Genetics and Genomic Regions Affecting Response to Newcastle Disease Virus Infection under Heat Stress in Layer Chickens
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Population
2.2. Phenotypic Measurements
2.3. Genotyping
2.4. Data Analysis
3. Results
3.1. Quantification of NDV Titers and Anti-NDV Antibody Levels in Hy-Line Brown Layer Chicks
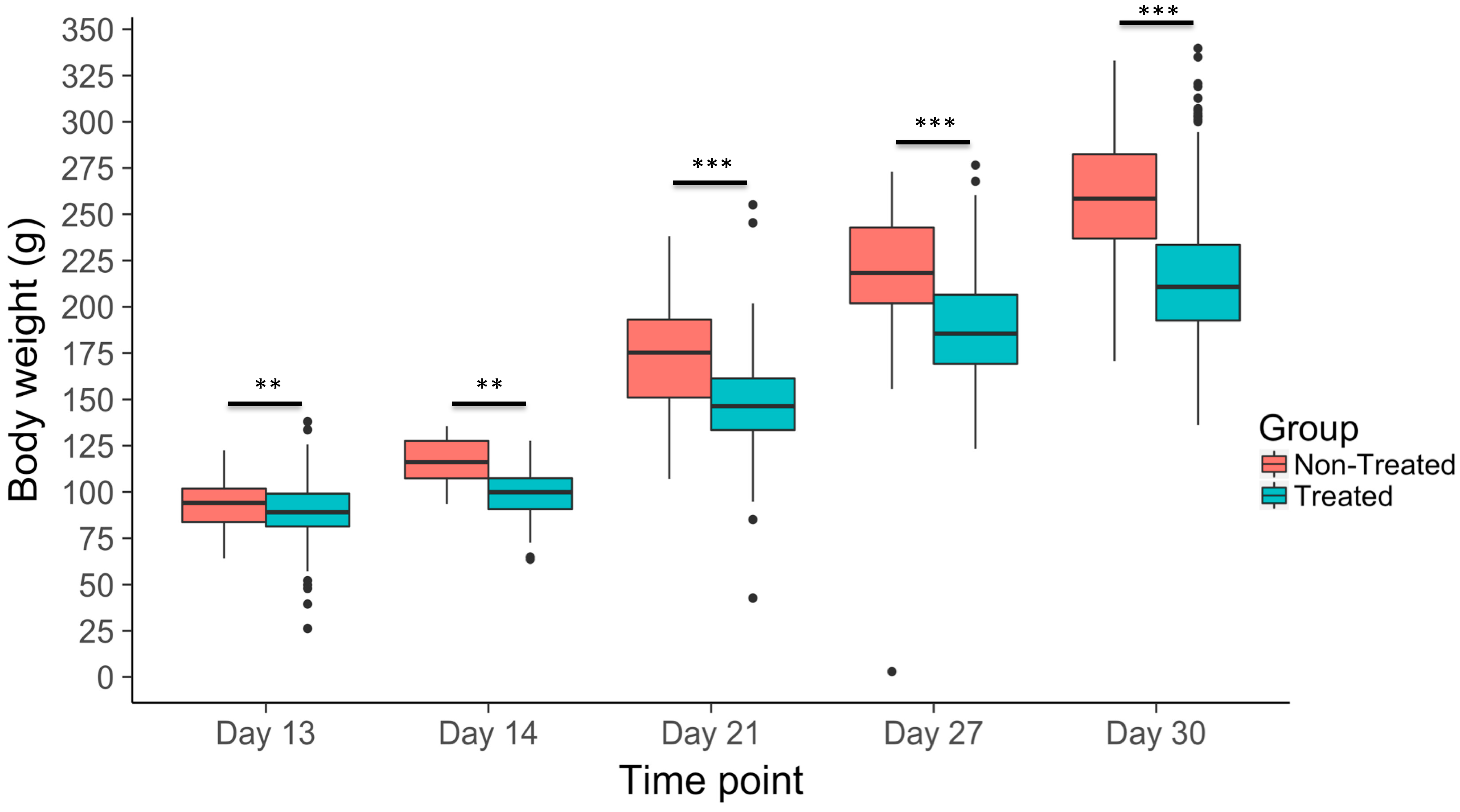
3.2. Body Weight Gain Across Time Points
3.3. Genetic Parameters
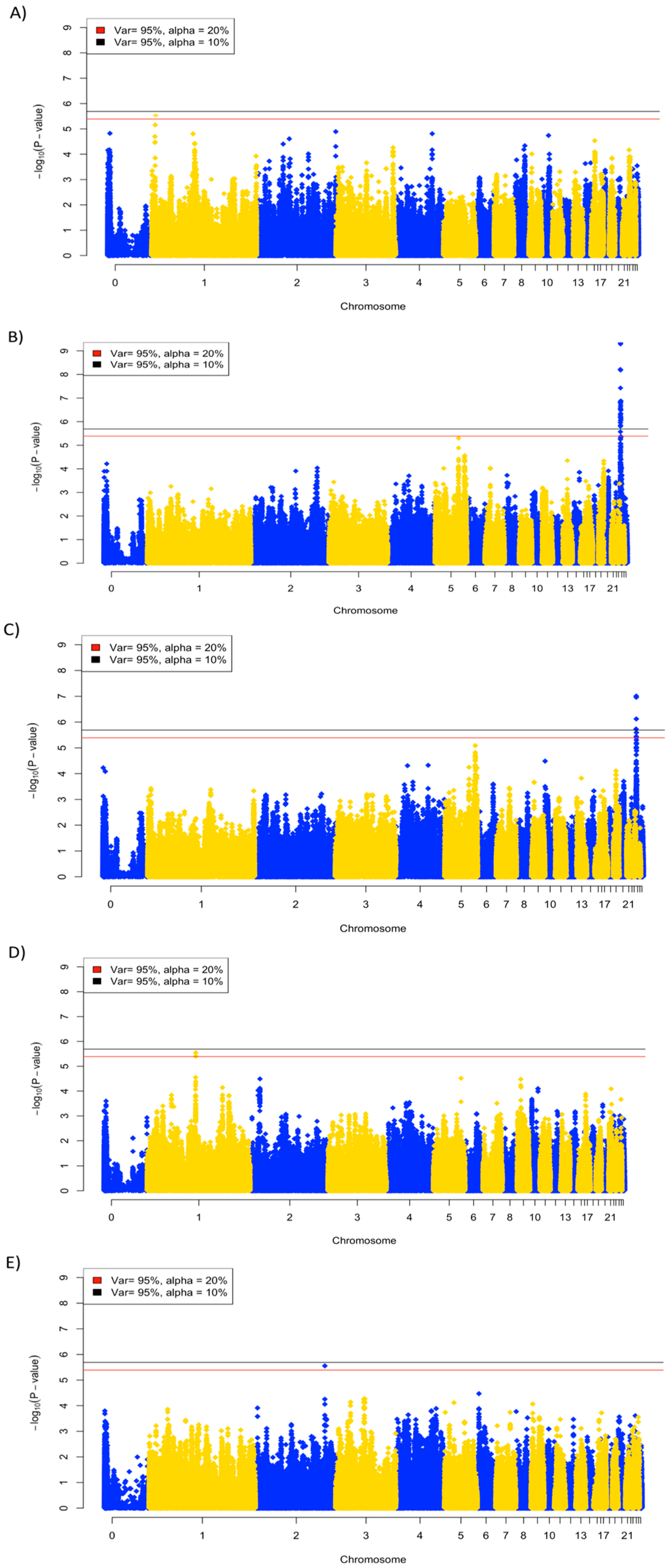
3.4. Genome-Wide Association Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kaleta, E.F.; Baldauf, C. Newcastle Disease in Free-Living and Pet Birds. In Newcastle Disease Developments in Veterinary Virology; Springer: Boston, MA, USA, 1988; Volume 8. [Google Scholar]
- World Bank. TAFS Forum World Livestock Disease Atlas: A Quantitative Analysis of Global Animal Health Data (2006–2009); World Bank: Washington, DC, USA, 2011. [Google Scholar]
- Bwala, D.G.; Clift, S.; Duncan, N.M.; Bisschop, S.P.R.; Oludayo, F.F. Determination of the distribution of lentogenic vaccine and virulent Newcastle disease virus antigen in the oviduct of SPF and commercial hen using immunohistochemistry. Res. Vet. Sci. 2012, 93, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.-H.; Kwon, H.-J.; Kim, T.-E.; Kim, J.-H.; Yoo, H.-S.; Park, M.-H.; Park, Y.-H.; Kim, S.-J. Characterization of a Recombinant Newcastle Disease Virus Vaccine Strain. Clin. Vaccine Immunol. 2008, 15, 1572–1579. [Google Scholar] [CrossRef] [PubMed]
- Albiston, H.E.; Gorrie, C.J.R. Newcastle disease in Victoria. Aust. Vet. J. 1942, 18, 75–79. [Google Scholar] [CrossRef]
- Alexander, D.J. Gordon Memorial Lecture. Newcastle disease. Br. Poult. Sci. 2001, 42, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Kelly, T.R.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Novel mechanisms revealed in the trachea transcriptome of resistant and susceptible chicken lines following infection with Newcastle disease virus. Clin. Vaccine Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Deist, M.S.; Gallardo, R.A.; Bunn, D.A.; Dekkers, J.C.M.; Zhou, H.; Lamont, S.J. Resistant and susceptible chicken lines show distinctive responses to Newcastle disease virus infection in the lung transcriptome. BMC Genom. 2017, 18, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Cole, R.K.; Hutt, F.B. Genetic differences in resistance to Newcastle disease. Avian Dis. 1961, 5, 205–214. [Google Scholar] [CrossRef]
- Lamon, S.J.; Cobl, D.J.; Bjorkquis, A.; Rothschil, M.F.; Persi, M.; Ashwel, C.; Schmid, C. Genomics of heat stress in chickens. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, August 2014. [Google Scholar]
- Lara, L.J.; Rostagno, M.H. Impact of Heat Stress on Poultry Production. Animals 2013, 3, 356–369. [Google Scholar] [CrossRef]
- Bartlett, J.R.; Smith, M.O.; Yan, Q.; Li, L. Effects of different levels of selenium on growth performance and immunocompetence of broilers under heat stress. Poult. Sci. 2003, 82, 1580–1588. [Google Scholar] [CrossRef]
- Slawinska, A.; Hsieh, J.C.; Schmidt, C.J.; Lamont, S.J. Heat stress and lipopolysaccharide stimulation of chicken macrophage-like cell line activates expression of distinct sets of genes. PLoS ONE 2016, 11, e0164575. [Google Scholar] [CrossRef]
- George, M.M. Epidemiology of Newcastle disease and the need to vaccinate local chickens in Uganda. In Proceedings of the International Workshop, Kuala Lumpur, Malaysia, 9–14 September 1991; pp. 155–158. [Google Scholar]
- Mishra, U. Present status of poultry in Nepal. In Proceedings of the 7th Tanzania Veterinary Association Scientific Conference, Aursha International Conference Center, Tanzania, 1989; pp. 403–407. [Google Scholar]
- Awan, M.A.; Otte, M.J.; James, A.D. The epidemiology of Newcastle disease in rural poultry: A review. Avian Pathol. 1994, 23, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Qu, H.; Ma, J.; Wang, J.; Li, C.; Yang, C.; Hu, X.; Li, N.; Shu, D. Genome-wide association study of antibody response to Newcastle disease virus in chicken. BMC Genet. 2013, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Yonash, N.; Cheng, H.H.; Hillel, J.; Heller, D.E.; Cahaner, A. DNA microsatellites linked to quantitative trait loci affecting antibody response and survival rate in meat-type chickens. Poult. Sci. 2001, 80, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, F.; Bovenhuis, H.; van Arendonk, J.A.; Parmentier, H.K.; Jungerius, A.P.; van der Poel, J.J. Across-line SNP association study of innate and adaptive immune response in laying hens. Anim. Genet. 2010, 41, 26–38. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Saelao, P.; Chanthavixay, K.; Gallardo, R.; Bunn, D.; Lamont, S.J.; Dekkers, J.M.; Kelly, T.; Zhou, H. Physiological responses to heat stress in two genetically distinct chicken inbred lines. Poult. Sci. 2018, 1–11. [Google Scholar] [CrossRef]
- R Core Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018.
- Gilmour, A. ASREML for testing fixed effects and estimating multiple trait variance components. Proc. Assoc. Adv. Anim. Breed. Genet. 1997, 12, 386–390. [Google Scholar]
- VanRaden, P.M. Efficient Methods to Compute Genomic Predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Aulchenko, Y.S.; Ripke, S.; Isaacs, A.; van Duijn, C.M. GenABEL: An R library for genome-wide association analysis. Bioinformatics 2007, 23, 1294–1296. [Google Scholar] [CrossRef]
- Waide, E.H.; Tuggle, C.K.; Serão, N.V.L.; Schroyen, M.; Hess, A.; Rowland, R.R.R.; Lunney, J.K.; Plastow, G.; Dekkers, J.C.M. Genomewide association of piglet responses to infection with one of two porcine reproductive and respiratory syndrome virus isolates. J. Anim. Sci. 2017, 95, 16–38. [Google Scholar] [CrossRef]
- Gao, X.; Starmer, J.; Martin, E.R. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet. Epidemiol. 2008, 32, 361–369. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Abasht, B.; Sandford, E.; Arango, J.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Hassen, A.; Habier, D.; Fernando, R.L.; Dekkers, J.C.M.; et al. Extent and consistency of linkage disequilibrium and identification of DNA markers for production and egg quality traits in commercial layer chicken populations. BMC Genom. 2009, 10, S2. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gocke, A.R.; Lovett-Racke, A.; Drew, P.D.; Racke, M.K. PPAR alpha regulation of the immune response and autoimmune encephalomyelitis. PPAR Res. 2008, 2008. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, K.; Plantinga, T.S.; Wong, J.; Monks, B.G.; Gay, N.J.; Netea, M.G.; Fitzgerald, K.A.; Golenbock, D.T. A TIR domain variant of MyD88 adapter-like (Mal)/TIRAP results in loss of MyD88 binding and reduced TLR2/TLR4 signaling. J. Biol. Chem. 2009, 284, 25742–25748. [Google Scholar] [CrossRef] [PubMed]
- Wasylyk, C.; Schlumberger, S.E.; Criqui-Filipe, P.; Wasylyk, B. Sp100 Interacts with ETS-1 and Stimulates Its Transcriptional Activity. Mol. Cell. Biol. 2002, 22, 2687–2702. [Google Scholar] [CrossRef]
- Bradshaw, S.; Zheng, W.J.; Tsoi, L.C.; Gilkeson, G.; Zhang, X.K. A role for Fli-1 in B cell proliferation: Implications for SLE pathogenesis. Clin. Immunol. 2008, 129, 19–30. [Google Scholar] [CrossRef]
- Wolc, A.; Arango, J.; Jankowski, T.; Dunn, I.; Settar, P.; Fulton, J.E.; O’Sullivan, N.P.; Preisinger, R.; Fernando, R.L.; Garrick, D.J.; et al. Genome-wide association study for egg production and quality in layer chickens. J. Anim. Breed. Genet. 2014, 131, 173–182. [Google Scholar] [CrossRef]
- Lwelamira, J.; Kifaro, G.C.; Gwakisa, P.S. Genetic parameters for body weights, egg traits and antibody response against Newcastle Disease Virus (NDV) vaccine among two Tanzania chicken ecotypes. Trop. Anim. Health Prod. 2009, 41, 51–59. [Google Scholar] [CrossRef]
- Rowland, K.; Wolc, A.; Gallardo, R.A.; Kelly, T.; Zhou, H.; Dekkers, J.C.M.; Lamont, S.J. Genetic Analysis of a Commercial Egg Laying Line Challenged with Newcastle Disease Virus. Front. Genet. 2018, 9, 362. [Google Scholar] [CrossRef] [PubMed]
- Charmantier, A.; Garant, D. Environmental quality and evolutionary potential: Lessons from wild populations. Proc. R. Soc. B Biol. Sci. 2005, 272, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Zhang, D.; Aravind, L. The eukaryotic translation initiation regulator CDC123 defines a divergent clade of ATP-grasp enzymes with a predicted role in novel protein modifications. Biol. Direct 2015, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Azad, A.K.; Chakrabarti, S.; Xu, Z.; Davidge, S.T.; Fu, Y.X. Coiled-coil domain containing 3 (CCDC3) represses tumor necrosis factor-α/nuclear factor κB-induced endothelial inflammation. Cell. Signal. 2014, 26, 2793–2800. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, Y.T.; Xia, J.; Yang, Q. MiR674 inhibits the neuraminidase-stimulated immune response on dendritic cells via down-regulated Mbnl3. Oncotarget 2016, 7, 48978–48994. [Google Scholar] [CrossRef]
- Kurt-Jones, E.A.; Popova, L.; Kwinn, L.; Haynes, L.M.; Jones, L.P.; Tripp, R.A.; Walsh, E.E.; Freeman, M.W.; Golenbock, D.T.; Anderson, L.J.; et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat. Immunol. 2000, 1, 398–401. [Google Scholar] [CrossRef]
- Yang, W.H.; Nussbaum, C.; Grewal, P.K.; Marth, J.D.; Sperandio, M. Coordinated roles of ST3Gal-VI and ST3Gal-IV sialyltransferases in the synthesis of selectin ligands. Blood 2012, 120, 1015–1026. [Google Scholar] [CrossRef]
- Guerin, A.; Stavropoulos, D.J.; Diab, Y.; Chénier, S.; Christensen, H.; Kahr, W.H.; Babul-Hirji, R.; Chitayat, D. Interstitial deletion of 11q-implicating the KIRREL3 gene in the neurocognitive delay associated with Jacobsen syndrome. Am. J. Med. Genet. A 2012, 158 A, 2551–2556. [Google Scholar] [CrossRef]

| Trait | Mean ± SE | h2 ± SE |
|---|---|---|
| Viral titer, 2 dpi | 4.77 ± 0.022 | 0.17 ± 0.096 |
| Viral titer, 6 dpi | 2.76 ± 0.026 | 0.11 ± 0.080 |
| Anti-NDV antibody | −0.10 ± 0.037 | 0.039 ± 0.063 |
| Viral clearance | 41.74 ± 0.57 | |
| Body weight change, day 13 | 89.39 ± 0.58 | 0.13 ± 0.076 |
| Body weight change, day 14 | 99.28 ± 0.98 | 0.18 ± 0.17 |
| Body weight change, day 21 | 147.54 ± 0.93 | 0.26 ± 0.11 |
| Body weight change, day 27 | 190.69 ± 1.53 | 0.22 ± 0.11 |
| Body weight change, day 30 | 215.89 ± 1.54 | 0.28 ± 0.094 |
| Viral Titer, 2 Dpi | Viral Titer, 6 Dpi | Anti-NDV Antibody | |
|---|---|---|---|
| Viral titer, 2 dpi | 0.14 ± 0.048 | 0.11 ± 0.05 | |
| Viral titer, 6 dpi | 0.51 ± 0.40 | 0.0199 ± 0.0493 | |
| Anti-NDV antibody | 0.35 ± 0.57 | −0.33 ± 0.79 |
| Trait | Number of Significant SNPs | |
|---|---|---|
| 10% Significance | 20% Significance | |
| Viral titer, 2 dpi | 0 | 1 |
| Viral titer, 6 dpi | 29 | 30 |
| Anti-NDV antibody | 0 | 3 |
| Viral clearance | 4 | 7 |
| Body weight change, day 13 | 0 | 1 |
| Body weight change, day 14 | 0 | 0 |
| Body weight change, day 21 | 0 | 0 |
| Body weight change, day 27 | 0 | 0 |
| Body weight change, day 30 | 0 | 0 |
| Trait | Number of SNPs | Chr: Mb | Genes |
|---|---|---|---|
| Viral titer, 2 dpi | 1 | 1: 6.1–7.1 | CDC123, CAMK1D, CCDC3, DHTKD1, BEND7, ECHDC3, PROSER2, OPTN, NUDT5, SEPHS1, gga-mir-1460, FRMD4A, MCM10, UPF2, USP6NL, PHYH, SEC61A2 |
| Viral titer, 6 dpi | 30 | 24: 0.1–1.9 | TIRAP, SRPRA, KIRREL3, ST3GAL4, EI24, FAM118B, DCPS, CDON, STT3A, CHEK1, FOXRED1, ETS1, FL11, KCNJ1, KCNJ5, 5S_rRNA, ARHGAP32, PANX3, PUS3, NRGN, RPUSD4, C2CD2L, VPS11, SLC37A2, MSANTD2, SIK2, DPAGT1, HMBS, PKNOX2, VSIG10L2, FEZ1, HYLS1, ESAM, HEPACAM, ROBO3, RF00001, BARX2, TMEM45B, APLP2, NFRKB, ST14, ZBTB44, ADAMTS8, ADAMTS15 |
| Viral clearance | 7 | 24: 0.1–1.1 | TIRAP, SRPRA, KIRREL3, ST3GAL4, EI24, FAM118B, DCPS, CDON, STT3A, CHEK1, FOXRED1, ETS1, FL11, KCNJ1, KCNJ5, 5S_rRNA, ARHGAP32 |
| Body weight change, day 13 | 1 | 2: 128.8–129.8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saelao, P.; Wang, Y.; Chanthavixay, G.; Gallardo, R.A.; Wolc, A.; Dekkers, J.C.M.; Lamont, S.J.; Kelly, T.; Zhou, H. Genetics and Genomic Regions Affecting Response to Newcastle Disease Virus Infection under Heat Stress in Layer Chickens. Genes 2019, 10, 61. https://doi.org/10.3390/genes10010061
Saelao P, Wang Y, Chanthavixay G, Gallardo RA, Wolc A, Dekkers JCM, Lamont SJ, Kelly T, Zhou H. Genetics and Genomic Regions Affecting Response to Newcastle Disease Virus Infection under Heat Stress in Layer Chickens. Genes. 2019; 10(1):61. https://doi.org/10.3390/genes10010061
Chicago/Turabian StyleSaelao, Perot, Ying Wang, Ganrea Chanthavixay, Rodrigo A. Gallardo, Anna Wolc, Jack C. M. Dekkers, Susan J. Lamont, Terra Kelly, and Huaijun Zhou. 2019. "Genetics and Genomic Regions Affecting Response to Newcastle Disease Virus Infection under Heat Stress in Layer Chickens" Genes 10, no. 1: 61. https://doi.org/10.3390/genes10010061
APA StyleSaelao, P., Wang, Y., Chanthavixay, G., Gallardo, R. A., Wolc, A., Dekkers, J. C. M., Lamont, S. J., Kelly, T., & Zhou, H. (2019). Genetics and Genomic Regions Affecting Response to Newcastle Disease Virus Infection under Heat Stress in Layer Chickens. Genes, 10(1), 61. https://doi.org/10.3390/genes10010061






