) | |
|---|---|---|---|---|---|---|
| NA4 | 70 | 12.5 | 1 | 12 | 40 | 6 |
| NA4Pt | 160 | 12.5 | 1 | 12 | 40 | 6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, M.M.; Provoost, A.; Maertens, L.; Leys, N.; Monsieurs, P.; Charlier, D.; Van Houdt, R. Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans. Genes 2019, 10, 63. https://doi.org/10.3390/genes10010063
Ali MM, Provoost A, Maertens L, Leys N, Monsieurs P, Charlier D, Van Houdt R. Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans. Genes. 2019; 10(1):63. https://doi.org/10.3390/genes10010063
Chicago/Turabian StyleAli, Md Muntasir, Ann Provoost, Laurens Maertens, Natalie Leys, Pieter Monsieurs, Daniel Charlier, and Rob Van Houdt. 2019. "Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans" Genes 10, no. 1: 63. https://doi.org/10.3390/genes10010063
APA StyleAli, M. M., Provoost, A., Maertens, L., Leys, N., Monsieurs, P., Charlier, D., & Van Houdt, R. (2019). Genomic and Transcriptomic Changes That Mediate Increased Platinum Resistance in Cupriavidus metallidurans. Genes, 10(1), 63. https://doi.org/10.3390/genes10010063






