The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Cells and Transient Transfection
2.3. Antibodies and Chemicals
2.4. Plasmids
2.5. SDS-PAGE and Western Blot Analysis
2.6. Indirect Immunofluorescence Analysis
2.7. Isolation of RcDNA and PreS1 Direct Sequencing
2.8. Rate-Zonal Sedimentation
2.9. Electron Microscopy
2.10. HBV Infection of Differentiated HepaRG Cells
2.11. Statistical Analysis
3. Results
3.1. Prevalence of PreS1 Start Codon Deletion Variants in European Patients Infected Chronically by GtA
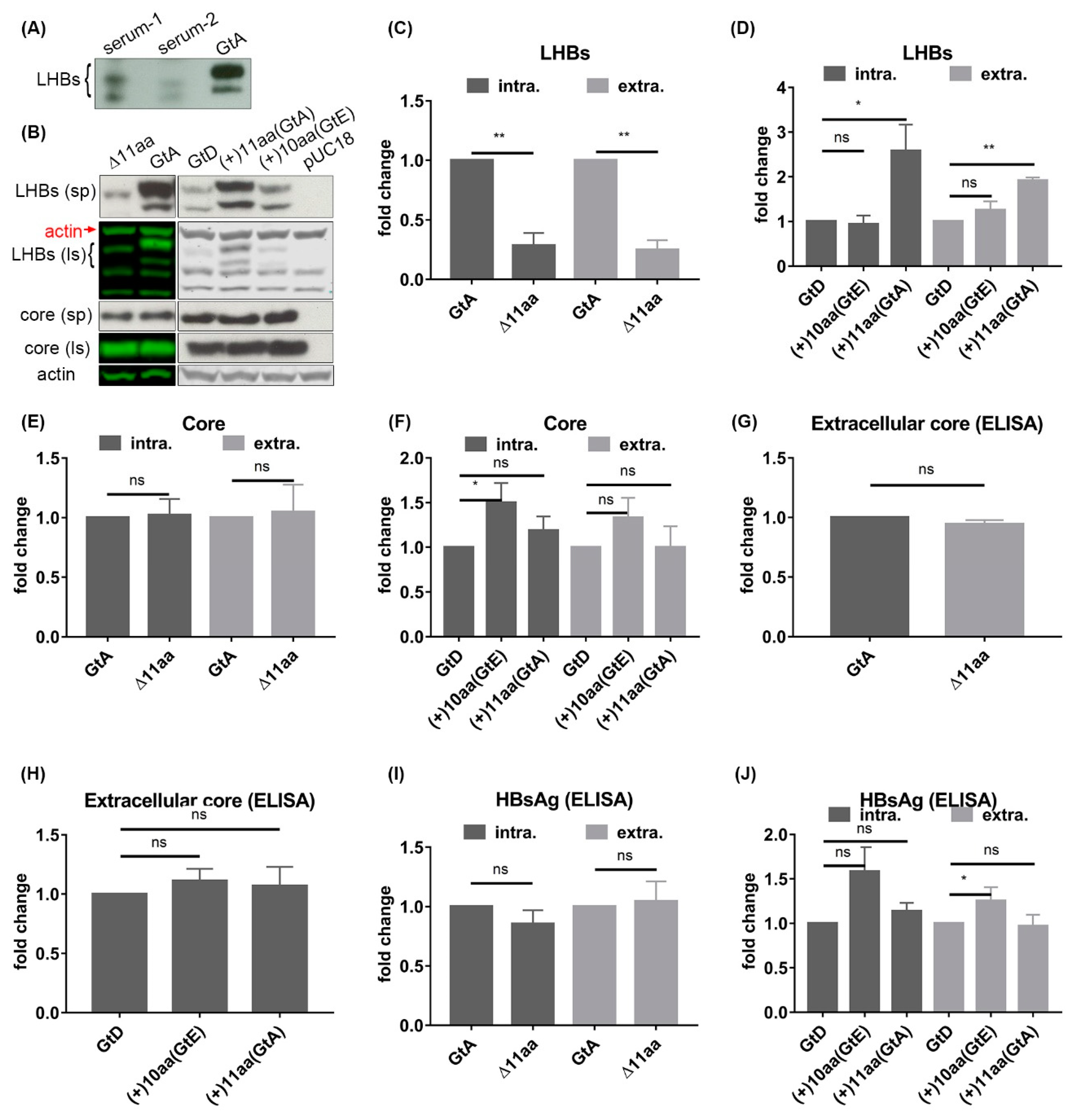
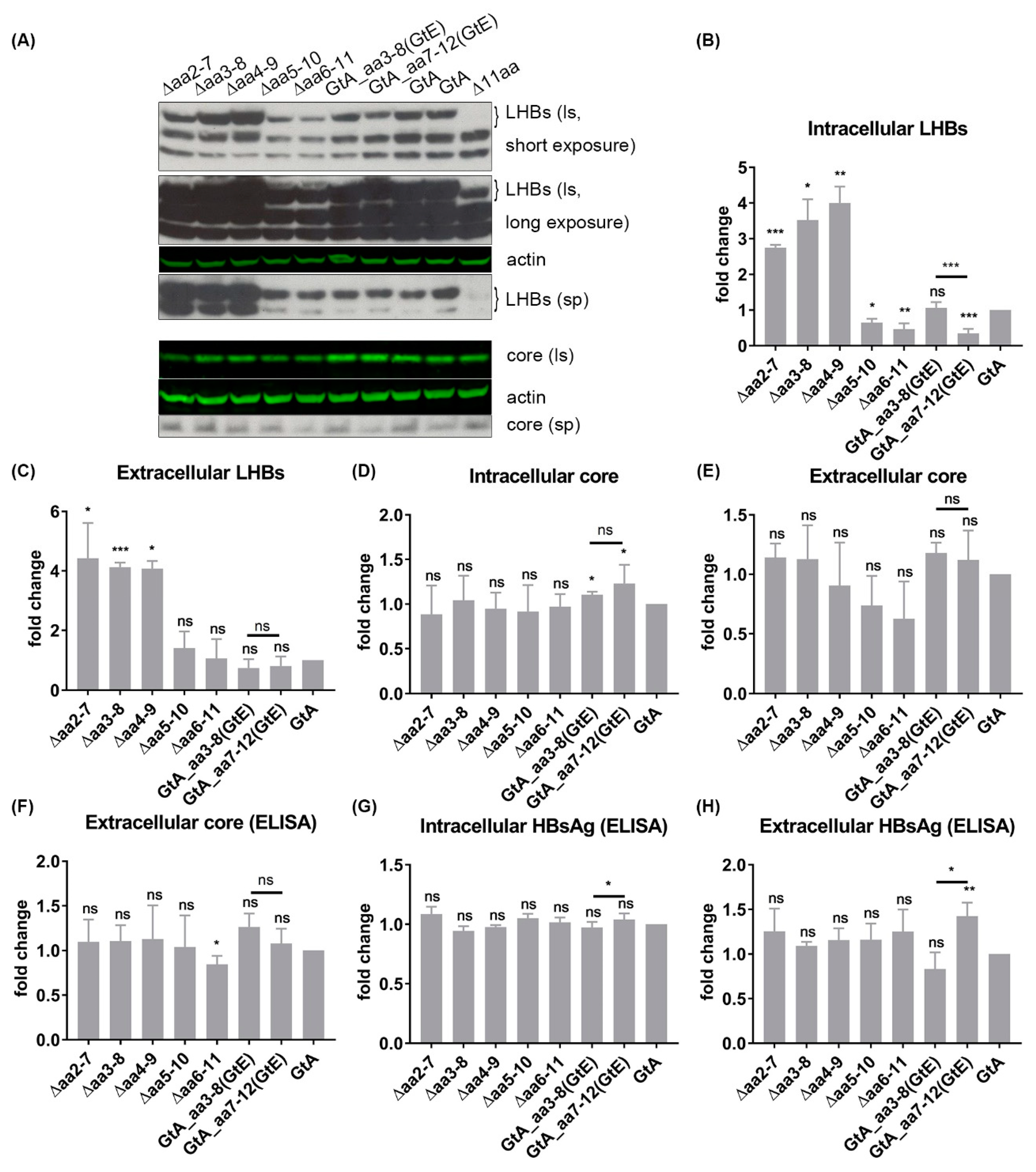
3.2. The N-Terminal 11aa in the PreS1 Domain of GtA Have an Impact on the Amount of Extra- and Intracellular LHBs
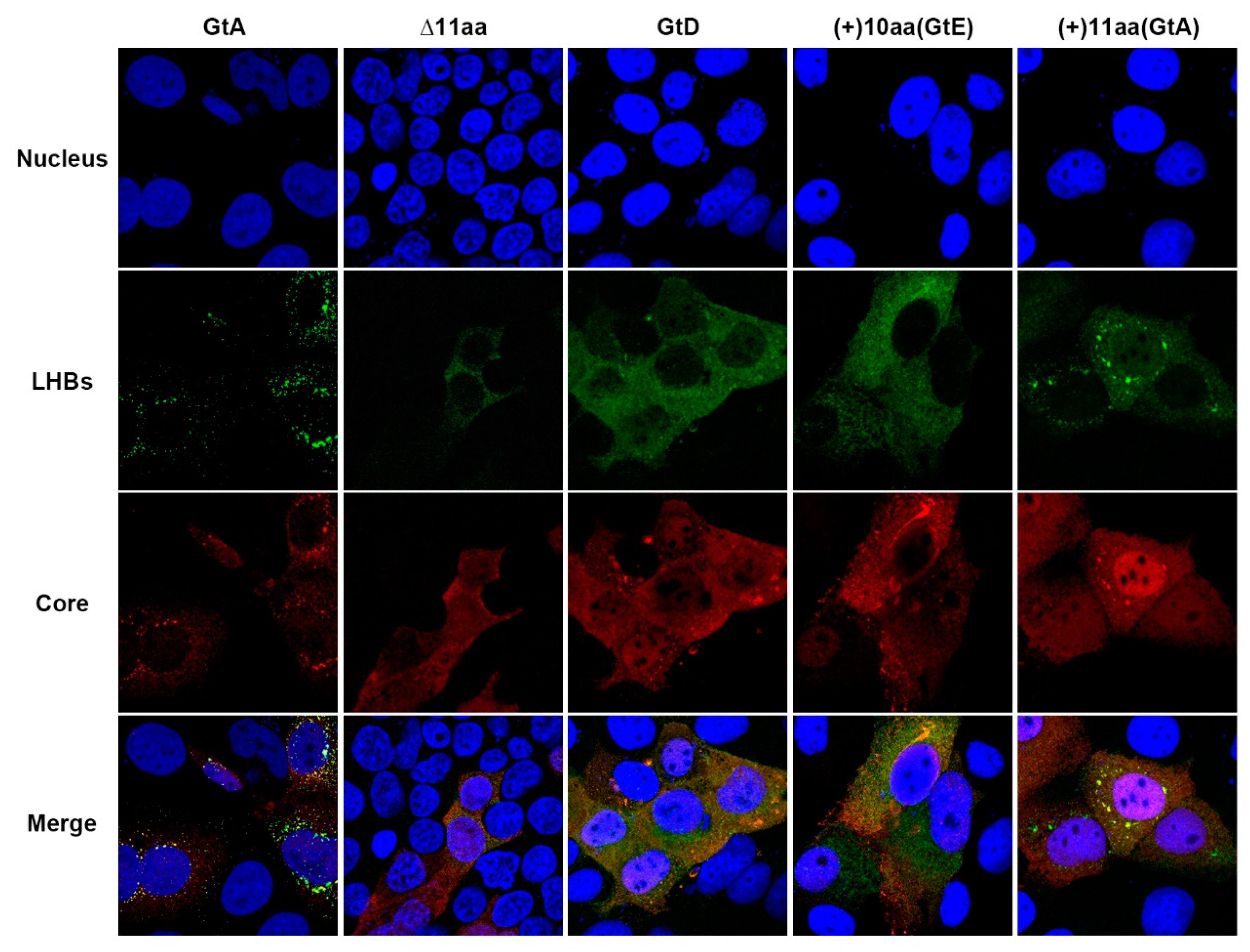
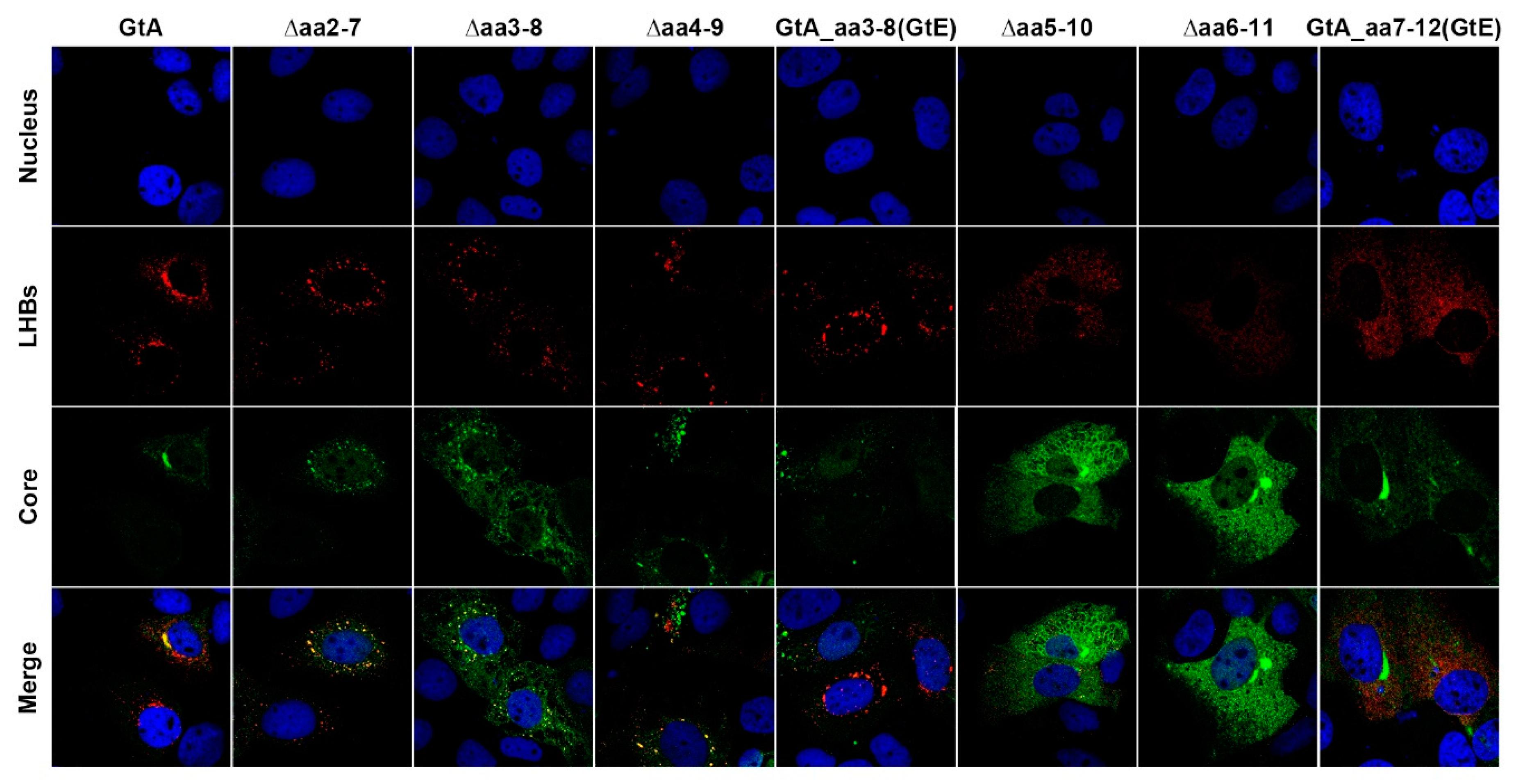
3.3. The Aminoterminal 11 aa in the PreS1-Domain of GtA Affect the Subcellular Distribution of LHBs
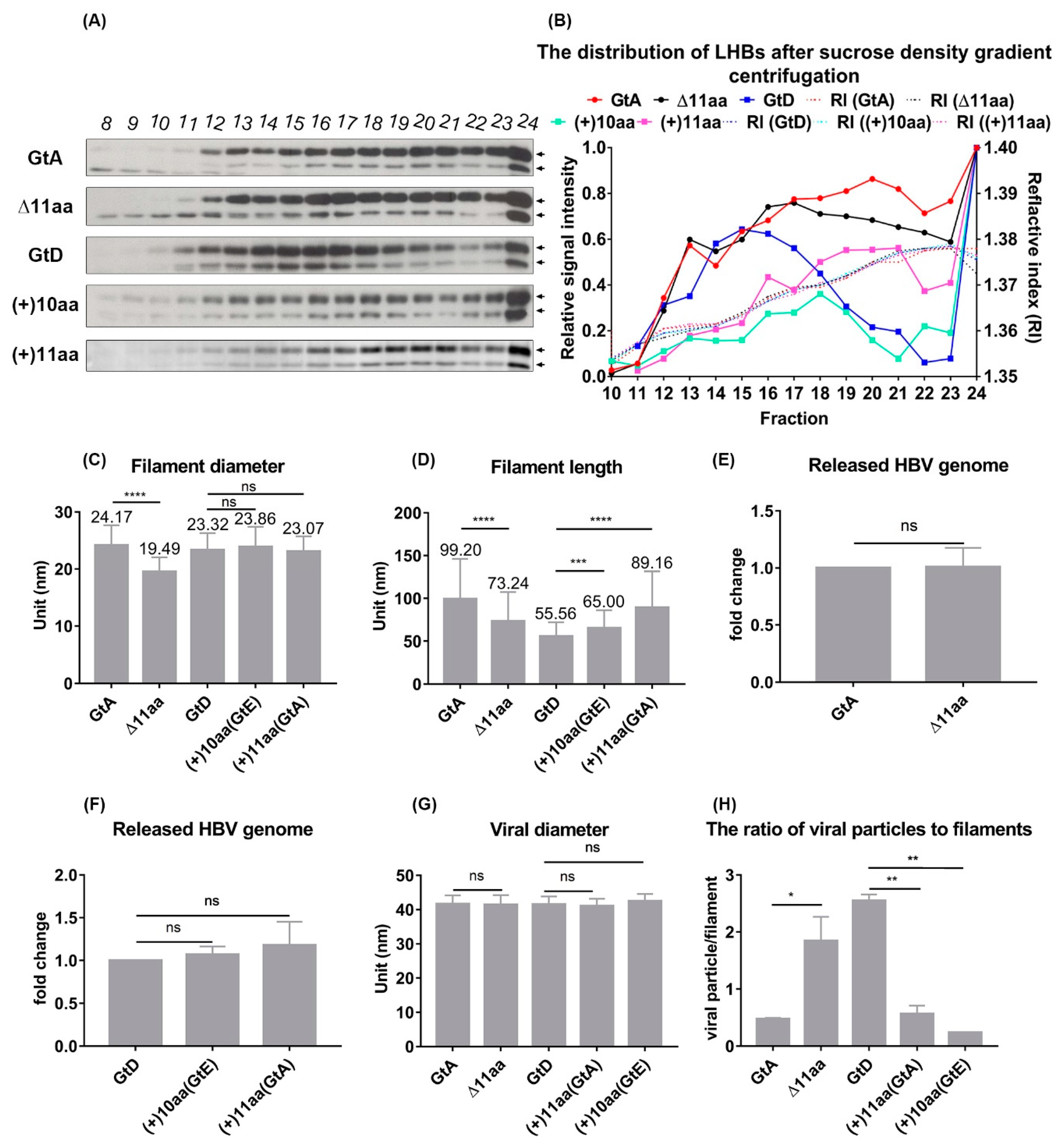
3.4. The N-Terminal Part of the PreS1-Domain of GtA Has an Impact on the Morphology of Filaments
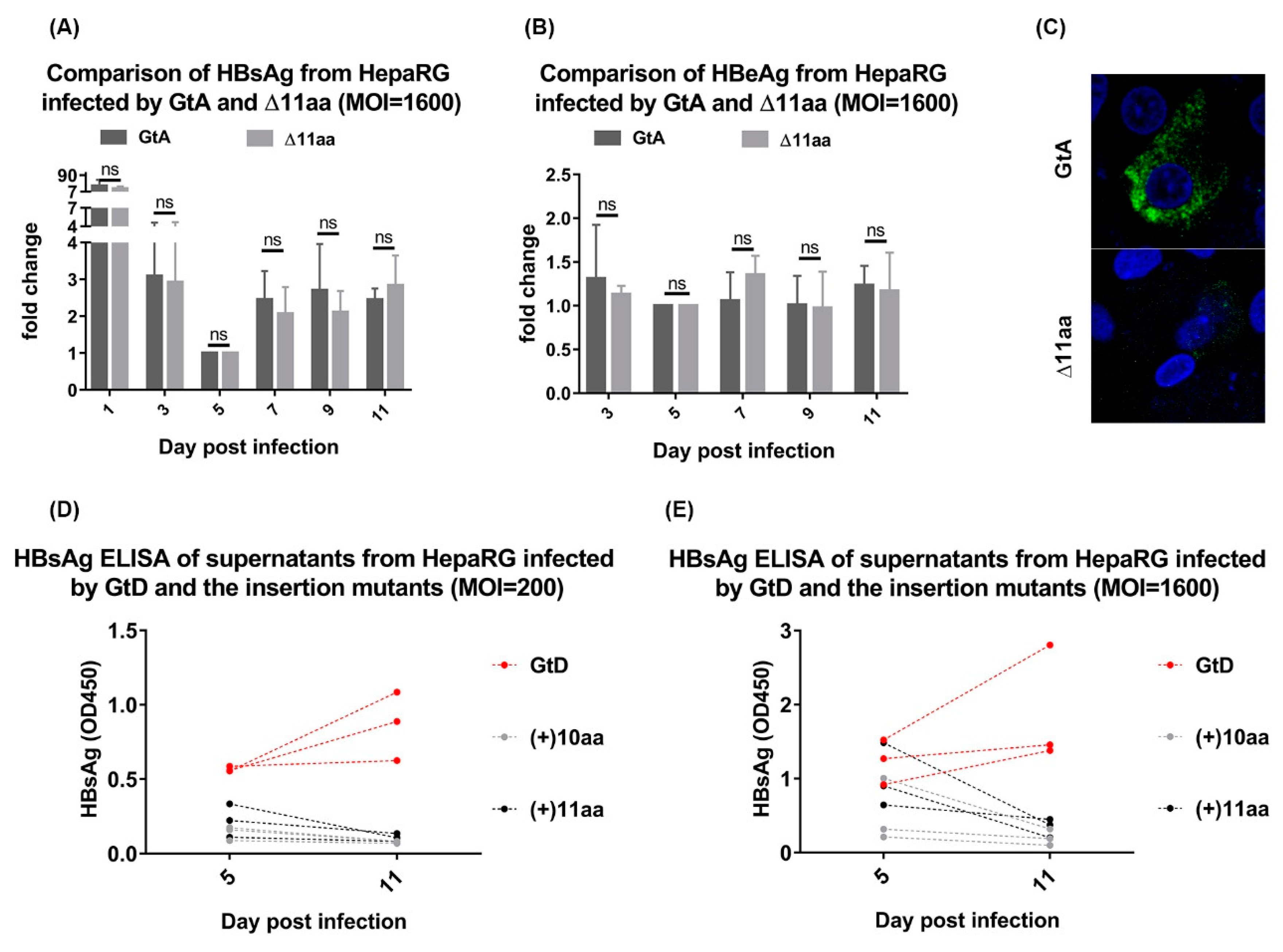
3.5. Deletion of 11 aa in the N-Terminal Part of GtA PreS1-Domain does not Affect the Viral Infectivity
3.6. The 11 aa in the N-Terminal Part of GtA PreS1-Domain Harbor a Positive and a Negative Element Regulating LHB Amount and Subcellular Distribution
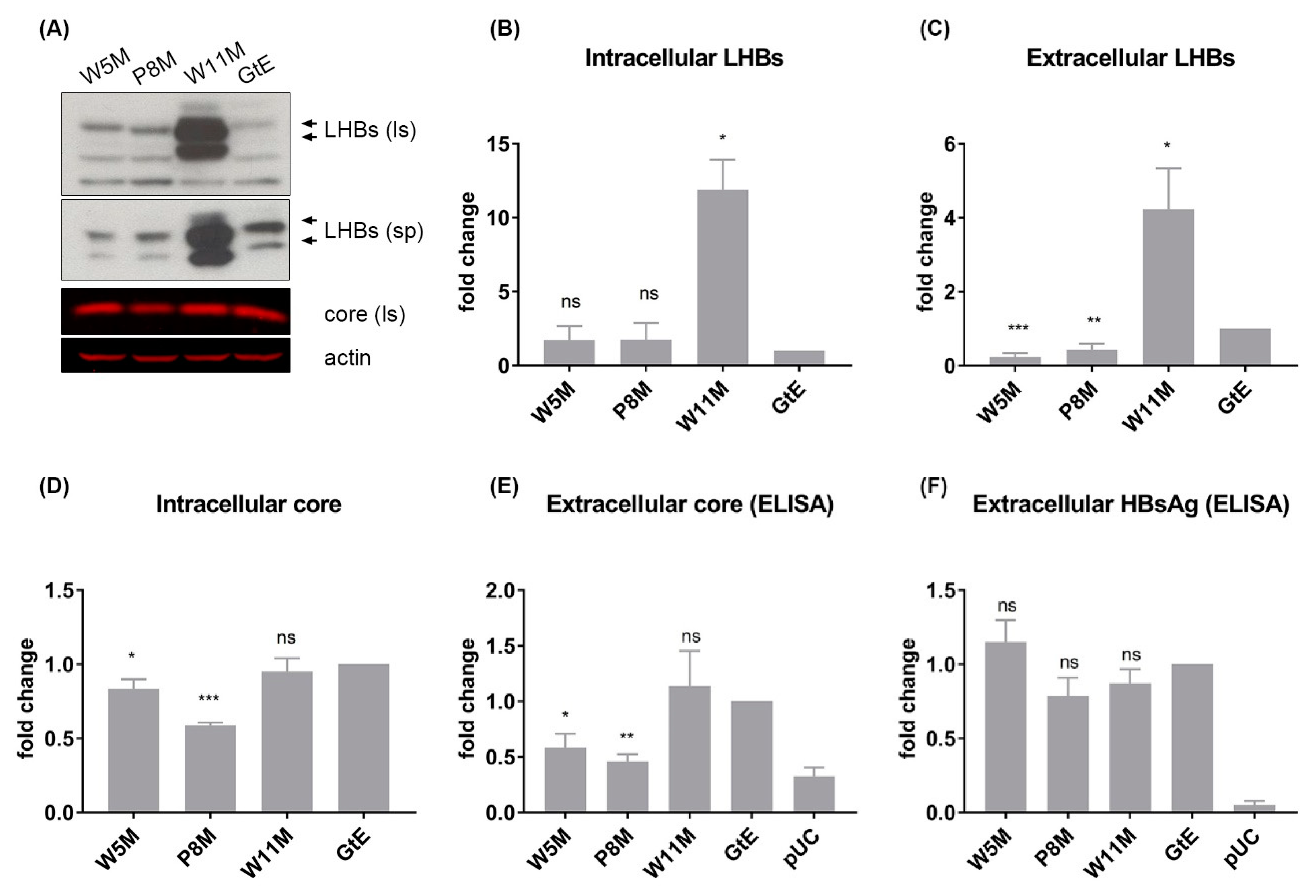
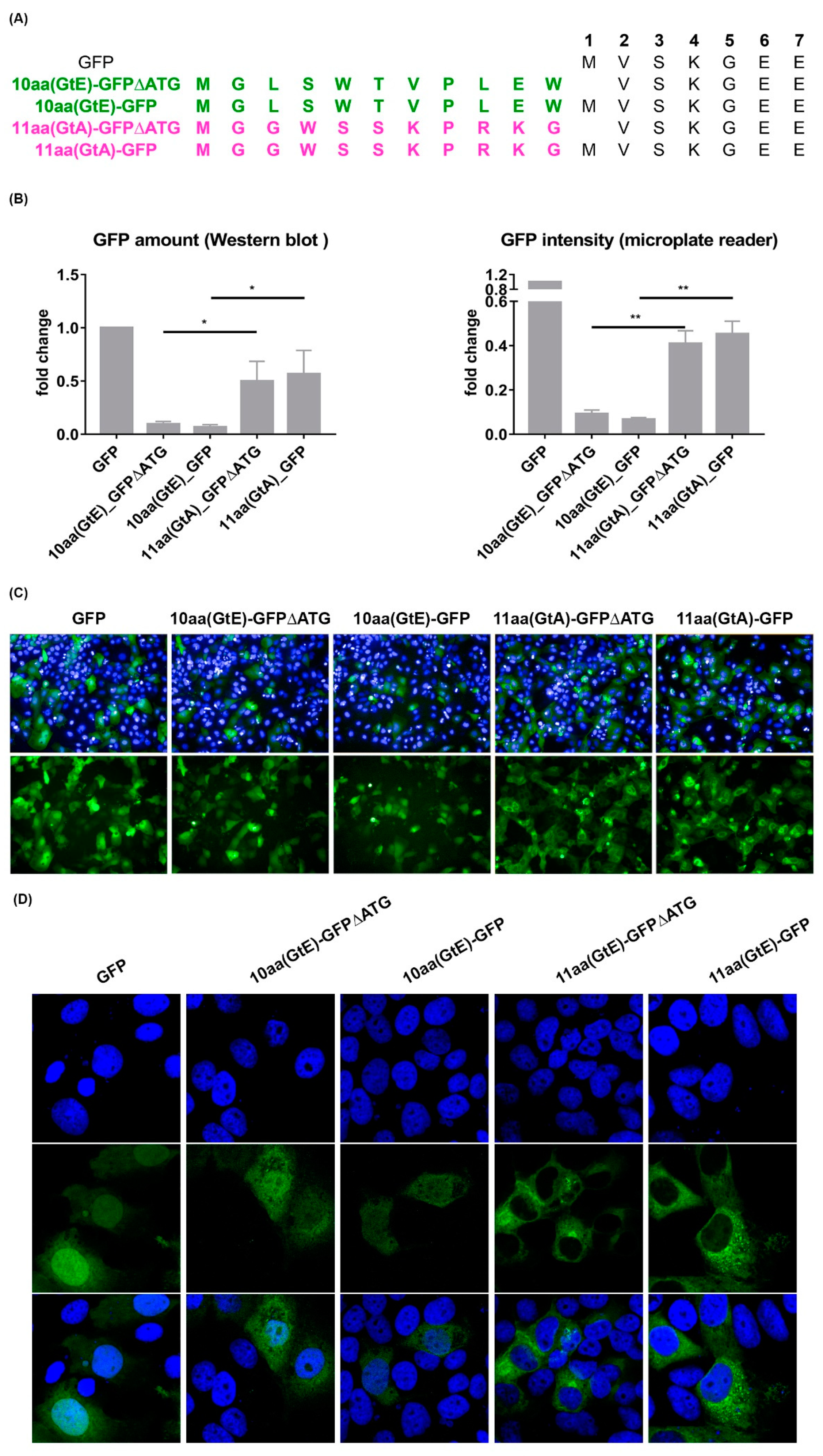
3.7. Deletion of 10 aa in the N-Terminal Part of GtE PreS1 Domain Increases the Production and Release of LHBs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zahn, T.; Akhras, S.; Spengler, C.; Murra, R.O.; Holzhauser, T.; Hildt, E. A new approach for therapeutic vaccination against chronic HBV infections. Vaccine 2020, 38, 3105–3120. [Google Scholar] [CrossRef] [PubMed]
- Heermann, K.-H.; Lu, X.; Xuanyong, L.; Bac, C. Functions of hepatitis B surface proteins. Ruminant Pestivirus Infect. 1992, 4, 129–132. [Google Scholar] [CrossRef]
- Heermann, K.H.; Goldmann, U.; Schwartz, W.; Seyffarth, T.; Baumgarten, H.; Gerlich, W.H. Large surface proteins of hepatitis B virus containing the pre-s sequence. J. Virol. 1984, 52, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V. Hepatitis B virus morphogenesis. World J. Gastroenterol. 2007, 13, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Le Seyec, J.; Chouteau, P.; Cannie, I.; Guguen-Guillouzo, C.; Gripon, P. Infection Process of the Hepatitis B Virus Depends on the Presence of a Defined Sequence in the Pre-S1 Domain. J. Virol. 1999, 73, 2052–2057. [Google Scholar] [CrossRef] [PubMed]
- Bruss, V.; Vieluf, K. Functions of the internal pre-S domain of the large surface protein in hepatitis B virus particle morphogenesis. J. Virol. 1995, 69, 6652–6657. [Google Scholar] [CrossRef] [PubMed]
- Chisari, F.V.; Klopchin, K.; Moriyama, T.; Pasquinelli, C.; Dunsford, H.A.; Sell, S.; Pinkert, C.A.; Brinster, R.L.; Palmiter, R.D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 1989, 59, 1145–1156. [Google Scholar] [CrossRef]
- Hildt, E.; Saher, G.; Bruss, V.; Hofschneider, P.H. The Hepatitis B Virus Large Surface Protein (LHBs) Is a Transcriptional Activator. Virol. 1996, 225, 235–239. [Google Scholar] [CrossRef]
- Ning, X.; Luckenbaugh, L.; Liu, K.; Bruss, V.; Sureau, C.; Hu, J. Common and Distinct Capsid and Surface Protein Requirements for Secretion of Complete and Genome-Free Hepatitis B Virions. J. Virol. 2018, 92, e00272-18. [Google Scholar] [CrossRef]
- Kramvis, A. Genotypes and Genetic Variability of Hepatitis B Virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.; Revill, P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 2016, 64, S4–S16. [Google Scholar] [CrossRef] [PubMed]
- Valsamakis, A. Molecular Testing in the Diagnosis and Management of Chronic Hepatitis B. Clin. Microbiol. Rev. 2007, 20, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Qin, Y.; Liu, Y.; Liao, H.; Lu, S.; Qiao, Y.; Xu, D. PreS deletion profiles of hepatitis B virus (HBV) are associated with clinical presentations of chronic HBV infection. J. Clin. Virol. 2016, 82, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Choe, W.H.; Kim, H.; Lee, S.; Choi, Y.; Kwon, S.Y.; Moon, H.W.; Hur, M. Three types of preS1 start codon deletion variants in the natural course of chronic hepatitis B infection. J. Gastroenterol. Hepatol. 2018, 33, 1370–1378. [Google Scholar] [CrossRef]
- Lee, S.-A.; Kim, K.-J.; Kim, H.; Choi, W.-H.; Won, Y.-S.; Kim, B.-J. Hepatitis B virus preS1 deletion is related to viral replication increase and disease progression. World J. Gastroenterol. 2015, 21, 5039–5048. [Google Scholar] [CrossRef]
- Kim, B.K.; Choi, S.H.; Ahn, S.H.; Chung, A.R.; Park, Y.K.; Han, K.-H.; Kim, S.; Kim, H.-S.; Park, J.H.; Kim, K.S.; et al. Pre-S mutations of hepatitis B virus affect genome replication and expression of surface antigens. J. Gastroenterol. Hepatol. 2014, 29, 843–850. [Google Scholar] [CrossRef]
- Kuhnhenn, L.; Jiang, B.; Kubesch, A.; Vermehren, J.; Knop, V.; Susser, S.; Dietz, J.; Carra, G.; Finkelmeier, F.; Grammatikos, G.; et al. Impact of HBV genotype and mutations on HBV DNA and qHBsAg levels in patients with HBeAg-negative chronic HBV infection. Aliment. Pharmacol. Ther. 2018, 47, 1523–1535. [Google Scholar] [CrossRef]
- Ehrhardt, C.; Schmolke, M.; Matzke, A.; Knoblauch, A.; Will, C.; Wixler, V.; Ludwig, S. Polyethylenimine, a cost-effective transfection reagent. Signal. Transduct. 2006, 6, 179–184. [Google Scholar] [CrossRef]
- Kucinskaite-Kodze, I.; Pleckaityte, M.; Bremer, C.M.; Seiz, P.L.; Zilnyte, M.; Bulavaite, A.; Mickiene, G.; Zvirblis, G.; Sasnauskas, K.; Glebe, D.; et al. New broadly reactive neutralizing antibodies against hepatitis B virus surface antigen. Virus Res. 2016, 211, 209–221. [Google Scholar] [CrossRef]
- Rost, M.; Mann, S.; Lambert, C.; Döring, T.; Thomé, N.; Prange, R. γ2-Adaptin, a Novel Ubiquitin-interacting Adaptor, and Nedd4 Ubiquitin Ligase Control Hepatitis B Virus Maturation. J. Boil. Chem. 2006, 281, 29297–29308. [Google Scholar] [CrossRef]
- Burckstummer, T.; Kriegs, M.; Lupberger, J.; Pauli, E.; Schmittel, S.; Hildt, E. Raf-1 kinase associates with Hepatitis C virus NS5A and regulates viral replication. FEBS Lett. 2005, 580, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, B.; Stockl, L.; Gutzeit, C.; Roos, M.; Lupberger, J.; Schwartlander, R.; Gelderblom, H.; Sauer, I.M.; Hofschneider, P.H.; Hildt, E. A novel system for efficient gene transfer into primary human hepatocytes via cell-permeable hepatitis B virus-like particle. Hepatology 2005, 42, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Himmelsbach, K.; Ren, H.; Boller, K.; Hildt, E. Subviral Hepatitis B Virus Filaments, like Infectious Viral Particles, Are Released via Multivesicular Bodies. J. Virol. 2015, 90, 3330–3341. [Google Scholar] [CrossRef] [PubMed]
- Chai, N.; Chang, H.E.; Nicolas, E.; Han, Z.; Jarnik, M.; Taylor, J.M. Properties of Subviral Particles of Hepatitis B Virus. J. Virol. 2008, 82, 7812–7817. [Google Scholar] [CrossRef]
- Gripon, P.; Cannie, I.; Urban, S. Efficient Inhibition of Hepatitis B Virus Infection by Acylated Peptides Derived from the Large Viral Surface Protein. J. Virol. 2005, 79, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Wu, Q.; Kuhnhenn, L.; Akhras, S.; Spengler, C.; Boller, K.; Peiffer, K.-H.; Hildt, E. Formation of semi-enveloped particles as a unique feature of a hepatitis B virus PreS1 deletion mutant. Aliment. Pharmacol. Ther. 2019, 50, 940–954. [Google Scholar] [CrossRef]
- Mun, H.-S.; Lee, S.-A.; Jee, Y.; Kim, H.; Park, J.-H.; Song, B.-C.; Yoon, J.-H.; Kim, Y.J.; Lee, H.-S.; Hyun, J.-W.; et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J. Med. Virol. 2008, 80, 1189–1194. [Google Scholar] [CrossRef] [PubMed]
- Song, B.-C.; Kim, H.; Kim, S.-H.; Cha, C.-Y.; Kook, Y.-H.; Kim, B.-J. Comparison of full length sequences of hepatitis B virus isolates in hepatocellular carcinoma patients and asymptomatic carriers of Korea. J. Med. Virol. 2004, 75, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Song, B.-C.; Kim, S.-H.; Kim, H.; Ying, Y.-H.; Kim, H.-J.; Kim, Y.J.; Yoon, J.-H.; Lee, H.-S.; Cha, C.-Y.; Kook, Y.-H.; et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J. Med. Virol. 2005, 76, 194–202. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Chen, S.; Yuan, Q.; Zhang, J.; Wu, J.; Jiang, Q.; Wang, Q.; Xia, N.; Zhang, J.; et al. Naturally occurring 5′ preS1 deletions markedly enhance replication and infectivity of HBV genotype B and genotype C. Gut 2020. [Google Scholar] [CrossRef]
- Radziwill, G.; Tucker, W.; Schaller, H. Mutational analysis of the hepatitis B virus P gene product: Domain structure and RNase H activity. J. Virol. 1990, 64, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, J.; Ryu, W.-S. Four Conserved Cysteine Residues of the Hepatitis B Virus Polymerase Are Critical for RNA Pregenome Encapsidation. J. Virol. 2009, 83, 8032–8040. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Schieck, A.; Ni, Y.; Mier, W.; Urban, S. Fine Mapping of Pre-S Sequence Requirements for Hepatitis B Virus Large Envelope Protein-Mediated Receptor Interaction. J. Virol. 2009, 84, 1989–2000. [Google Scholar] [CrossRef] [PubMed]
- Persing, D.H.; Varmus, H.E.; Ganem, D. The preS1 protein of hepatitis B virus is acylated at its amino terminus with myristic acid. J. Virol. 1987, 61, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Le Seyec, J.; Rumin, S.; Guguen-Guillouzo, C. Myristylation of the Hepatitis B Virus Large Surface Protein Is Essential for Viral Infectivity. Virology 1995, 213, 292–299. [Google Scholar] [CrossRef]
- Bruss, V.; Hagelstein, J.; Gerhardt, E.; Galle, P.R. Myristylation of the Large Surface Protein Is Required for Hepatitis B Virusin VitroInfectivity. Virology 1996, 218, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Utsumi, T.; Nakano, K.; Funakoshi, T.; Kayano, Y.; Nakao, S.; Sakurai, N.; Iwata, H.; Ishisaka, R. Vertical-scanning mutagenesis of amino acids in a model N-myristoylation motif reveals the major amino-terminal sequence requirements for protein N-myristoylation. JBIC J. Boil. Inorg. Chem. 2004, 271, 863–874. [Google Scholar] [CrossRef]
- Abou-Jaoudé, G.; Molina, S.; Maurel, P.; Sureau, C. Myristoylation signal transfer from the large to the middle or the small HBV envelope protein leads to a loss of HDV particles infectivity. Virology 2007, 365, 204–209. [Google Scholar] [CrossRef]

| Parameter | n | Age (Year, Mean ± SD) | Male Gender | HBV DNA (Mean log IU/mL ± SD) | qHBsAg (Mean log IU/mL ± SD) | ALT (Mean U/L ± SD) | Ethnicity | ||
|---|---|---|---|---|---|---|---|---|---|
| White | Asian | African-American | |||||||
| Total, n (%) | 115 | 42.2 ± 12.5 | 44 (38) | 2.7 ± 0.7 | 3.6 ± 0.9 | 29.1 ± 15.4 | 97 | 7 | 11 |
| Deletions Covering the First Methionine | 2 | 41.5 ± 5.5 | 0 (0) | 2.6 ± 0.22 | 4 ± 0.9 | 24.5 ± 4.5 | 2 | 0 | 0 |
| GtA | ∆11aa | GtD | (+)10aa | (+)11aa | |
|---|---|---|---|---|---|
| Sucrose Fraction | F20 | F17 | F15 | F18 | F21 |
| Diameter (nm) | 24.1 ± 0.3 | 19.4 ± 0.2 | 23.3 ± 0.3 | 23.8 ± 0.3 | 23.0 ± 0.2 |
| Length (nm) | 99.2 ± 4.6 | 73.2 ± 2.9 | 55.5 ± 1.7 | 65.0 ± 2.0 | 89.1 ± 4.7 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, B.; Wen, X.; Wu, Q.; Bender, D.; Carra, G.; Basic, M.; Kubesch, A.; Peiffer, K.-H.; Boller, K.; Hildt, E. The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles. Cells 2020, 9, 1898. https://doi.org/10.3390/cells9081898
Jiang B, Wen X, Wu Q, Bender D, Carra G, Basic M, Kubesch A, Peiffer K-H, Boller K, Hildt E. The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles. Cells. 2020; 9(8):1898. https://doi.org/10.3390/cells9081898
Chicago/Turabian StyleJiang, Bingfu, Xingjian Wen, Qingyan Wu, Daniela Bender, Gert Carra, Michael Basic, Alica Kubesch, Kai-Henrik Peiffer, Klaus Boller, and Eberhard Hildt. 2020. "The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles" Cells 9, no. 8: 1898. https://doi.org/10.3390/cells9081898
APA StyleJiang, B., Wen, X., Wu, Q., Bender, D., Carra, G., Basic, M., Kubesch, A., Peiffer, K.-H., Boller, K., & Hildt, E. (2020). The N-Terminus Makes the Difference: Impact of Genotype-Specific Disparities in the N-Terminal Part of The Hepatitis B Virus Large Surface Protein on Morphogenesis of Viral and Subviral Particles. Cells, 9(8), 1898. https://doi.org/10.3390/cells9081898






