Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of PAL Genes in the Common Walnut (Juglans Regia)
2.2. Analysis of Protein Sequence Properties
2.3. Phylogenetic Analysis
2.4. Conserved Domain and Motifs Displayed in J. Regia PAL Proteins
2.5. Analysis of Gene Exon–Intron Structures
2.7. Chromosome Location of Common Walnut PAL Genes
2.8. miRNA Predicted in Common Walnut PAL Family Genes
2.9. Plant Materials, Treatments, Collections, and RNA Isolation and Analysis of Gene Expression Profiles
3. Results
3.1. Identification and Characterization of Common Walnut PAL Genes
3.2. Phylogenetic Relationship of Common Walnut and Other Four Plants in the PAL Gene Family
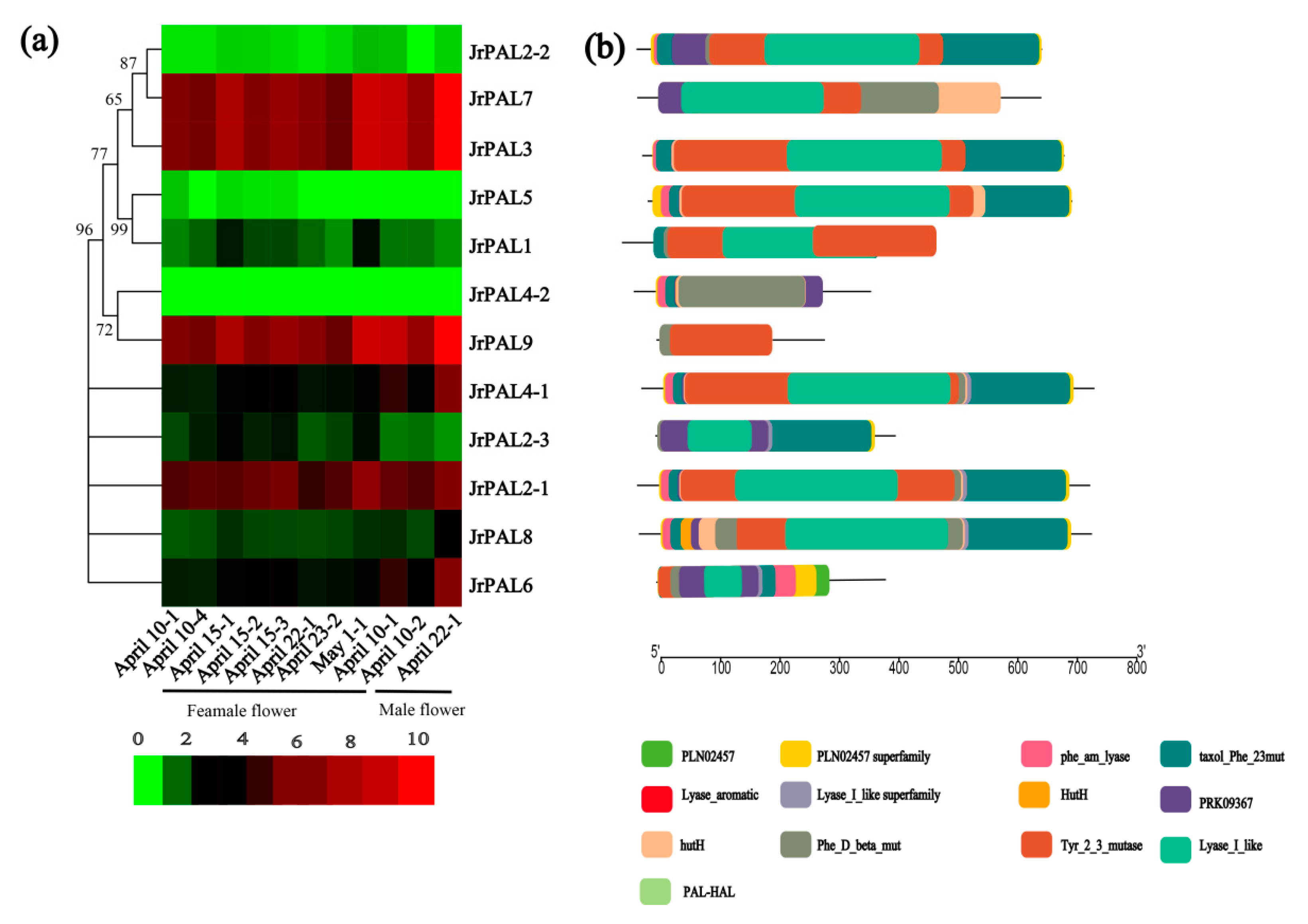
3.3. Position, Conserved Motifs, and Exon–Intron of Common Walnut PALs
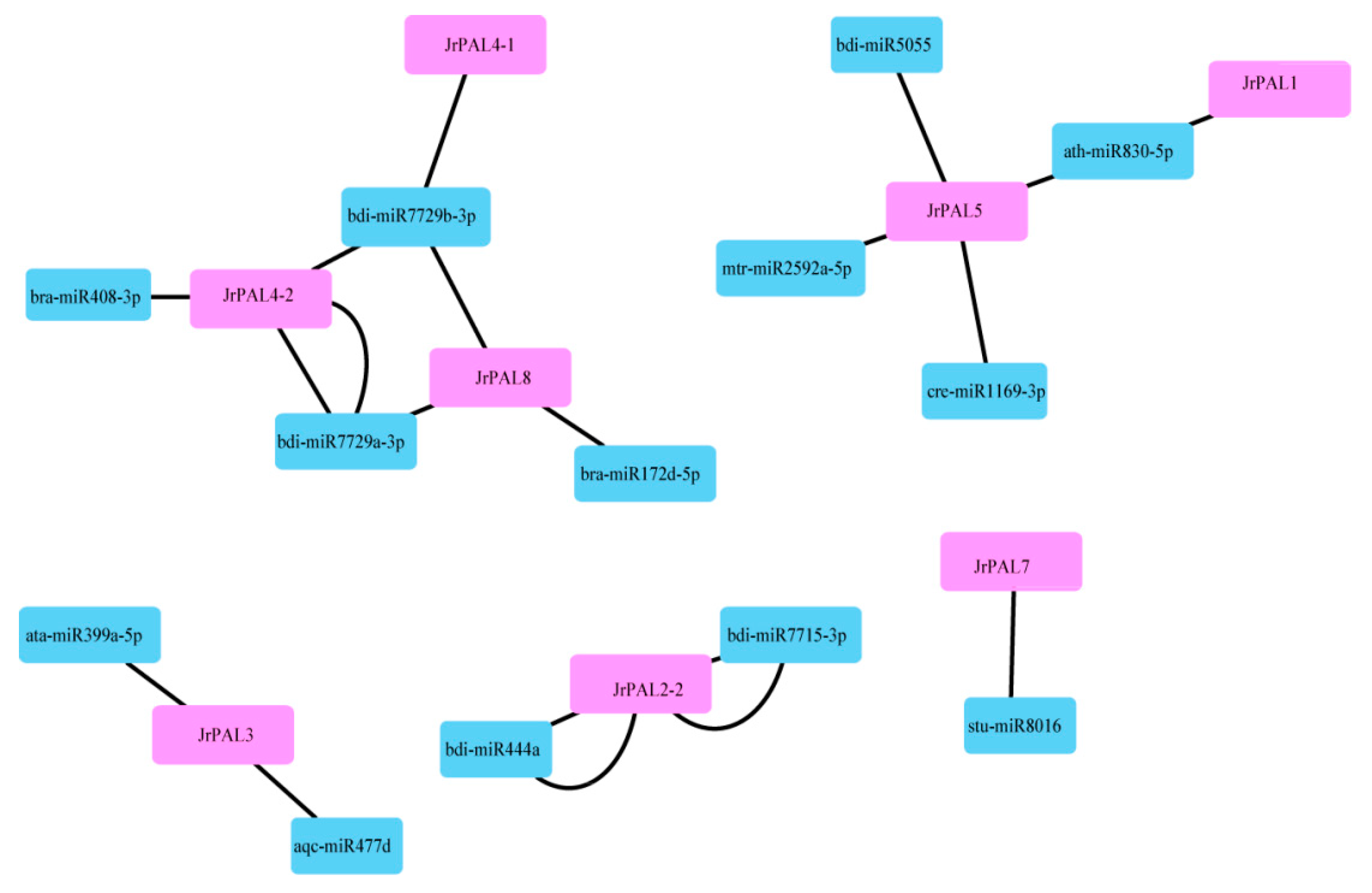
3.4. MicroRNA Targeting and Expression Profile Analysis of Common Walnut PAL Genes
4. Discussion
4.1. Characteristics of Phenylalanine Ammonia-Lyase Gene in J. Regia
4.2. Comparisons of PAL Gene Family in Plants; Expansion and Evolution of Common Walnut PALs
4.3. Comprehensive Analysis of microRNAs Targeting Common Walnut PAL Genes and Expression Levels of PALs in Common Walnut Flowers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Huang, J.; Gu, M.; Lai, Z.; Fan, B.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Chen, Z. Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol. 2010, 153, 1526–1538. [Google Scholar] [CrossRef]
- Wu, P.; Guo, Q.Q.; Qin, Z.W. The fungicide propamocarb increases lignin by activating the phenylpropanoid pathway in Cucumis sativus L. Hortic. Environ. Biotechnol. 2016, 57, 511–518. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085. [Google Scholar] [CrossRef]
- Camera, S.L.; Gouzerh, G.; Dhondt, S.; Hoffmann, L.; Fritig, B.; Legrand, M.; Heitz, T. Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunol. Rev. 2010, 198, 267–284. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Koukol, J.; Conn, E.E. The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J. Boil. Chem. 1966, 236, 2692–2698. [Google Scholar]
- Lois, R.; Dietrich, A.; Hahlbrock, K.; Schulz, W. A phenylalanine ammonia-lyase gene from parsley: Structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989, 8, 1641–1648. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, M.J.; D’Cunha, G.B. A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol. 2007, 85, 273. [Google Scholar] [CrossRef]
- Cochrane, F.C.; Davin, L.B.; Lewis, N.G. The Arabidopsis phenylalanine ammonia lyase gene family: Kinetic characterization of the four PAL isoforms. Phytochemistry 2004, 65, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Rawal, H.C.; Singh, N.K.; Sharma, T.R. Conservation, divergence, and genome-wide distribution of PAL and POX A gene families in plants. Int. J. of Genom. 2013, 2013, 678–969. [Google Scholar]
- Hou, X.; Shao, F.; Ma, Y.; Lu, S. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: Genome-wide characterization, molecular cloning and expression analysis. Mol. Biol. Rep. 2013, 40, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Bagal, U.R. The phenylalanine ammonia lyase (PAL) gene family shows a gymnosperm-specific lineage. BMC Genom. 2012, 13, S1. [Google Scholar]
- De, J.F.; Hanley, S.J.; Beale, M.H.; Karp, A. Characterisation of the willow phenylalanine ammonia-lyase (PAL) gene family reveals expression differences compared with poplar. Phytochemistry 2015, 117, 90–97. [Google Scholar]
- Wang, Z.B.; Chen, X.; Wang, W.; Cheng, K.D.; Kong, J.Q. Transcriptome-wide identification and characterization of Ornithogalum saundersiae phenylalanine ammonia lyase gene family. RSC Adv. 2014, 4, 27159–27175. [Google Scholar] [CrossRef]
- Wanner, L.A.; Li, G.; Ware, D.; Somssich, I.E.; Davis, K.R. The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol. Biol. 1995, 27, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Raes, J.; Rohde, A.; Christensen, J.H.; Van, Y.D.P.; Boerjan, W. Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol. 2003, 133, 1051–1071. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Sun, Y.H.; Li, Q.; Heber, S.; Sederoff, R.; Chiang, V.L. Towards a systems approach for lignin biosynthesis in Populus trichocarpa: Transcript abundance and specificity of the monolignol biosynthetic genes. Plant Cell Physiol. 2010, 51, 144–163. [Google Scholar] [CrossRef]
- Hui, X.; Park, N.I.; Li, X.; Yong, K.K.; Lee, S.Y.; Sang, U.P. Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour. Technol. 2010, 101, 9715–9722. [Google Scholar]
- Lepelley, M.; Mahesh, V.; Mccarthy, J.; Rigoreau, M.; Crouzillat, D.; Chabrillange, N.; De, K.A.; Campa, C. Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora pierre (Rubiaceae). Planta 2012, 236, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.J.; Ning, C.; Zhang, Z.G.; Shang, Q.M. Phenylalanine ammonia-lyase gene families in cucurbit species: Structure, evolution, and expression. J. Integr. Agric. 2016, 15, 1239–1255. [Google Scholar] [CrossRef]
- Shang, Q.M.; Li, L.; Dong, C.J. Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus L. Planta 2012, 236, 1093–1105. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463. [Google Scholar] [PubMed]
- Wada, K.C.; Mizuuchi, K.; Koshio, A.; Kaneko, K.; Mitsui, T.; Takeno, K. Stress enhances the gene expression and enzyme activity of phenylalanine ammonia-lyase and the endogenous content of salicylic acid to induce flowering in pharbitis. J. Plant Physiol. 2014, 171, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhuang, Y.; Xu, T.; Li, Y.; Li, Y.; Lin, W. Changes in rice allelopathy and rhizosphere microflora by inhibiting rice phenylalanine ammonia-lyase gene expression. J. Chem. Ecol. 2013, 39, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Pollegioni, P.; Woeste, K.E.; Chiocchini, F.; Lungo, S.D.; Olimpieri, I.; Tortolano, V.; Clark, J.; Hemery, G.E.; Mapelli, S.; Malvolti, M.E. Ancient humans influenced the current spatial genetic structure of common walnut populations in Asia. PLoS ONE 2015, 10, e0135980. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Woeste, K.E.; Hu, Y.; Dang, M.; Zhang, T.; Gao, X.X.; Zhou, H.; Feng, X.; Zhao, G.; Zhao, P. Genetic diversity and population structure of common walnut (Juglans regia) in China based on EST-SSRs and the nuclear gene phenylalanine ammonia-lyase (PAL). Tree Genet. Genomes 2016, 12, 111. [Google Scholar] [CrossRef]
- Panth, N.; Paudel, K.R.; Karki, R. Phytochemical profile and biological activity of Juglans regia. Chin J. Integr. Med. 2016, 14, 359–373. [Google Scholar] [CrossRef]
- Martínez-García, P.J.; Crepeau, M.W.; Puiu, D.; Gonzalez-Ibeas, D.; Whalen, J.; Stevens, K.A.; Paul, R.; Butterfield, T.S.; Britton, M.T.; Reagan, R.L.; et al. The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J. 2016, 87, 507–532. [Google Scholar] [CrossRef]
- Tsoukas, M.A.; Ko, B.J.; Witte, T.R.; Dincer, F.; Hardman, W.E.; Mantzoros, C.S. Dietary walnut suppression of colorectal cancer in mice: Mediation by miRNA patterns and fatty acid incorporation. J. Nutr. Biochem. 2015, 26, 776–783. [Google Scholar] [CrossRef]
- Xu, F.; Deng, G.; Cheng, S.; Zhang, W.; Huang, X.; Li, L.; Cheng, H.; Rong, X.; Li, J. Molecular cloning, characterization and expression of the phenylalanine ammonia-lyase gene from Juglans regia. Molecules 2012, 17, 7810–7823. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, M.V.; Tsantili, E. Participation of phenylalanine ammonia-lyase (PAL) in increased phenolic compounds in fresh cold stressed walnut (Juglans regia L.) kernels. Postharvest Biol. Technol. 2015, 104, 17–25. [Google Scholar] [CrossRef]
- Persic, M.; Mikulicpetkovsek, M.; Halbwirth, H.; Solar, A.; Veberic, R.; Slatnar, A. Red walnut: Characterization of the phenolic profiles, activities and gene expression of selected enzymes related to the phenylpropanoid pathway in pellicle during walnut development. J. Agric. Food Chem. 2018, 66, 2742–2748. [Google Scholar] [CrossRef]
- Beritognolo, I.; Magel, E.; Abdellatif, A.; Charpentier, J.P.; Jayallemand, C.; Breton, C. Expression of genes encoding chalcone synthase, flavanone 3-hydroxylase and dihydroflavonol 4-reductase correlates with flavanol accumulation during heartwood formation in Juglans nigra. Tree Physiol. 2002, 22, 291. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Meng, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome wide identification, evolutionary, and expression analysis of VQ genes from two Pyrus species. Genes 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Penelope, C.; Eberhardt, R.Y.; Eddy, S.R.; Jaina, M.; Mitchell, A.L.; Potter, S.C.; Marco, P.; Matloob, Q.; Amaia, S.V. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Doerks, T.; Bork, P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, 302–305. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Lobo, I. Basic local alignment search tool (BLAST). J. Mol. Biol. 2012, 215, 403–410. [Google Scholar]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Ma, M.; Ming, L.; Jian, C.; Song, Y.; Wang, S.; Li, P. Molecular and biological characterization of Chinese Sacbrood virus LN isolate. Comp. Funct. Genom. 2011, 2011, 386–409. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, CE.; Noble, W.S. The MEME suite. Nucleic Acids Res. 2015, 43, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Harding, S.A.; Tschaplinski, T.J.; Lindroth, R.L.; Yuan, Y. Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol. 2010, 172, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2014, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, W.; He, Q.; Daud, M.K.; Chen, J.; Zhu, S. Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PLoS ONE 2015, 10, e0123281. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39, W155–W159. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhu, H.; Han, X.; Lv, J.; Zhao, L.; Xu, X.; Zhang, T.; Guo, W. Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. hirsutum. BMC Plant Biol. 2011, 11, 11–40. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.A.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, Y.; Liu, Z.; Cheng, H.; Xue, Y. HEMI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef]
- Likun, W.; Zhixing, F.; Xi, W.; Xiaowo, W.; Xuegong, Z. Degseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Piao, Y.; Shon, H.S.; Ryu, K.H. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-seq data. BMC Bioinform. 2015, 16, 347. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Love, M.I.; Anders, S.; Huber, W. DESeq2: Differential gene expression analysis based on the negative binomial distribution. 2014. Available online: https://rdrr.io/bioc/DESeq2/ (accessed on 24 September 2018).
- Conesa, A.; Götz, S.; Garcíagómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Achnine, L.; Blancaflor, E.B.; Rasmussen, S.; Dixon, R.A. Colocalization of l-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 2004, 16, 3098–3109. [Google Scholar] [CrossRef]
- Ma, R.F.; Liu, Q.Z.; Xiao, Y.; Zhang, L.; Li, Q.; Yin, J.; Chen, W.S. The phenylalanine ammonia-lyase gene family in Isatis indigotica fort.: Molecular cloning, characterization, and expression analysis. Chin. J. Nat. Med. 2016, 14, 801–812. [Google Scholar] [CrossRef]
- Lei, L.; Zhou, S.L.; Ma, H.; Zhang, L.S. Expansion and diversification of the set domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Duan, L.; Liu, D.; Guo, J.; Ge, S.; Dicks, J.; ÓMáille, P.; Osbourn, A.; Qi, X. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012, 193, 1022–1038. [Google Scholar] [CrossRef] [PubMed]
- Ober, D. Seeing double: Gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 2005, 10, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.J.; Shang, Q.M. Genome-wide characterization of phenylalanine ammonia-lyase gene family in watermelon (Citrullus lanatus). Planta 2013, 238, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Pereira, P.A.; Slotkin, R.K.; Martienssen, R.A.; Becker, J.D. MicroRNA activity in the Arabidopsis male germline. J. Exp. Bot. 2011, 62, 3699. [Google Scholar] [CrossRef] [PubMed]
- Jeong, D.H.; Green, P.J. Comprehensive investigation of microRNAs enhanced by analysis of sequence variants, expression patterns, argonaute loading, and target cleavage. Plant Physiol. 2013, 162, 1225–1245. [Google Scholar] [CrossRef] [PubMed]
- Baranwal, V.K.; Negi, N.; Khurana, P. Auxin response factor genes repertoire in Mulberry: Identification, and structural, functional and evolutionary analyses. Genes 2017, 8, 202. [Google Scholar] [CrossRef] [PubMed]
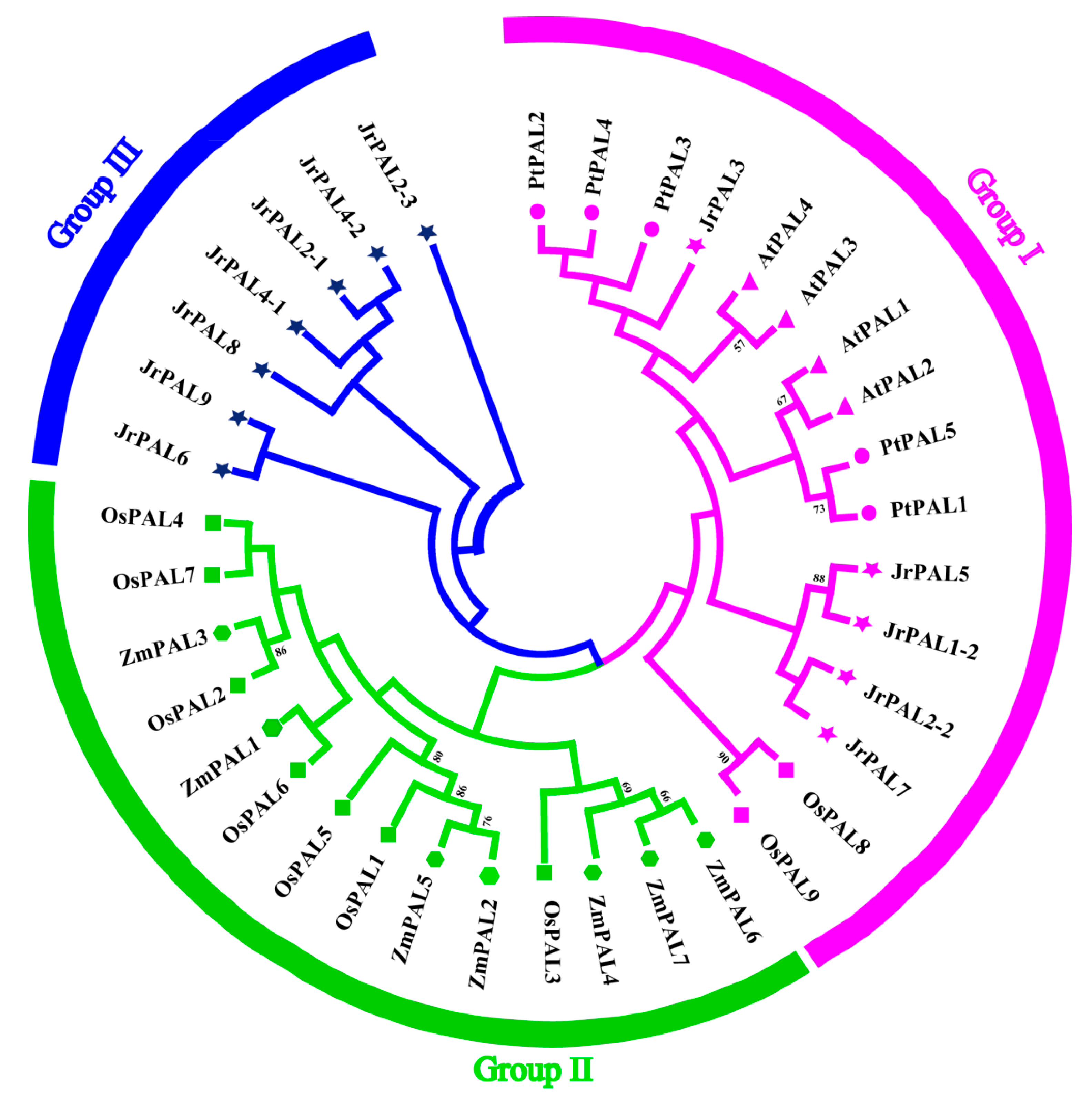
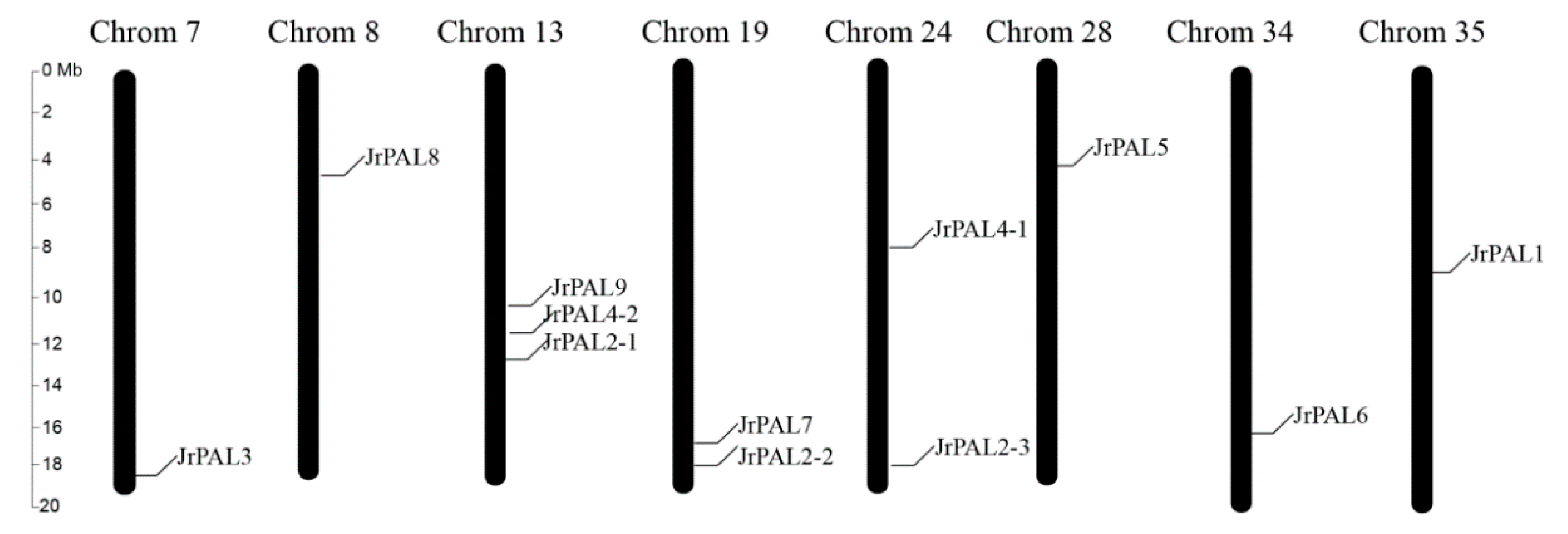
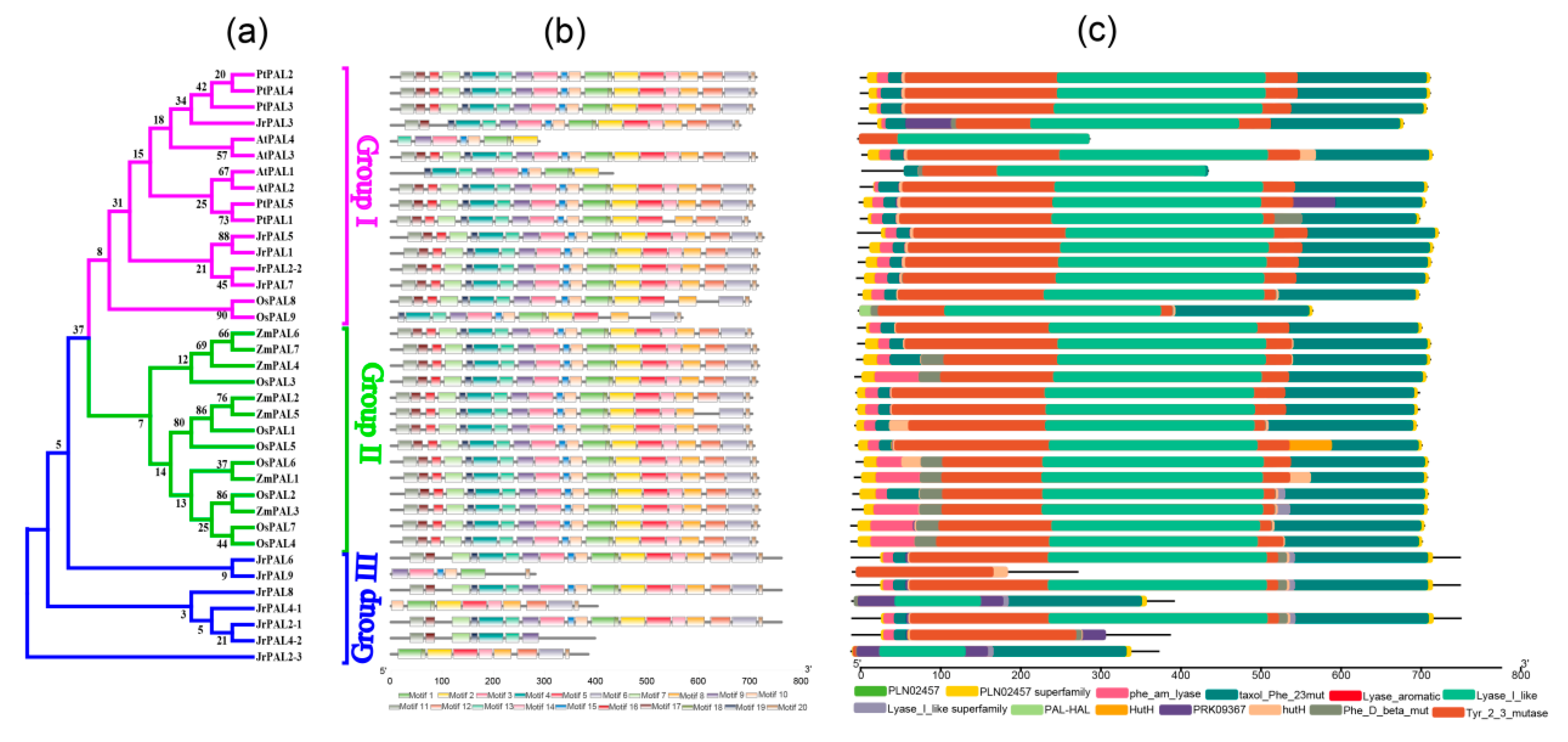
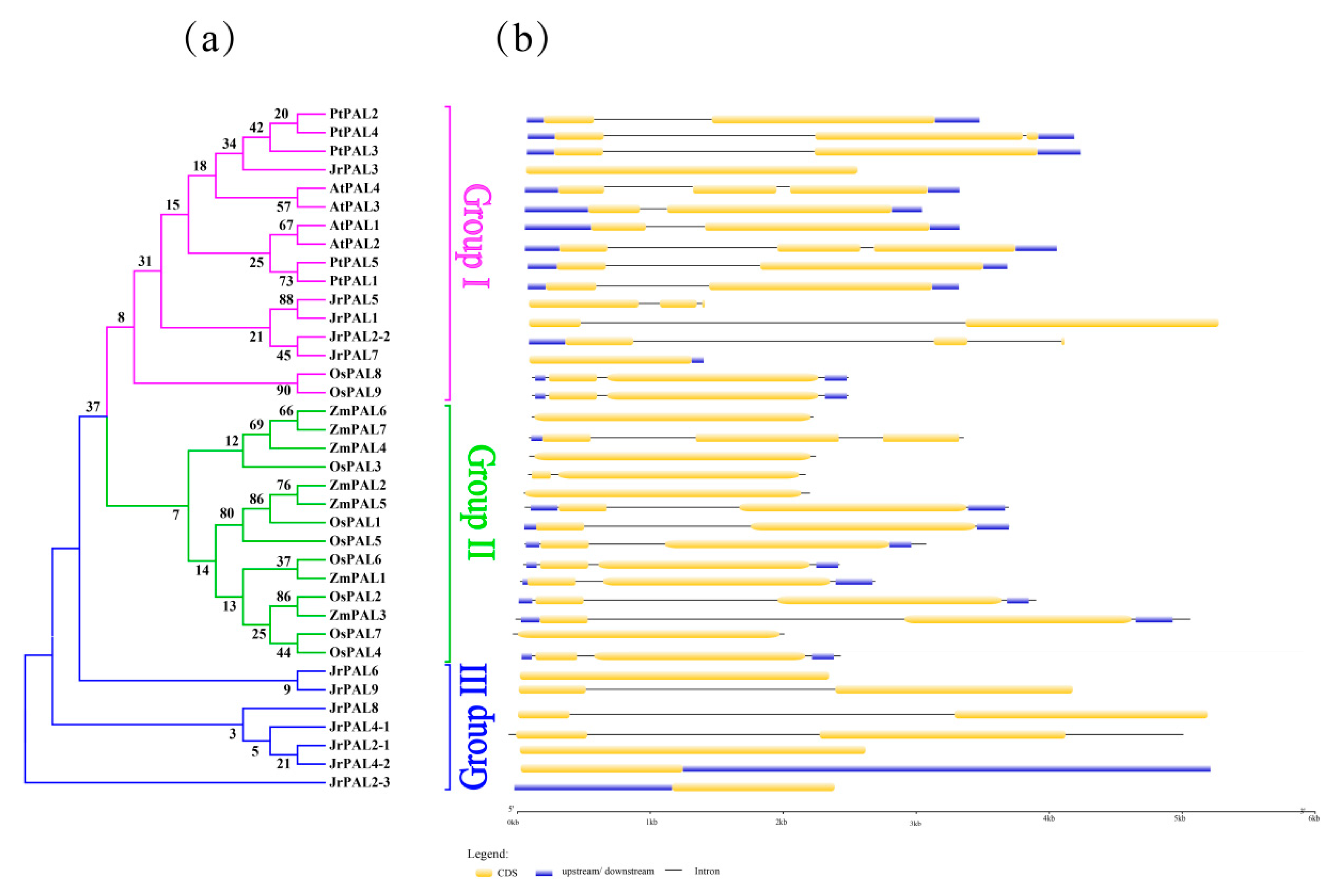
| Gene Name | Protein ID | Gene ID | CDS ID | Subcellular Location | Amino Acids (aa) | Molecular Weight (kDa) | Theoretical pI | Chromosome | Chromosome Length | Gene Position | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Start | End | ||||||||||
| JrPAL3 | XP_018828772.1 | NW_017443600.1:c859630-855244 | XM_018973227.1 | Cytoplasm | 708 | 77.04 | 5.96 | Chr7 | 19,001,705 | 18,431,863 | 1,8427,477 |
| JrPAL5 | XP_018859391.1 | NW_017437924.1:10925-15347 | XM_019003846.1 | Cytoplasm | 712 | 77.71 | 6.06 | Chr28 | 18,296,634 | 2,940,954 | 2,945,376 |
| JrPAL2-2 | XP_018844813.1 | NW_017389549.1:c62915-57410 | XM_018989268.1 | Cytoplasm | 680 | 74.11 | 5.91 | Chr19 | 18,508,379 | 1,8341,787 | 18,337,338 |
| JrPAL2-1 | XP_018827035.1 | NW_017443587.1:471936-474695 | XM_018971490.1 | Cytoplasm | 760 | 83.67 | 6.16 | Chr13 | 18,490,500 | 1,2030,270 | 12,033,029 |
| JrPAL4-1 | XP_018845411.1 | NW_017389589.1:c87043-84575 | XM_018989866.1 | Cytoplasm | 760 | 83.92 | 6.38 | Chr24 | 18,356,466 | 3,643,363 | 3,640,895 |
| JrPAL8 | XP_018817312.1 | NW_017443031.1:261176-263621 | XM_018961767.1 | Cytoplasm | 760 | 83.97 | 6.35 | Chr8 | 19,363,960 | 5,898,473 | 5,900,918 |
| JrPAL1 | XP_018853318.1 | NW_017394290.1:c1301-6 | XM_018997773.1 | Cytoplasm | 432 | 47.05 | 6.57 | Chr35 | 18,286,198 | 9,845,144 | 9,843,849 |
| JrPAL2-3 | XP_018845408.1 | NW_017389589.1:c6632-4075 | XM_018989863.1 | Cytoplasm | 402 | 44.94 | 5.74 | Chr24 | 18,356,466 | 3,562,298 | 3,560,395 |
| JrPAL4-2 | XP_018827000.1 | NW_017443587.1:336894-338297 | XM_018971455.1 | Cytoplasm | 397 | 44.11 | 8.75 | Chr13 | 18,490,500 | 11,895,228 | 11,896,204 |
| JrPAL6 | XP_018855337.1 | NW_017419648.1:c5073-3779 | XM_018989863.1 | Cytoplasm | 384 | 42.92 | 5.85 | Chr34 | 18,288,579 | 16,505,050 | 16,503,756 |
| JrPAL7 | AAX18624.1 | NW_017389549.1:c62915-57410 | XM_018989268.1 | Cytoplasm | 289 | 31.19 | 5.58 | Chr19 | 18,508,379 | 18,341,787 | 18,337,338 |
| JrPAL9 | XP_018827002.1 | NW_017443587.1:394996-399270 | XM_018971457.1 | Cytoplasm | 281 | 31.47 | 6.40 | Chr13 | 18,490,500 | 11,954,327 | 11,956,842 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, F.; Li, H.; Zhao, P. Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes 2019, 10, 46. https://doi.org/10.3390/genes10010046
Yan F, Li H, Zhao P. Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes. 2019; 10(1):46. https://doi.org/10.3390/genes10010046
Chicago/Turabian StyleYan, Feng, Huaizhu Li, and Peng Zhao. 2019. "Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.)" Genes 10, no. 1: 46. https://doi.org/10.3390/genes10010046
APA StyleYan, F., Li, H., & Zhao, P. (2019). Genome-Wide Identification and Transcriptional Expression of the PAL Gene Family in Common Walnut (Juglans Regia L.). Genes, 10(1), 46. https://doi.org/10.3390/genes10010046






