The Diversity of Muscles and Their Regenerative Potential across Animals
Abstract
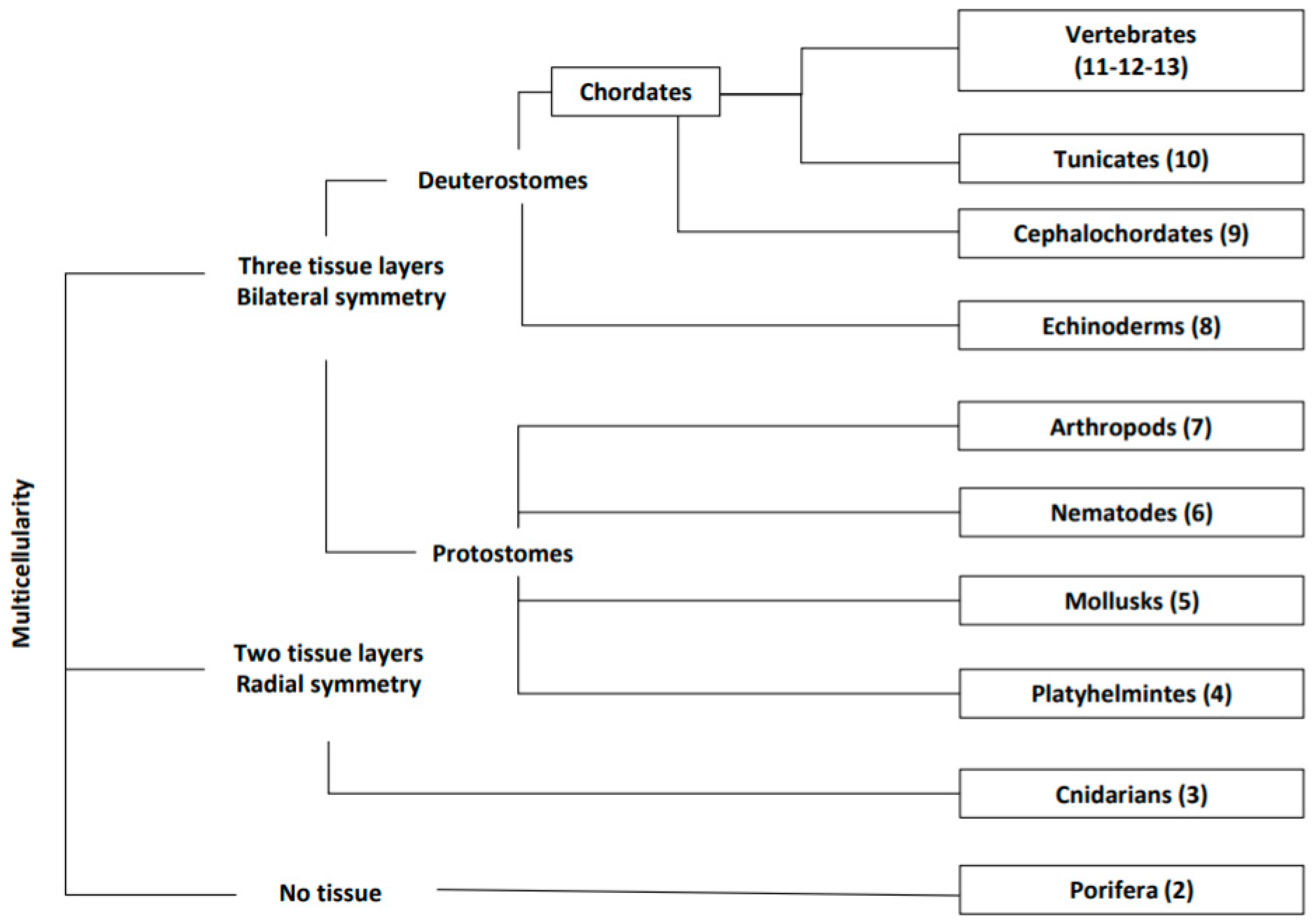
:1. Introduction
2. Porifera: Low Body Complexity with High Regenerative Capabilities
Muscle-Like Cells
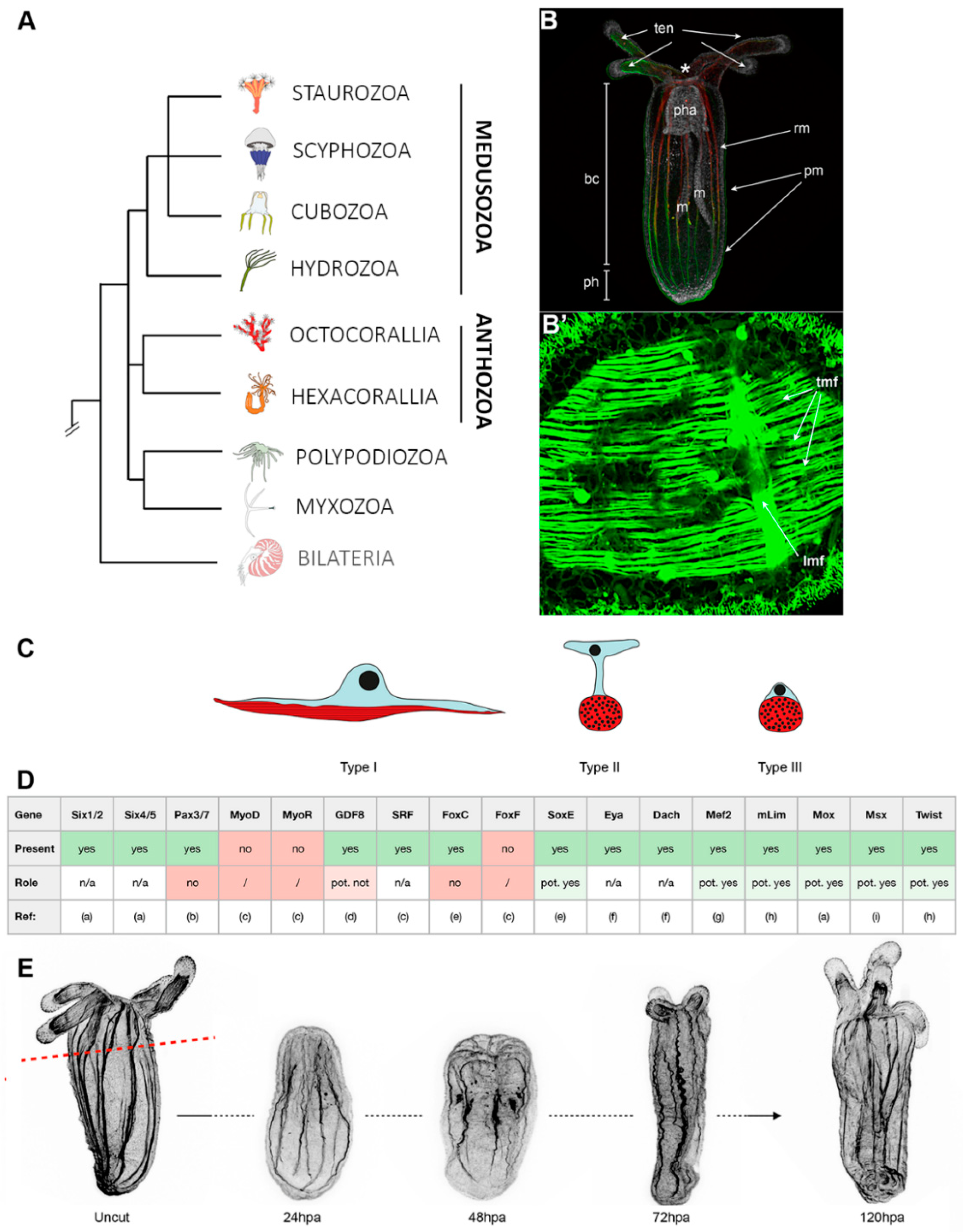
3. Cnidarians: The Starlet Sea Anemone, Nematostella vectensis (Anthozoa)
3.1. Muscle Types, Organization, and Myogenic Genes
3.2. Muscle Regeneration and Role of Epitheliomuscular Cells in the Regenerative Process
4. Platyhelminthes: The Freshwater Planarian
4.1. Muscle Types
4.2. Muscle Regeneration and Homeostasis
5. Mollusks: The Cephalopods
5.1. Muscle Types
5.2. Muscle Regeneration
6. Nematodes: The Caenorabditis elegans Model
Muscle Type and Homeostasis
7. Artropods: The Insect Drosophila melanogaster
Muscle Type and Homeostasis
8. Echinoderms: A Compendium of Regeneration Strategies
8.1. Echinoderm Muscles
8.2. Echinoderm Muscle Regeneration
9. Cephalochordates, the Basal Chordates: Amphioxus
9.1. Structure of Amphioxus Muscles
9.2. Muscle Regeneration in the Amphioxus Tail
10. Tunicates: The Sister Group of Vertebrates
10.1. Myogenesis during Embryogenesis and Metamorphoses
10.2. Myogenesis during Budding and Regeneration
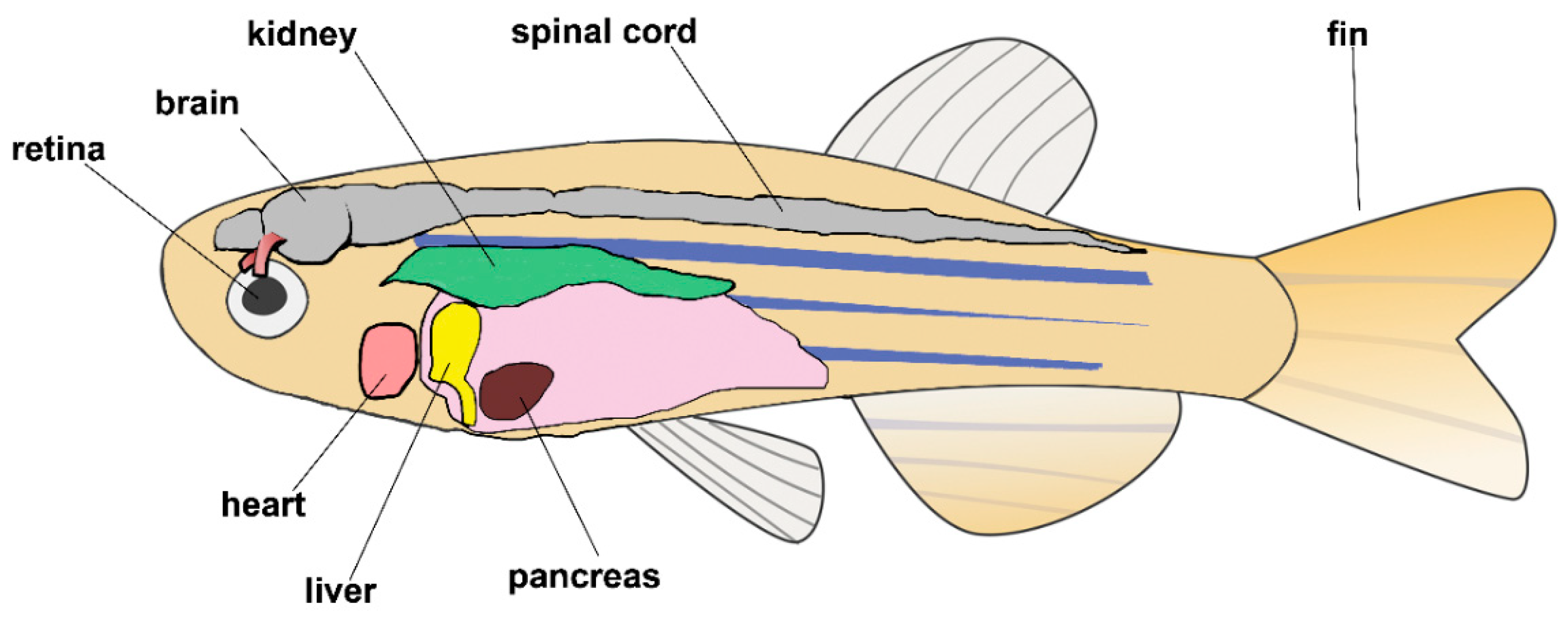
11. Vertebrates: The Zebrafish
11.1. Zebrafish Skeletal Muscle Regeneration
11.2. Zebrafish Heart Regeneration
11.3. Zebrafish as a Model of Human Regeneration
12. Mammals: Cell Therapy for Skeletal Muscle Regeneration
13. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ciciliot, S.; Schiaffino, S. Regeneration of mammalian skeletal muscle. Basic mechanisms and clinical implications. Curr. Pharm. Des. 2010, 16, 906–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Musaro, A. The basis of muscle regeneration. Adv. Biol. 2014, 2014, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Maden, M. The evolution of regeneration—Where does that leave mammals? Int. J. Dev. Biol. 2018, 62, 369–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiozzo, S.; Copley, R. Reconsidering regeneration in metazoans: An evo-devo approach. Front. Ecol. Evol. 2015, 3, 67. [Google Scholar] [CrossRef] [Green Version]
- Steinmetz, P.R.; Kraus, J.E.; Larroux, C.; Hammel, J.U.; Amon-Hassenzahl, A.; Houliston, E.; Worheide, G.; Nickel, M.; Degnan, B.M.; Technau, U. Independent evolution of striated muscles in cnidarians and bilaterians. Nature 2012, 487, 231–234. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Yang, H.; Zhong, T. Regeneration across metazoan phylogeny: Lessons from model organisms. J. Genet. Genom. 2015, 42. [Google Scholar] [CrossRef]
- Baghdadi, M.; Tajbakhsh, S. Regulation and phylogeny of skeletal muscle regeneration. Dev. Biol. 2017, 433. [Google Scholar] [CrossRef]
- Hejnol, A. Muscle’s dual origins. Nature 2012, 487, 181–182. [Google Scholar] [CrossRef]
- Telford, M.J.; Moroz, L.L.; Halanych, K.M. Evolution: A sisterly dispute. Nature 2016, 529, 286–287. [Google Scholar] [CrossRef]
- Brunet, T.; Fischer, A.H.L.; Steinmetz, P.R.H.; Lauri, A.; Bertucci, P.; Arendt, D. The evolutionary origin of bilaterian smooth and striated myocytes. eLife 2016, 5, e19607. [Google Scholar] [CrossRef]
- Sikes, J.M.; Newmark, P.A. Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 2013, 500, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Selck, C.; Friedrich, B.; Lutz, R.; Vila-Farré, M.; Dahl, A.; Brandl, H.; Lakshmanaperumal, N.; Henry, I.; Rink, J.C. Reactivating head regrowth in a regeneration-deficient planarian species. Nature 2013, 500, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Umesono, Y.; Tasaki, J.; Nishimura, Y.; Hrouda, M.; Kawaguchi, E.; Yazawa, S.; Nishimura, O.; Hosoda, K.; Inoue, T.; Agata, K. The molecular logic for planarian regeneration along the anterior-posterior axis. Nature 2013, 500, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Mokalled, M.H.; Poss, K.D. A Regeneration Toolkit. Dev. Cell 2018, 47, 267–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruppert, E.E.; Fox, R.S.; Barnes, R.D. Invertebrate Zoology: A Functional Evolutionary Approach, 7th ed.; Thomson-Brooks/Cole: Pacific Grove, CA, USA, 2004; pp. 662–664. [Google Scholar]
- Dorit, R.L.; Walker, W.F.; Barnes, R.D. Zoology; Saunders College Publishers: New York, NY, USA, 1991; pp. 154–196. [Google Scholar]
- Nickel, M.; Scheer, C.; Hammel, J.U.; Herzen, J.; Beckmann, F. The contractile sponge epithelium sensu lato--body contraction of the demosponge Tethya wilhelma is mediated by the pinacoderm. J. Exp. Biol. 2011, 214, 1692–1698. [Google Scholar] [CrossRef] [Green Version]
- Borisenko, I.; Adamska, M.; Tokina, D.; Ereskovsky, A. Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera). PeerJ 2015, 3, e1211. [Google Scholar] [CrossRef]
- Funayama, N. The stem cell system in demosponges: Insights into the origin of somatic stem cells. Dev. Growth Differ. 2010, 52, 1–14. [Google Scholar] [CrossRef]
- Lorenz, B.; Bohnensack, R.; Gamulin, V.; Steffen, R.; Muller, W.E. Regulation of motility of cells from marine sponges by calcium ions. Cell. Signal. 1996, 8, 517–524. [Google Scholar] [CrossRef]
- Katz, A.M. Review of calcium in muscle contraction, cellular, and molecular physiology, by J. C. Rüegg. J. Mol. Cell. Cardiol. 1994, 26. [Google Scholar] [CrossRef]
- Müller, W. Sponges (Porifera); Springer: New York, NY, USA, 2003; Volume 3, pp. 154–196. [Google Scholar]
- Padua, A.; Klautau, M. Regeneration in calcareous sponges (Porifera). J. Mar. Biol. Assoc. UK 2015, 96, 553–558. [Google Scholar] [CrossRef]
- Kürn, U.; Rendulic, S.; Tiozzo, S.; Lauzon, R. Asexual propagation and regeneration in colonial ascidians. Biol. Bull. 2011, 221, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Basile, G.; Cerrano, C.; Radjasa, O.K.; Povero, P.; Zocchi, E. ADP-ribosyl cyclase and abscisic acid are involved in the seasonal growth and in post-traumatic tissue regeneration of Mediterranean sponges. J. Exp. Mar. Biol. Ecol. 2009, 381, 10–17. [Google Scholar] [CrossRef]
- Pozzolini, M.; Gallus, L.; Ghignone, S.; Ferrando, S.; Candiani, S.; Bozzo, M.; Bertolino, M.; Costa, G.; Bavestrello, G.; Scarfì, S. Insights into the evolution of metazoan regenerative mechanisms: Roles of TGF superfamily members in tissue regeneration of the marine sponge Chondrosia reniformis. J. Exp. Biol. 2019, 222, jeb207894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zapata, F.; Goetz, F.; Smith, S.; Howison, M.; Siebert, S.; Church, S.; Sanders, S.; Ames, C.L.; McFadden, C.; France, S.; et al. Phylogenomic analyses support traditional relationships within cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclere, L.; Rottinger, E. Diversity of cnidarian muscles: Function, Anatomy, development and regeneration. Front. Cell Dev. Biol. 2016, 4, 157. [Google Scholar] [CrossRef]
- Holstein, T.W.; Hobmayer, E.; Technau, U. Cnidarians: An evolutionarily conserved model system for regeneration? Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2003, 226, 257–267. [Google Scholar] [CrossRef]
- Renfer, E.; Amon-Hassenzahl, A.; Steinmetz, P.R.; Technau, U. A muscle-specific transgenic reporter line of the sea anemone, nematostella vectensis. Proc. Natl. Acad. Sci. USA 2010, 107, 104–108. [Google Scholar] [CrossRef] [Green Version]
- Jahnel, S.M.; Walzl, M.; Technau, U. Development and epithelial organisation of muscle cells in the sea anemone Nematostella vectensis. Front. Zool. 2014, 11, 44. [Google Scholar] [CrossRef] [Green Version]
- Ryan, J.F.; Mazza, M.E.; Pang, K.; Matus, D.Q.; Baxevanis, A.D.; Martindale, M.Q.; Finnerty, J.R. Pre-bilaterian origins of the Hox cluster and the Hox code: Evidence from the sea anemone, nematostella vectensis. PLoS ONE 2007, 2, e153. [Google Scholar] [CrossRef] [Green Version]
- Matus, D.Q.; Pang, K.; Daly, M.; Martindale, M.Q. Expression of Pax gene family members in the anthozoan cnidarian, Nematostella vectensis. Evol. Dev. 2007, 9, 25–38. [Google Scholar] [CrossRef]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V.; et al. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saina, M.; Genikhovich, G.; Renfer, E.; Technau, U. BMPs and chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl. Acad. Sci. USA 2009, 106, 18592–18597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magie, C.R.; Pang, K.; Martindale, M.Q. Genomic inventory and expression of Sox and Fox genes in the cnidarian Nematostella vectensis. Dev. Genes Evol. 2005, 215, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, P.R.H.; Aman, A.; Kraus, J.E.M.; Technau, U. Gut-like ectodermal tissue in a sea anemone challenges germ layer homology. Nat. Ecol. Evol. 2017, 1, 1535–1542. [Google Scholar] [CrossRef]
- Genikhovich, G.; Technau, U. Complex functions of Mef2 splice variants in the differentiation of endoderm and of a neuronal cell type in a sea anemone. Development 2011, 138, 4911–4919. [Google Scholar] [CrossRef] [Green Version]
- Martindale, M.Q.; Pang, K.; Finnerty, J.R. Investigating the origins of triploblasty: ‘mesodermal’ gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 2004, 131, 2463–2474. [Google Scholar] [CrossRef] [Green Version]
- Ryan, J.F.; Burton, P.M.; Mazza, M.E.; Kwong, G.K.; Mullikin, J.C.; Finnerty, J.R. The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: Evidence from the starlet sea anemone, Nematostella vectensis. Genome Biol. 2006, 7, R64. [Google Scholar] [CrossRef] [Green Version]
- Layden, M.J.; Rentzsch, F.; Rottinger, E. The rise of the starlet sea anemone nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 408–428. [Google Scholar] [CrossRef] [Green Version]
- Hand, C.; Uhlinger, K.R. The culture, sexual and asexual reproduction, and growth of the sea anemone nematostella vectensis. Biol. Bull. 1992, 182, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.F.; Guerlais, V.; Amiel, A.R.; Johnston, H.; Nedoncelle, K.; Rottinger, E. NvERTx: A gene expression database to compare embryogenesis and regeneration in the sea anemone nematostella vectensis. Development 2018, 145. [Google Scholar] [CrossRef] [Green Version]
- Sebe-Pedros, A.; Saudemont, B.; Chomsky, E.; Plessier, F.; Mailhe, M.P.; Renno, J.; Loe-Mie, Y.; Lifshitz, A.; Mukamel, Z.; Schmutz, S.; et al. Cnidarian Cell type diversity and regulation revealed by whole-organism single-cell RNA-Seq. Cell 2018, 173, 1520–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rottinger, E.; Dahlin, P.; Martindale, M.Q. A framework for the establishment of a cnidarian gene regulatory network for "endomesoderm" specification: The inputs of ss-catenin/TCF signaling. PLoS Genet. 2012, 8, e1003164. [Google Scholar] [CrossRef] [PubMed]
- Layden, M.J.; Rottinger, E.; Wolenski, F.S.; Gilmore, T.D.; Martindale, M.Q. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, nematostella vectensis. Nat. Protoc. 2013, 8, 924–934. [Google Scholar] [CrossRef] [PubMed]
- Ikmi, A.; McKinney, S.A.; Delventhal, K.M.; Gibson, M.C. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 2014, 5, 5486. [Google Scholar] [CrossRef]
- Amiel, A.R.; Foucher, K.; Ferreira, S. Synergic coordination of stem cells is required to induce a regenerative response in anthozoan cnidarians. bioRxiv 2019. [Google Scholar] [CrossRef]
- Lin, Y.C.J.; Spencer, A. Localisation of intracellular calcium stores in the striated muscles of the jellyfish Polyorchis penicillatus: Possible involvement in excitation-contraction coupling. J. Exp. Biol. 2001, 204, 3727–3736. [Google Scholar]
- Hyman, L. The invertebrates. (Scientific books: The invertebrates: Protozoa through ctenophora). Science 1940, 92, 219–220. [Google Scholar]
- Boero, F.; Gravili, C.; Pagliara, P.; Piraino, S.; Bouillon, J.; Schmid, V. The cnidarian premises of metazoan evolution: From triploblasty, to coelom formation, to metamery. Ital. J. Zool. 1998, 65, 5–9. [Google Scholar] [CrossRef]
- Satterlie, R.; Thomas, K.; Gray, C. Muscle organization of the cubozoan jellyfish tripedalia cystophora conant 1897. Biol. Bull. 2005, 209, 154–163. [Google Scholar] [CrossRef]
- Helm, R.; Tiozzo, S.; Lilley, M.; Fabien, L.; Dunn, C. Comparative muscle development of scyphozoan jellyfish with simple and complex life cycles. EvoDevo 2015, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Andrikou, C.; Pai, C.Y.; Su, Y.H.; Arnone, M.I. Logics and properties of a genetic regulatory program that drives embryonic muscle development in an echinoderm. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Hand, C.; Uhlinger, K.R. Asexual reproduction by transverse fission and some anomalies in the sea anemone nematostella vectensis. Invertebr. Biol. 1995, 114, 9–18. [Google Scholar] [CrossRef]
- Reitzel, A.; Burton, P.; Krone, C.; Finnerty, J. Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: Embryogenesis, regeneration, and two forms of asexual fission. Invertebr. Biol. 2007, 126, 99–112. [Google Scholar] [CrossRef]
- Passamaneck, Y.; Martindale, M. Cell proliferation is necessary for the regeneration of oral structures in the anthozoan cnidarian nematostella vectensis. Bmc Dev. Biol. 2012, 12, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiel, A.; Johnston, H.; Nedoncelle, K.; Warner, J.; Ferreira, S.; Röttinger, E. Characterization of morphological and cellular events underlying oral regeneration in the sea anemone, nematostella vectensis. Int. J. Mol. Sci. 2015, 2015, 28449–28471. [Google Scholar] [CrossRef]
- Warner, J.; Amiel, A.; Johnston, H.; Röttinger, E. Regeneration is a partial Redeployment of the embryonic Gene Network. bioRxiv 2019. [Google Scholar] [CrossRef] [Green Version]
- Morgan, T.H. Regeneration and liability to injury. Science 1901, 14, 235–248. [Google Scholar] [CrossRef]
- Bossert, P.E.; Dunn, M.P.; Thomsen, G.H. A staging system for the regeneration of a polyp from the aboral physa of the anthozoan Cnidarian Nematostella vectensis. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2013, 242, 1320–1331. [Google Scholar] [CrossRef]
- Cebria, F. Planarian body-wall muscle: Regeneration and function beyond a simple skeletal support. Front. Cell Dev. Biol. 2016, 4, 8. [Google Scholar] [CrossRef] [Green Version]
- Moumen, M.; Chiche, A.; Cagnet, S.; Petit, V.; Raymond, K.; Faraldo, M.M.; Deugnier, M.A.; Glukhova, M.A. The mammary myoepithelial cell. Int. J. Dev. Biol. 2011, 55, 763–771. [Google Scholar] [CrossRef] [Green Version]
- Newmark, P.; Sánchez Alvarado, A. Not your father’s planarian: A classic model enters the era of functional genomics. Nat. Rev. Genet. 2002, 3, 210–219. [Google Scholar] [CrossRef]
- Giacomotto, J.; Ségalat, L. High-throughput screening and small animal models, where are we? Br. J. Pharmacol. 2010, 160, 204–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, J.D.; Marchant, J.S. Pharmacological and functional genetic assays to manipulate regeneration of the planarian Dugesia japonica. J. Vis. Exp. 2011. [Google Scholar] [CrossRef] [PubMed]
- King, R.S.; Newmark, P.A. Whole-mount in situ hybridization of planarians. Methods Mol. Biol. 2018, 1774, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Ivankovic, M.; Haneckova, R.; Thommen, A.; Grohme, M.A.; Vila-Farré, M.; Werner, S.; Rink, J.C. Model systems for regeneration: Planarians. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scimone, M.L.; Cote, L.E.; Reddien, P.W. Orthogonal muscle fibres have different instructive roles in planarian regeneration. Nature 2017, 551, 623–628. [Google Scholar] [CrossRef]
- Witchley, J.N.; Mayer, M.; Wagner, D.E.; Owen, J.H.; Reddien, P.W. Muscle cells provide instructions for planarian regeneration. Cell Rep. 2013, 4, 633–641. [Google Scholar] [CrossRef] [Green Version]
- Cote, L.E.; Simental, E.; Reddien, P.W. Muscle functions as a connective tissue and source of extracellular matrix in planarians. Nat. Commun. 2019, 10, 1592. [Google Scholar] [CrossRef]
- Baguñà, J. The planarian neoblast: The rambling history of its origin and some current black boxes. Int. J. Dev. Biol. 2012, 56, 19–37. [Google Scholar] [CrossRef] [Green Version]
- Kreshchenko, N.D. Pharynx regeneration in planarians. Ontogenez 2009, 40, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Adler, C.E.; Seidel, C.W.; McKinney, S.A.; Sanchez Alvarado, A. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife 2014, 3, e02238. [Google Scholar] [CrossRef] [PubMed]
- Guedelhoefer, O.C., IV; Alvarado, A.S. Planarian immobilization, partial irradiation, and tissue transplantation. JoVE 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, A.; Li, H.; Guo, L.; Gao, X.; McKinney, S.; Wang, Y.; Yu, Z.; Park, J.; Semerad, C.; Ross, E.; et al. Prospectively isolated tetraspanin. Cell 2018, 173, 1593–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dattani, A.; Sridhar, D.; Aziz Aboobaker, A. Planarian flatworms as a new model system for understanding the epigenetic regulation of stem cell pluripotency and differentiation. Semin. Cell Dev. Biol. 2019, 87, 79–94. [Google Scholar] [CrossRef]
- Felix, D.; Gutiérrez-Gutiérrez, Ó.; Espada, L.; Thems, A.; González-Estévez, C. It is not all about regeneration: Planarians striking power to stand starvation. Semin. Cell Dev. Biol. 2018, 87. [Google Scholar] [CrossRef]
- Pellettieri, J.; Sánchez Alvarado, A. Cell turnover and adult tissue homeostasis: From humans to planarians. Annu. Rev. Genet. 2007, 41, 83–105. [Google Scholar] [CrossRef]
- Karami, A.; Tebyanian, H.; Goodarzi, V.; Shiri, S. Planarians: An in vivo model for regenerative medicine. Int. J. Stem. Cells 2015, 8, 128–133. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, L.; Alejo-Plata, C. Arms regeneration in lolliguncula panamensis (Mollusca: Cephalopoda). Lat. Am. J. Aquat. Res. 2019, 47, 356–360. [Google Scholar] [CrossRef] [Green Version]
- Imperadore, P.; Fiorito, G. Cephalopod tissue regeneration: Consolidating over a century of knowledge. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Tressler, J.; Maddox, F.; Goodwin, E.; Zhang, Z.; Tublitz, N. Arm regeneration in two species of cuttlefish Sepia officinalis and Sepia pharaonis. Invertebr. Neurosci. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Zullo, L.; Imperadore, P. Regeneration and healing. In Handbook of Pathogens and Diseases in Cephalopods; Gestal, C., Pascual, S., Guerra, Á., Fiorito, G., Vieites, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 193–199. [Google Scholar] [CrossRef] [Green Version]
- Fossati, S.; Benfenati, F.; Zullo, L. Morphological characterization of the Octopus vulgaris arm. Front. Cell Dev. Biol. 2011, 61, 191–195. [Google Scholar]
- Zullo, L.; Fossati, S.M.; Benfenati, F. Transmission of sensory responses in the peripheral nervous system of the arm of Octopus vulgaris. Front. Cell Dev. Biol. 2011, 61, 197–201. [Google Scholar]
- Kier, W.; Stella, M. The arrangement and function of octopus arm musculature and connective tissue. J. Morphol. 2007, 268, 831–843. [Google Scholar] [CrossRef]
- Zullo, L.; Eichenstein, H.; Maiole, F.; Hochner, B. Motor control pathways in the nervous system of Octopus vulgaris arm. Front. Cell Dev. Biol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Nödl, M.T.; Fossati, S.M.; Domingues, P.; Sánchez, F.J.; Zullo, L. The making of an octopus arm. Front. Cell Dev. Biol. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Ochiai, Y. Structural and phylogenetic profiles of muscle actins from cephalopods. J. Basic Appl. Sci. 2013. [Google Scholar] [CrossRef]
- Motoyama, K.; Ishizaki, S.; Nagashima, Y.; Shiomi, K. Cephalopod tropomyosins: Identification as major allergens and molecular cloning. Food Chem. Toxicol. 2007, 44, 1997–2002. [Google Scholar] [CrossRef]
- Zullo, L.; Fossati, S.M.; Imperadore, P.; Nödl, M.-T. Molecular determinants of cephalopod muscles and their implication in muscle regeneration. Front. Cell Dev. Biol. 2017, 5, 53. [Google Scholar] [CrossRef] [Green Version]
- Kier, W.M. The musculature of coleoid cephalopod arms and tentacles. Front. Cell Dev. Biol. 2016, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Matzner, H.; Gutfreund, Y.; Hochner, B. Neuromuscular system of the flexible arm of the octopus: Physiological characterization. J. Neurophysiol. 2000, 83, 1315–1328. [Google Scholar] [CrossRef] [PubMed]
- Rokni, D.; Hochner, B. Ionic currents underlying fast action potentials in the obliquely striated muscle cells of the octopus arm. J. Neurophysiol. 2003, 88, 3386–3397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nesher, N.; Maiole, F.; Shomrat, T.; Hochner, B.; Zullo, L. From synaptic input to muscle contraction: Arm muscle cells of Octopus vulgaris show unique neuromuscular junction and excitation-contraction coupling properties. Proc. R. Soc. B 2019, 286. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.; Sampaio, L.; Gobin, A. Building new hearts: A review of trends in cardiac tissue engineering. Am. J. Transplant. 2014, 14. [Google Scholar] [CrossRef] [PubMed]
- Sommese, L.; Zullo, A.; Schiano, C.; Mancini, F.; Napoli, C. Possible muscle repair in the human cardiovascular system. Stem. Cell Rev. Rep. 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Gilly, W.; Renken, C.; Rosenthal, J.; Kier, W. Specialization for rapid excitation in fast squid tentacle muscle involves action potentials absent in slow arm muscle. J. Exp. Biol. 2020, 223, jeb.218081. [Google Scholar] [CrossRef]
- Kang, R.; Guglielmino, E.; Zullo, L.; Branson, D.T.; Godage, I.; Caldwell, D.G. Embodiment design of soft continuum robots. Adv. Mech. Eng. 2016, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zullo, L.; Hochner, B. A new perspective on the organization of an invertebrate brain. Commun. Integr. Biol. 2011, 4, 26–29. [Google Scholar] [CrossRef]
- Zullo, L.; Sumbre, G.; Agnisola, C.; Flash, T.; Hochner, B. Nonsomatotopic organization of the higher motor centers in octopus. Curr. Biol. 2009, 19, 1632–1636. [Google Scholar] [CrossRef] [Green Version]
- Grimaldi, A.; Tettamanti, G.; Rinaldi, L.; Brivio, M.; Castellani, D.; Eguileor, M. Muscle differentiation in tentacles of Sepia officinalis (Mollusca) is regulated by muscle regulatory factors (MRF) related proteins. Dev. Growth Differ. 2004, 46, 83–95. [Google Scholar] [CrossRef] [Green Version]
- Albertin, C.B.; Simakov, O.; Mitros, T.; Wang, Z.Y.; Pungor, J.R.; Edsinger-Gonzales, E.; Brenner, S.; Ragsdale, C.W.; Rokhsar, D.S. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 2015, 524, 220–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimaldi, A.; Tettamanti, G.; Acquati, F.; Bossi, E.; Guidali, M.L.; Banfi, S.; Monti, L.; Valvassori, R.; de Eguileor, M. A hedgehog homolog is involved in muscle formation and organization of Sepia officinalis (mollusca) mantle. Dev. Dyn. 2008, 237, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Lange, M.M. On the regeneration and finer structure of the arms of the cephalopods. J. Exp. Zool. 1920, 31, 1–57. [Google Scholar] [CrossRef]
- Shaw, T.J.; Osborne, M.; Ponte, G.; Fiorito, G.; Andrews, P.L.R. Mechanisms of wound closure following acute arm injury in Octopus vulgaris. Zool. Lett. 2016, 2, 8. [Google Scholar] [CrossRef] [Green Version]
- Fossati, S.M.; Carella, F.; De Vico, G.; Benfenati, F.; Zullo, L. Octopus arm regeneration: Role of acetylcholinesterase during morphological modification. J. Exp. Mar. Biol. Ecol. 2013, 447, 93–99. [Google Scholar] [CrossRef]
- Fossati, S.M.; Candiani, S.; Nödl, M.T.; Maragliano, L.; Pennuto, M.; Domingues, P.; Benfenati, F.; Pestarino, M.; Zullo, L. Identification and expression of acetylcholinesterase in octopus vulgaris arm development and regeneration: A conserved role for ACHE? Mol. Neurobiol. 2015, 52, 45–56. [Google Scholar] [CrossRef]
- Vibert, L.; Daulny, A.; Jarriault, S. Wound healing, cellular regeneration and plasticity: The elegans way. Int. J. Dev. Biol. 2018, 62, 491–505. [Google Scholar] [CrossRef]
- Benian, G.; Epstein, H. Caenorhabditis elegans muscle A Genetic and molecular model for protein interactions in the heart. Circ. Res. 2011, 109, 1082–1095. [Google Scholar] [CrossRef] [Green Version]
- Moerman, D.; Fire, A. Muscle: Structure, Function, and Development, 2nd ed.; Springer: New York, NY, USA, 1997; Volume 33, pp. 154–196. [Google Scholar]
- Kiontke, K.; Sudhaus, W. Ecology of caenorhabditis species. WormBook 2006. [Google Scholar] [CrossRef]
- Bird, A.; Bird, J. The Structure of Nematodes, 2nd ed.; Academic Press: Boston, MA, USA, 1991; pp. 10–317. [Google Scholar]
- Corsi, A.; Wightman, B.; Chalfie, M. A transparent window into biology: A primer on caenorhabditis elegans. WormBook 2015, 200, 1–31. [Google Scholar] [CrossRef]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode caenorhabditis elegans. Philos. Trans. R. Soc. 1986, 275, 327–348. [Google Scholar]
- Gieseler, K.; Qadota, H.; Benian, G. Development, structure, and maintenance of C. elegans body wall muscle. WormBook 2016, 2017, 1–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stetina, S.E. Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: Boston, MA, USA, 2013; pp. 469–476. [Google Scholar]
- Brouilly, N.; Lecroisey, C.; Martin, E.; Pierson, L.; Mariol, M.C.; Qadota, H.; Labouesse, M.; Streichenberger, N.; Mounier, N.; Gieseler, K. Ultra-structural time-course study in the C. elegans model for Duchenne muscular dystrophy highlights a crucial role for sarcomere-anchoring structures and sarcolemma integrity in the earliest steps of the muscle degeneration process. Hum. Mol. Genet. 2015, 24, 6428–6445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamberlain, J.S.; Benian, G.M. Muscular dystrophy: The worm turns to genetic disease. Curr. Biol. CB 2000, 10, R795–R797. [Google Scholar] [CrossRef] [Green Version]
- Marden, J.H. Variability in the size, composition, and function of insect flight muscles. Annu. Rev. Physiol. 2000, 62, 157–178. [Google Scholar] [CrossRef] [PubMed]
- Bhakthan, N.; Nair, K.; Borden, J. Fine structure of degenerating and regenerating flight muscles in a bark beetle, Ips confusus. II. Regeneration. Can. J. Zool. 1971, 49, 85–89. [Google Scholar] [CrossRef]
- Bernstein, S.; O’Donnell, P.; Cripps, R. Molecular genetic analysis of muscle development, structure, and function in drosophila. Int. Rev. Cytol. 1993, 143, 63–152. [Google Scholar] [CrossRef]
- Josephson, R.; Malamud, J.; Stokes, D. Asynchronous muscle: A primer. J. Exp. Biol. 2000, 203, 2713–2722. [Google Scholar]
- Osborne, M.P. Supercontraction in the muscles of the blowfly larva: An ultrastructural study. J. Insect Physiol. 1967, 13, 1471–1482. [Google Scholar] [CrossRef]
- Goldstein, M.; Burdette, W. Striated visceral muscle of Drosophila melanogaster. J. Morphol. 1971, 134, 315–334. [Google Scholar] [CrossRef]
- Goldstein, M.A. An ultrastructural study of supercontraction in the body wall muscles of Drosophila melanogaster larvae. Anat. Rec. 1971, 169, 326. [Google Scholar]
- Hardie, J. The tension/length relationship of an insect (Calliphora erythrocephala) supercontracting muscle. Cell. Mol. Life Sci. CMLS 1976, 32, 714–716. [Google Scholar] [CrossRef]
- Herrel, A.; Meyers, J.; Aerts, P.; Nishikawa, K. Functional implications of supercontracting muscle in the chameleon tongue retractors. J. Exp. Biol. 2001, 204, 3621–3627. [Google Scholar] [PubMed]
- Beramendi, A.; Peron, S.; Megighian, A.; Reggiani, C.; Cantera, R. The IκB ortholog cactus is necessary for normal neuromuscular function in drosophila melanogaster. Neuroscience 2005, 134, 397–406. [Google Scholar] [CrossRef]
- Staehling, K.; Hoffmann, F.; Baylies, M.; Rushton, E.; Bate, M. Dpp induces mesodermal gene expression in drosophila. Nature 1994, 372, 783–786. [Google Scholar] [CrossRef]
- Azpiazu, N.; Lawrence, P.A.; Vincent, J.-P.; Frasch, M. Segmentation and specification of the drosophila mesoderm. Genes Dev. 1997, 10, 3183–3194. [Google Scholar] [CrossRef] [Green Version]
- Baylies, M.; Bate, M. twist: A Myogenic Switch in Drosophila. Science 1996, 272, 1481–1484. [Google Scholar] [CrossRef]
- Chaturvedi, D.; Reichert, H.; Gunage, R.; Vijayraghavan, K. Identification and functional characterization of muscle satellite cells in Drosophila. eLife 2017, 6. [Google Scholar] [CrossRef]
- Postigo, A.A.; Ward, E.; Skeath, J.B.; Dean, D.C. zfh-1, the Drosophila homologue of ZEB, is a transcriptional repressor that regulates somatic myogenesis. Mol. Cell. Biol. 1999, 19, 7255–7263. [Google Scholar] [CrossRef] [Green Version]
- Carnevali, M.D.C. Regeneration in echinoderms: Repair, regrowth, cloning. Invertebr. Surviv. J. 2006, 3, 64–76. [Google Scholar]
- Dupont, S.; Thorndyke, M. Bridging the regeneration gap: Insights from echinoderm models. Nat. Rev. Genet. 2007, 8, 320. [Google Scholar] [CrossRef]
- Ziegler, A.; Schröder, L.; Ogurreck, M.; Faber, C.; Stach, T. Evolution of a Novel Muscle Design in Sea Urchins (Echinodermata: Echinoidea). PLoS ONE 2012, 7, e37520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz-Pineda, P.; Ramirez-Gomez, F.; Pérez-Ortiz, J.; González-Díaz, S.; Jesús, F.; Hernández-Pasos, J.; Avila, C.; Rojas-Cartagena, C.; Suárez-Castillo, E.; Tossas, K.; et al. Gene expression profiling of intestinal regeneration in the sea cucumber. BMC Genom. 2009, 10, 262. [Google Scholar] [CrossRef] [Green Version]
- García-Arrarás, J.E.; Dolmatov, I.Y. Echinoderms: Potential model systems for studies on muscle regeneration. Curr. Pharm. Des. 2010, 16, 942–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quiñones, J.L.; Rosa, R.; Ruiz, D.L.; García-Arrarás, J.E. Extracellular matrix remodeling and metalloproteinase involvement during intestine regeneration in the sea cucumber holothuria glaberrima. Dev. Biol. 2002, 250, 181–197. [Google Scholar] [CrossRef] [Green Version]
- Dolmatov, I.Y.; Eliseikina, M.G.; Ginanova, T.T.; Lamash, N.E.; Korchagin, V.P.; Bulgakov, A.A. Muscle regeneration in the holothurian Stichopus japonicus. Roux’s Arch. Dev. Biol. 1996, 205, 486–493. [Google Scholar] [CrossRef]
- Holland, L.; Albalat, R.; Azumi, K.; Benito Gutierrez, E.; Blow, M.; Bronner-Fraser, M.; Brunet, F.; Butts, T.; Candiani, S.; Dishaw, L.; et al. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 2008, 18, 1100–1111. [Google Scholar] [CrossRef] [Green Version]
- Somorjai, I.; Escrivà, H.; GarciaFernandez, J. Amphioxus makes the cut—Again. Commun. Integr. Biol. 2012, 5, 499–502. [Google Scholar] [CrossRef] [Green Version]
- Somorjai, I.; Somorjai, R.; GarciaFernandez, J.; Escrivà, H. Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proc. Natl. Acad. Sci. USA 2011, 109, 517–522. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Rathnayake, D.; Huang, S.; Pathirana, A.; Xu, Q.; Zhang, S. BMP signaling is required for amphioxus tail regeneration. Development 2019, 146, dev.166017. [Google Scholar] [CrossRef] [Green Version]
- Kaneto, S.; Wada, H. Regeneration of amphioxus oral cirri and its skeletal rods: Implications for the origin of the vertebrate skeleton. J. Exp. Zool. Part. B Mol. Dev. Evol. 2011, 316, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Flood, P.; Guthrie, D.; Banks, J. Paramyosin muscle in the notochord of amphioxus. Nature 1969, 222, 87–88. [Google Scholar] [CrossRef] [PubMed]
- Peachey, L. Structure of the longitudinal body muscles of Amphioxus. J. Biophys. Biochem. Cytol. 1961, 10, 159–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hagiwara, S.; Henkart, M.; Kidokoro, Y. Excitation-contraction coupling in amphioxus muscle cells. J. Physiol. 1972, 219, 233–251. [Google Scholar] [CrossRef] [PubMed]
- Welsch, U. The fine structure of the pharynx, cyrtopodocytes and digestive caecum of amphioxus (Branchiostoma lanceolatum). Symp. Zool. Soc. Lond. 1975, 36, 17–41. [Google Scholar]
- Aldea, D.; Subirana, L.; Keime, C.; Meister, L.; Maeso, I.; Marcellini, S.; Gómez-Skarmeta, J.; Bertrand, S.; Escrivà, H. Genetic regulation of amphioxus somitogenesis informs the evolution of the vertebrate head mesoderm. Nat. Ecol. Evol. 2019, 3. [Google Scholar] [CrossRef]
- Meulemans, D.; Bronner-Fraser, M.; Holland, L.; Holland, N. Differential mesodermal expression of two amphioxus MyoD family members (AmphiMRF1 and AmphiMRF2). Gene Expr. Patterns GEP 2003, 3, 199–202. [Google Scholar] [CrossRef]
- Bertrand, S.; Camasses, A.; Somorjai, I.; Belgacem, M.; Chabrol, O.; Escande, M.-L.; Pontarotti, P.; Escrivà, H. Amphioxus FGF signaling predicts the acquisition of vertebrate morphological traits. Proc. Natl. Acad. Sci. USA 2011, 108, 9160–9165. [Google Scholar] [CrossRef] [Green Version]
- Onai, T.; Aramaki, T.; Inomata, H.; Hirai, T.; Kuratani, S. On the origin of vertebrate somites. Zool. Lett. 2015, 1. [Google Scholar] [CrossRef] [Green Version]
- Shenkar, N.; Swalla, B. Global Diversity of Ascidiacea. PLoS ONE 2011, 6, e20657. [Google Scholar] [CrossRef]
- Delsuc, F.; Brinkmann, H.; Chourrout, D.; Philippe, H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 2006, 439, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Tiozzo, S.; Brown, F.; de Tomaso, A. Regeneration and stem cells in ascidians. In Stem Cells; Springer: New York, NY, USA, 2008; pp. 95–112. [Google Scholar]
- Alié, A.; Hiebert, L.; Scelzo, M.; Tiozzo, S. The eventful history of non-embryonic development in tunicates. J. Exp. Zool. Part. B Mol. Dev. Evol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Nakauchi, M. Asexual development of ascidians: Its biological significance, diversity, and morphogenesis. Am. Zool. 1982, 22. [Google Scholar] [CrossRef] [Green Version]
- Razy-Krajka, F.; Stolfi, A. Regulation and evolution of muscle development in tunicates. EvoDevo 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Jeffery, W. Progenitor targeting by adult stem cells in ciona homeostasis, injury, and regeneration. Dev. Biol. 2018, 448. [Google Scholar] [CrossRef]
- Degasperi, V.; Gasparini, F.; Shimeld, S.; Sinigaglia, C.; Burighel, P.; Manni, L. Muscle differentiation in a colonial ascidian: Organisation, gene expression and evolutionary considerations. BMC Dev. Biol. 2009, 9, 48. [Google Scholar] [CrossRef] [Green Version]
- Meedel, T.; Chang, P.; Yasuo, H. Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev. Biol. 2007, 302, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Anderson, H.E.; Christiaen, L. Ciona as a simple chordate model for heart development and regeneration. J. Cardiovasc. Dev. Dis. 2016, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Jeffery, W. Closing the wounds: One hundred and twenty five years of regenerative biology in the ascidian ciona intestinalis. Genesis 2015, 53. [Google Scholar] [CrossRef] [Green Version]
- Christiaen, L.; Tolkin, T. Rewiring of an ancestral Tbx1/10-Ebf-Mrf network for pharyngeal muscle specification in distinct embryonic lineages. BioRxiv 2016. [Google Scholar] [CrossRef] [Green Version]
- Razy-Krajka, F.; Lam, K.; Wang, W.; Stolfi, A.; Joly, M.; Bonneau, R.; Christiaen, L. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev. Cell 2014, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, B. Ciona intestinalis as a model for cardiac development. Semin. Cell Dev. Biol. 2007, 18, 16–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricci, L.; Cabrera, F.; Lotito, S.; Tiozzo, S. Re-deployment of germ layers related TFs shows regionalized expression during two non-embryonic developments. Dev. Biol. 2016, 416. [Google Scholar] [CrossRef] [PubMed]
- Prünster, M.M.; Ricci, L.; Brown, F.; Tiozzo, S. De novo neurogenesis in a budding chordate: Co-option of larval anteroposterior patterning genes in a transitory neurogenic organ. Dev. Biol. 2018, 448. [Google Scholar] [CrossRef]
- Prünster, M.M.; Ricci, L.; Brown, F.; Tiozzo, S. Modular co-option of cardiopharyngeal genes during non-embryonic myogenesis. EvoDevo 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Voskoboynik, A.; Simon-Blecher, N.; Soen, Y.; de Tomaso, A.; Ishizuka, K.; Weissman, I. Striving for normality: Whole body regeneration through a series of abnormal generations. FASEB J. 2007, 21, 1335–1344. [Google Scholar] [CrossRef]
- Freeman, G. The role of blood cells in the process of asexual reproduction in the tunicate Perophora. J. Exp. Zool. 1964, 156, 157–183. [Google Scholar] [CrossRef]
- Kassmer, S.; Langenbacher, A.; de Tomaso, A. Primordial Blasts, a Population of Blood Borne Stem Cells Responsible for Whole Body Regeneration in a basal Chordate; Springer: New York, NY, USA, 2019. [Google Scholar] [CrossRef] [Green Version]
- Marques, I.J.; Lupi, E.; Mercader, N. Model systems for regeneration: Zebrafish. Development 2019, 146. [Google Scholar] [CrossRef] [Green Version]
- Gemberling, M.; Bailey, T.; Hyde, D.; Poss, K. The zebrafish as a model for complex tissue regeneration. TIG 2013, 29. [Google Scholar] [CrossRef] [Green Version]
- Berberoglu, M.; Gallagher, T.; Morrow, Z.; Talbot, J.; Hromowyk, K.; Tenente, I.; Langenau, D.; Amacher, S. Satellite-like cells contribute to pax7-dependent skeletal muscle repair in adult zebrafish. Dev. Biol. 2017, 424. [Google Scholar] [CrossRef]
- Keenan, S.R.; Currie, P.D. The developmental phases of zebrafish myogenesis. J. Dev. Biol. 2019, 7, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, S.L.; Marin-Juez, R.; Moura, P.L.; Kuenne, C.; Lai, J.K.H.; Tsedeke, A.T.; Guenther, S.; Looso, M.; Stainier, D.Y. Reciprocal analyses in zebrafish and medaka reveal that harnessing the immune response promotes cardiac regeneration. eLife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Charvet, B.; Malbouyres, M.; Pagnon-Minot, A.; Ruggiero, F.; le Guellec, D. Development of the zebrafish myoseptum with emphasis on the myotendinous junction. Cell Tissue Res. 2011, 346, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Hammond, C.L.; Hinits, Y.; Osborn, D.P.; Minchin, J.E.; Tettamanti, G.; Hughes, S.M. Signals and myogenic regulatory factors restrict pax3 and pax7 expression to dermomyotome-like tissue in zebrafish. Dev. Biol. 2007, 302, 504–521. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, D.B.; Nguyen, P.D.; Siegel, A.L.; Ehrlich, O.V.; Sonntag, C.; Phan, J.M.; Berger, S.; Ratnayake, D.; Hersey, L.; Berger, J.; et al. Asymmetric division of clonal muscle stem cells coordinates muscle regeneration in vivo. Science 2016, 353, aad9969. [Google Scholar] [CrossRef]
- Rossi, G.; Messina, G. Comparative myogenesis in teleosts and mammals. Cell. Mol. Life Sci. 2014, 71, 3081–3099. [Google Scholar] [CrossRef] [Green Version]
- González-Rosa, J.M.; Burns, C.; Burns, C. Zebrafish heart regeneration: 15 years of discoveries. Regeneration 2017, 4. [Google Scholar] [CrossRef]
- Poss, K.D.; Wilson, L.G.; Keating, M.T. Heart regeneration in zebrafish. Science 2002, 298, 2188–2190. [Google Scholar] [CrossRef]
- Beffagna, G. Zebrafish as a smart model to understand regeneration after heart injury: How fish could help humans. Front. Cardiovasc. Med. 2019, 6. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, K.; Holdway, J.; Werdich, A.; Anderson, R.; Fang, Y.; Egnaczyk, G.; Evans, T.; Macrae, C.; Stainier, D.; Poss, K. Primary contribution to zebrafish heart regeneration by Gata4 cardiomyocytes. Nature 2010, 464, 601–605. [Google Scholar] [CrossRef]
- Jopling, C.; Sleep, E.; Raya, M.; Marti, M.; Raya, A.; Izpisua Belmonte, J.C. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef] [PubMed]
- Sande-Melón, M.; Marques, I.; Galardi-Castilla, M.; Langa Oliva, X.; Pérez-López, M.; Botos, M.-A.; Sánchez-Iranzo, H.; Guzmán-Martínez, G.; Francisco, D.; Pavlinic, D.; et al. Adult sox10+ Cardiomyocytes Contribute to Myocardial Regeneration in the Zebrafish. Cell Rep. 2019, 29, 1041–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.-C.; Yang, C.-C.; Chen, I.H.; Liu, L.Y.-m.; Chang, S.J.; Chuang, Y.-J. Treatment of glucocorticoids inhibited early immune responses and impaired cardiac repair in adult zebrafish. PLoS ONE 2013, 8, e66613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Preux Charles, A.-S.; Bise, T.; Baier, F.; Marro, J.; Jazwinska, A. Distinct effects of inflammation on preconditioning and regeneration of the adult zebrafish heart. Open Biol. 2016, 6, 160102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, M.; Bussmann, J.; Huang, Y.; Zhao, L.; Osorio, A.; Burns, C.; Burns, C.; Sucov, H.; Siekmann, A.; Lien, C.-L. Chemokine-guided angiogenesis directs coronary vasculature formation in zebrafish. Dev. Cell 2015, 33, 442–454. [Google Scholar] [CrossRef] [Green Version]
- González-Rosa, J.M.; Martín, V.; Peralta, M.; Torres, M.; Mercader, N. Extensive scar formation and regression during heart regeneration after cryoinjury in zebrafish. Development 2011, 138, 1663–1674. [Google Scholar] [CrossRef] [Green Version]
- Schnabel, K.; Wu, C.C.; Kurth, T.; Weidinger, G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS ONE 2011, 6, e18503. [Google Scholar] [CrossRef]
- Marín-Juez, R.; Marass, M.; Gauvrit, S.; Rossi, A.; Lai, S.-L.; Materna, S.; Black, B.; Stainier, D. Fast revascularization of the injured area is essential to support zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2016, 113. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Iranzo, H.; Galardi-Castilla, M.; Sanz-Morejón, A.; González-Rosa, J.M.; Costa, R.; Ernst, A.; Sainz de Aja, J.; Langa Oliva, X.; Mercader, N. Transient fibrosis resolves via fibroblast inactivation in the regenerating zebrafish heart. Proc. Natl. Acad. Sci. USA 2018, 115, 201716713. [Google Scholar] [CrossRef] [Green Version]
- Cao, J.; Poss, K. The epicardium as a hub for heart regeneration. Nat. Rev. Cardiol. 2018, 15. [Google Scholar] [CrossRef]
- Lavine, K.; Yu, K.; White, A.; Zhang, X.; Smith, C.; Partanen, J.; Ornitz, D. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev. Cell 2005, 8, 85–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Cao, J.; Dickson, A.; Poss, K. Epicardial regeneration is guided by cardiac outflow tract and Hedgehog signalling. Nature 2015, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepilina, A.; Coon, A.N.; Kikuchi, K.; Holdway, J.E.; Roberts, R.W.; Burns, C.G.; Poss, K.D. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell 2006, 127, 607–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Rosa, J.M.; Peralta, M.; Mercader, N. Pan-epicardial lineage tracing reveals that epicardium derived cells give rise to myofibroblasts and perivascular cells during zebrafish heart regeneration. Dev. Biol. 2012, 370, 173–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Harrison, M.; Osorio, A.; Kim, J.; Baugh, A.; Duan, C.; Sucov, H.; Lien, C.-L. Igf Signaling is required for cardiomyocyte proliferation during zebrafish heart development and regeneration. PLoS ONE 2013, 8, e67266. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Wu, Q.; Zhang, Y.; Wiens, K.; Huang, Y.; Rubin, N.; Shimada, H.; Handin, R.; Chao, M.; Tuan, T.-L.; et al. PDGF signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc. Natl. Acad. Sci. USA 2010, 107, 17206–17210. [Google Scholar] [CrossRef] [Green Version]
- Chablais, F.; Jazwinska, A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development 2012, 139, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Chablais, F.; Jazwinska, A. Induction of myocardial infarction in adult zebrafish using cryoinjury. J. Vis. Exp. JoVE 2012, 62. [Google Scholar] [CrossRef] [Green Version]
- Dogra, D.; Ahuja, S.; Kim, H.-T.; Rasouli, S.J.; Stainier, D.; Reischauer, S. Opposite effects of Activin type 2 receptor ligands on cardiomyocyte proliferation during development and repair. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Wu, C.C.; Kruse, F.; Dalvoy, M.; Junker, J.; Zebrowski, D.C.; Fischer, K.; Noel, E.; Grün, D.; Berezikov, E.; Engel, F.; et al. Spatially resolved genome-wide transcriptional profiling identifies BMP signaling as essential regulator of zebrafish cardiomyocyte regeneration. Dev. Cell 2015, 36. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Borikova, A.; Ben-Yair, R.; Guner-Ataman, B.; Macrae, C.; Lee, R.; Burns, C.; Burns, C. Notch signaling regulates cardiomyocyte proliferation during zebrafish heart regeneration. Proc. Natl. Acad. Sci. USA 2014, 111, 1403–1408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, V.; Thomson, J.; Thummel, R.; Hyde, D.; Hammond, S.; Poss, K. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008, 22, 728–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, C.-L.; Hou, Y.; Xu, C.; Chang, N.; Wang, F.; Hu, K.; He, A.; Luo, Y.; Wang, J.; Peng, J.; et al. Chromatin-remodelling factor Brg1 regulates myocardial proliferation and regeneration in zebrafish. Nat. Commun. 2016, 7, 13787. [Google Scholar] [CrossRef] [PubMed]
- Keßler, M.; Rottbauer, W.; Just, S. Recent progress in the use of zebrafish for novel cardiac drug discovery. Expert Opin. Drug Discov. 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Moro, E.; Vettori, A.; Porazzi, P.; Schiavone, M.; Rampazzo, E.; Casari, A.; Ek, O.; Facchinello, N.; Astone, M.; Zancan, I.; et al. Generation and application of signaling pathway reporter lines in zebrafish. Mol. Genet. Genom. 2013, 288, 231–242. [Google Scholar] [CrossRef] [Green Version]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Beltrami, A.P.; Urbanek, K.; Kajstura, J.; Yan, S.-M.; Finato, N.; Bussani, R.; Nadal-Ginard, B.; Silvestri, F.; Leri, A.; Beltrami, C.A.; et al. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 2001, 344, 1750–1757. [Google Scholar] [CrossRef]
- Pasumarthi, K.B.; Field, L.J. Cardiomyocyte cell cycle regulation. Circ. Res. 2002, 90, 1044–1054. [Google Scholar] [CrossRef]
- Bassett, D.I.; Currie, P.D. The zebrafish as a model for muscular dystrophy and congenital myopathy. Hum. Mol. Genet. 2003, 12, R265–R270. [Google Scholar] [CrossRef] [Green Version]
- Otten, C.; Abdelilah-Seyfried, S. Laser-inflicted injury of zebrafish embryonic skeletal muscle. JoVE 2013. [Google Scholar] [CrossRef] [Green Version]
- Saera-Vila, A.; Kish, P.E.; Kahana, A. Fgf regulates dedifferentiation during skeletal muscle regeneration in adult zebrafish. Cell. Signal. 2016, 28, 1196–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Naranjo, J.; Dziki, J.; Badylak, S. Regenerative medicine approaches for age-related muscle loss and sarcopenia: A mini-review. Gerontology 2017, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Saul, D.; Böker, K.; Ernst, J.; Lehmann, W.; Schilling, A. Current methods for skeletal muscle tissue repair and regeneration. Biomed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Zullo, A.; Mancini, F.P.; Schleip, R.; Wearing, S.; Yahia, L.H.; Klingler, W. The interplay between fascia, skeletal muscle, nerves, adipose tissue, inflammation and mechanical stress in musculo-fascial regeneration. J. Gerontol. Geriatr. 2017, 65, 271–283. [Google Scholar]
- Forcina, L.; Cosentino, M.; Musaro, A. Mechanisms regulating muscle regeneration: Insights into the interrelated and time-dependent phases of tissue healing. Cells 2020, 9, 1297. [Google Scholar] [CrossRef]
- Sicherer, S.; Grasman, J. Recent trends in injury models to study skeletal muscle regeneration and repair. Bioengineering 2020, 7, 76. [Google Scholar] [CrossRef]
- Khodabukus, A.; Prabhu, N.; Wang, J.; Bursac, N. In vitro tissue-engineered skeletal muscle models for studying muscle physiology and disease. Adv. Healthc. Mater. 2018, 7, 1701498. [Google Scholar] [CrossRef]
- Mueller, A.; Bloch, R. Skeletal muscle cell transplantation: Models and methods. J. Muscle Res. Cell Motil. 2019. [Google Scholar] [CrossRef]
- Marg, A.; Escobar, H.; Karaiskos, N.; Grunwald, S.; Metzler, E.; Kieshauer, J.; Sauer, S.; Pasemann, D.; Malfatti, E.; Mompoint, D.; et al. Human muscle-derived CLEC14A-positive cells regenerate muscle independent of PAX7. Nat. Commun. 2019, 10, 5776. [Google Scholar] [CrossRef] [Green Version]
- Lingjun, R.; Qian, Y.; Khodabukus, A.; Ribar, T.; Bursac, N. Engineering human pluripotent stem cells into a functional skeletal muscle tissue. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Chan, S.S.-K.; Arpke, R.; Filareto, A.; Xie, N.; Pappas, M.; Penaloza, J.; Perlingeiro, R.; Kyba, M. Skeletal muscle stem cells from PSC-derived teratomas have functional regenerative capacity. Cell Stem Cell 2018, 23, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Costela, M.C.O.; López, M.G.; López, V.C.; Gallardo, M.E. iPSCs: A powerful tool for skeletal muscle tissue engineering. J. Cell. Mol. Med. 2019, 23. [Google Scholar] [CrossRef]
- Hall, M.; Hall, J.; Cadwallader, A.; Pawlikowski, B.; Doles, J.; Elston, T.; Olwin, B. Transplantation of skeletal muscle stem cells. Methods Mol. Biol. 2017, 1556, 237–244. [Google Scholar] [PubMed]
- Quattrocelli, M.; Swinnen, M.; Giacomazzi, G.; Camps, J.; Barthélémy, I.; Ceccarelli, G.; Caluwé, E.; Grosemans, H.; Thorrez, L.; Pelizzo, G.; et al. Mesodermal iPSC–derived progenitor cells functionally regenerate cardiac and skeletal muscle. J. Clin. Investig. 2015, 125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maffioletti, S.; Sarcar, S.; Henderson, A.; Mannhardt, I.; Pinton, L.; Moyle, L.; Steele, H.; Cappellari, O.; Wells, K.; Ferrari, G.; et al. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danišovič, Ľ.; Galambosova, M.; Csobonyeiová, M. Induced pluripotent stem cells for duchenne muscular dystrophy modeling and therapy. Cells 2018, 7, 253. [Google Scholar] [CrossRef] [Green Version]
- Choi, I.; Lim, H.; Estrellas, K.; Mula, J.; Cohen, T.; Zhang, Y.; Donnelly, C.; Richard, J.-P.; Kim, Y.J.; Kim, H.; et al. Concordant but Varied phenotypes among duchenne muscular dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell Rep. 2016, 15, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Webster, M.; Fan, C. c-MET regulates myoblast motility and myocyte fusion during adult skeletal muscle regeneration. PLoS ONE 2013, 8, e81757. [Google Scholar] [CrossRef] [Green Version]
- Wal, E.; Herrero-Hernandez, P.; Wan, R.; Broeders, M.; Groen, S.; Gestel, T.; Van Ijcken, W.; Cheung, T.; Ploeg, A.; Schaaf, G.; et al. Large-scale expansion of human iPSC-derived skeletal muscle cells for disease modeling and cell-based therapeutic strategies. Stem Cell Rep. 2018, 10. [Google Scholar] [CrossRef]
- Ong, C.S.; Yesantharao, P.; Huang, C.-Y.; Mattson, G.; Boktor, J.; Fukunishi, T.; Zhang, H.; Hibino, N. 3D bioprinting using stem cells. Pediatr. Res. 2017, 83. [Google Scholar] [CrossRef] [Green Version]
- Mandrycky, C.; Wang, D.Z.; Kim, K.; Kim, D.-H. 3D Bioprinting for engineering complex tissues. Biotechnol. Adv. 2015, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollot, B.; Rathbone, C.; Wenke, J.; Guda, T. Natural polymeric hydrogel evaluation for skeletal muscle tissue engineering: Skeletal muscle engineering natural hydrogels. J. Biomed. Mater. Res. Part. B Appl. Biomater. 2017, 106. [Google Scholar] [CrossRef] [PubMed]
- Fuoco, C.; Petrilli, L.L.; Cannata, S.; Gargioli, C. Matrix scaffolding for stem cell guidance toward skeletal muscle tissue engineering. J. Orthop. Surg. Res. 2016, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, A.; Moerk, C.; Penland, N.; Perla, M.; Kim, J.; Smith, A.; Murry, C.; Kim, D.-H. Regulation of skeletal myotube formation and alignment by nanotopographically controlled cell-secreted extracellular matrix. J. Biomed. Mater. Res. Part. A 2018, 106. [Google Scholar] [CrossRef]
- Matthews, B.; Vosshall, L. How to turn an organism into a model organism in 10 ‘easy’ steps. J. Exp. Biol. 2020, 223, jeb218198. [Google Scholar] [CrossRef] [Green Version]
- Dickinson, M.; Vosshall, L.; Dow, J. Genome editing in non-model organisms opens new horizons for comparative physiology. J. Exp. Biol. 2020, 223, jeb221119. [Google Scholar] [CrossRef] [Green Version]
- Chargé, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Hardy, D.; Besnard, A.; Latil, M.; Jouvion, G.; Briand, D.; Thépenier, C.; Pascal, Q.; Guguin, A.; Gayraud-Morel, B.; Cavaillon, J.-M.; et al. Comparative study of injury models for studying muscle regeneration in mice. PLoS ONE 2016, 11, e0147198. [Google Scholar] [CrossRef]
- Hardy, D.; Latil, M.; Gayraud-Morel, B.; Briand, D.; Jouvion, G.; Rocheteau, P.; Chrétien, F. Choosing the appropriate model for studying muscle regeneration in mice: A comparative study of classical protocols. Morphologie 2015, 99, 168. [Google Scholar] [CrossRef]
- Yun, M. Changes in Regenerative Capacity through Lifespan. Int. J. Mol. Sci. 2015, 16, 25392–25432. [Google Scholar] [CrossRef] [Green Version]
- Alvarado, A.S. Regeneration in the metazoans: Why does it happen? BioEssays 2000, 22, 578–590. [Google Scholar] [CrossRef]
- Imperadore, P.; Shah, S.B.; Makarenkova, H.P.; Fiorito, G. Nerve degeneration and regeneration in the cephalopod mollusc Octopus vulgaris: The case of the pallial nerve. Sci. Rep. 2017, 7, 46564. [Google Scholar] [CrossRef] [PubMed]


| Animal Species | Muscle Type * | REG | Cell Precursor | Known Signaling Pathways and Molecular Players |
|---|---|---|---|---|
| IPSC ** | Any | n/a | n/a | MyoD, Pax7/3, Wnt and BMPs |
| Vertebrates | Striated; Cardiac | yes | Satellite stem cell; Pre-existing muscle cells | Notch, BMP, TGF-β, IGF, FGF family, Ngr1; Pax3/7, Met |
| Tunicates | Striated, smooth-like, cardiac | yes | Stem cells-like precursors (?) | Notch, Nk4, Tbx1/10, MRF |
| Cephalocordates | Striated, mononucleated | yes | De-differentiation, Multipotent cells | Pax3/7, Wnt/β-catenin and BMP |
| Echinoderms | Smooth-like, striated, mononucleated | yes | De-differentiation | BMP/TGFB, HOX, Ependymin, etc. |
| Arthropods | Striated | no *** | Adult muscle precursor (AMP) | Notch-Delta, Transcription factor Zhf1 |
| Nematodes | Striated, mononucleated | no | --- | --- |
| Mollusks | Striated, mononucleated | yes | Sarcoblasts (?) | AChE, Growth factors (EGF, FGFs and VEGF)?? |
| Platyhelmintes | Combine features of both vertebrate skeletal and smooth muscle cells | yes | Neoblasts: Adult pluripotent stem cells | Many known signaling pathways such as PCGs, Wnt/β-catenin, FGF family, insulin/IGF-1, Pax3/7, TGF-β, Hox genes, etc. (see text for references) |
| Cnidarians | Epitheliomuscular, smooth | yes | Hydrozoan: i-cells in Hydractinia, epithelial stem cells in Hydra Anthozoan: yet to be determined | The myogenic gene repertoire is present in cnidarians, but no experimental evidence relate them to the myogenic trajectory |
| Porifera | Myocytes | yes | Adult stem cells (ASC) Totypotent, pluripotent cells | ADPRC (ADP-ribosyl cyclase), TGF-β |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zullo, L.; Bozzo, M.; Daya, A.; Di Clemente, A.; Mancini, F.P.; Megighian, A.; Nesher, N.; Röttinger, E.; Shomrat, T.; Tiozzo, S.; et al. The Diversity of Muscles and Their Regenerative Potential across Animals. Cells 2020, 9, 1925. https://doi.org/10.3390/cells9091925
Zullo L, Bozzo M, Daya A, Di Clemente A, Mancini FP, Megighian A, Nesher N, Röttinger E, Shomrat T, Tiozzo S, et al. The Diversity of Muscles and Their Regenerative Potential across Animals. Cells. 2020; 9(9):1925. https://doi.org/10.3390/cells9091925
Chicago/Turabian StyleZullo, Letizia, Matteo Bozzo, Alon Daya, Alessio Di Clemente, Francesco Paolo Mancini, Aram Megighian, Nir Nesher, Eric Röttinger, Tal Shomrat, Stefano Tiozzo, and et al. 2020. "The Diversity of Muscles and Their Regenerative Potential across Animals" Cells 9, no. 9: 1925. https://doi.org/10.3390/cells9091925
APA StyleZullo, L., Bozzo, M., Daya, A., Di Clemente, A., Mancini, F. P., Megighian, A., Nesher, N., Röttinger, E., Shomrat, T., Tiozzo, S., Zullo, A., & Candiani, S. (2020). The Diversity of Muscles and Their Regenerative Potential across Animals. Cells, 9(9), 1925. https://doi.org/10.3390/cells9091925








