Abstract
Cellular DNA is constantly being damaged by numerous internal and external mutagenic factors. Probably the most severe type of insults DNA could suffer are the double-strand DNA breaks (DSBs). They sever both DNA strands and compromise genomic stability, causing deleterious chromosomal aberrations that are implicated in numerous maladies, including cancer. Not surprisingly, cells have evolved several DSB repair pathways encompassing hundreds of different DNA repair proteins to cope with this challenge. In eukaryotic cells, DSB repair is fulfilled in the immensely complex environment of the chromatin. The chromatin is not just a passive background that accommodates the multitude of DNA repair proteins, but it is a highly dynamic and active participant in the repair process. Chromatin alterations, such as changing patterns of histone modifications shaped by numerous histone-modifying enzymes and chromatin remodeling, are pivotal for proficient DSB repair. Dynamic chromatin changes ensure accessibility to the damaged region, recruit DNA repair proteins, and regulate their association and activity, contributing to DSB repair pathway choice and coordination. Given the paramount importance of DSB repair in tumorigenesis and cancer progression, DSB repair has turned into an attractive target for the development of novel anticancer therapies, some of which have already entered the clinic.
1. Introduction
DNA carries all hereditary genetic instructions dictating and regulating cellular functions and fate. Following the discovery of its notable double helical structure, DNA was considered a fairly stable molecule. However, it did not take much time before scientists discovered that DNA is subject to the damaging effects of multiple mutagens that generate a remarkable diversity of harmful lesions [1]. On the one hand, the emergence of mutations in DNA, which generate new alleles or change the position or the number of the genes in the genome, is the driving force of evolution since these mutations are the “raw material” upon which evolution exerts selection [2]. However, on the other hand, most mutations are deleterious and disrupt the function(s) of the damaged genes, leading to various pathologies. Accumulation of DNA lesions has been implicated in cell cycle arrest, cell senescence and death, aging, tumorigenesis, diverse developmental defects, and neurodegenerative diseases [3,4]. It has been estimated that every nucleated cell in the human body suffers approximately 70,000 DNA lesions every day, and external sources of mutation only increase this number. Taking into account the huge number of mutations arising constantly and their potential to inflict detrimental effects, it is logical that for billions of years of evolution, cells have been developing mechanisms for the detection and repair of DNA lesions [2,3,5].
Research in many laboratories over the years revealed an intricate network of hundreds of proteins and protein complexes organized in discrete pathways that recognize and repair specific types of DNA lesions [6]. These DNA repair pathways are precisely coordinated with the progression of the cell cycle and function in the immensely complicated and dynamic context of the chromatin [7,8,9]. The importance of the DNA damage response (DDR) is demonstrated by the fact that almost all types of cancer exhibit some kind of DNA repair deficiency [10] and mutations in genes coding for essential DNA repair factors are often associated with considerably increased predisposition to cancer and/or premature aging [3].
In this review, we discuss the mechanisms that eukaryotic cells employ to repair double-strand DNA breaks (DSBs)—the most toxic type of DNA lesions. Especially, we focus on the elaborate network of dynamic chromatin modifications and extensive chromatin remodeling, which facilitate and regulate DSB repair pathway choice and implementation. Being of paramount importance to the process of tumorigenesis, DSB repair mechanisms and DSB repair-associated chromatin changes have turned into an attractive target for the development of novel therapeutic strategies for treating cancer patients [11,12]. We consider the numerous tactics that have emerged to target DSB repair pathways with an emphasis on the therapeutic paradigm of synthetic lethality.
2. Tying up Loose Ends—Overview of DNA Double-Strand Break Repair Pathways
2.1. Sources of Damage
Mutagens can broadly be divided into endogenous and exogenous by their source of origin. Endogenous mutagens are generated inside the cells of the organism and are generally chemical in nature comprising Reactive Oxygen Species (ROS) generated in the oxidative reactions of the metabolism and other highly reactive byproducts of certain enzymatic reactions such as formaldehyde formed during histone demethylation [13,14]. Exogenous mutagens include a large cohort of DNA-damaging compounds and high energy electromagnetic radiations such as UV-, X-ray, and gamma-radiation that are able to damage DNA either directly or indirectly [3].
It has been shown that ROS generated during oxidative metabolism are among the primary sources of double-strand DNA breaks in eukaryotic cells. Oxygen-containing radicals such as hydroxyl radicals (●OH) and superoxide anion radicals (●O2−) are extremely reactive particles capable of damaging molecules in their immediate vicinity, including DNA, creating a significant DSB burden if their production is left unattended [13,15,16]. In addition, tumorigenesis requires the activation of oncogenes, which in turn stimulate the uncontrolled proliferation of the cells. To achieve this, oncogenes increase DNA replication origin firing leading to replication stress due to altered nucleotide levels and replication fork speed. Replication stress may cause replication fork stalling and collapse, generating copious amounts of DSBs during the S phase of the cell cycle [17,18]. Moreover, increased replication rates could lead to transcription-replication conflicts resulting in DNA breaks that fuel genomic instability [19]. Both DNA and RNA polymerases lead to the generation of superhelical strain in DNA during transactions such as DNA replication, repair, and transcription. This tension is relieved by the action of a specific group of enzymes dubbed topoisomerases, which induce the formation of either single- or double-strand breaks in DNA to relax the strained DNA molecules [20]. Lesions that are present on DNA molecules during DNA-based processes increase the probability for abortive topoisomerase reactions that could lead to the formation of DSBs [2,20,21]. Certain cellular processes include the controlled formation of DNA breaks. For example, V(D)J and immunoglobulin heavy chain class switch recombination entail the formation of DSBs in order for B- and T-lymphocytes to form their extensive repertoire of antibodies or cell surface antigen receptors [22]. Meiotic crossing over, one of the main processes creating genetic diversity during sexual reproduction, also involves the regulated induction of DNA breaks in order for homologous chromosomes to exchange genetic material in gametogenesis [23].
Environmental factors such as ionizing radiation and certain chemicals can induce the formation of excessive numbers of DSBs. High energy X-ray and gamma-radiation can lead to DSBs either directly or indirectly through the generation of ROS. Low doses of radiation received during procedures such as medical imaging or even air travel can inflict damage and increase the basal levels of DSBs [3]. Mutagenic chemical compounds contained in cigarette smoke (benzo[a]pyrene) or biological toxins such as the aflatoxins that are present in the spores of Aspergillus fungi covalently bind to DNA and induce the formation of DSBs following replication fork stalling and collapse [24,25]. Acetaldehyde produced during ethanol oxidation can induce inter-strand DNA crosslinks (IPCs) and DNA-protein crosslinks (DPCs) that may lead to DSB formation during DNA repair or transcription [14,26].
Having in mind the complexity of DSB induction and the consequences organisms suffer from untimely and mutagenic DSBs, cells must have evolved specific molecular mechanisms to cope with the genomic instability posed by double-strand DNA breaks.
2.2. Pathways for Double-Strand DNA Breaks Repair
Four different DSB repair pathways have been described thus far (Figure 1). Although these pathways are very specific in their mechanisms and outcomes, they share many common proteins and are substantially interdependent [18]. The two main DSB repair pathways are the classical non-homologous end joining (c-NHEJ) pathway and homologous recombination (HR). c-NHEJ proteins are rapidly assembled on the ends of broken DNA molecules, and following limited or no processing of the DSBs, those ends are ligated (Figure 1a). c-NHEJ does not require broken ends to contain homology, although the presence of microhomology of up to 4bp can speed up the process and increase its accuracy [27]. c-NHEJ entailing end processing of the breaks is typically error-prone and may lead to chromosome rearrangements such as deletions and translocations, especially if many DSBs are formed in close proximity. HR, on the other hand, is a usually error-free DSB repair mechanism that requires extensive end processing of the DNA breaks (5′-end resection) in order to use the long ssDNA regions that have been generated to invade a homologous DNA sequence that is used as a template to achieve accurate repair of the DSB (Figure 1d). The choice between these two major pathways for DSB repair is linked to the progression of the cell cycle. Since c-NHEJ does not require homology, it is active throughout all phases of the cell cycle. The need for extensive homology for HR restricts this mechanism to the S and G2 phases of the cell cycle when following DNA replication, an identical DNA sequence that can serve as a template for error-free DSB repair is present in the nucleus [28]. The complicated cell cycle-dependent sequence of events, which leads to DSB repair pathway choice, entails extensive modifications and remodeling of the chromatin in the vicinity of the break that are thoroughly discussed below.
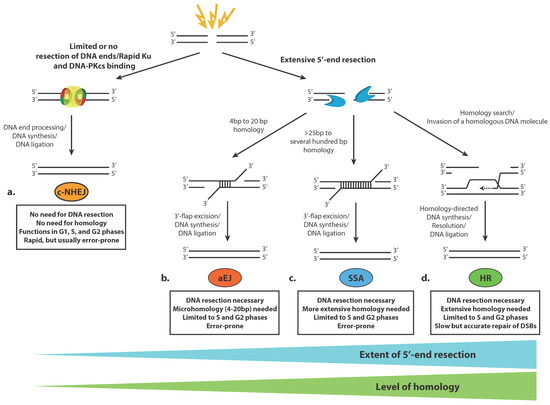
Figure 1.
General overview of double-strand DNA break repair pathways in eukaryotic cells. (a) Classical non-homologous end joining (c-NHEJ), (b) Alternative end joining (aEJ), (c) Single-strand annealing (SSA), (d) Homologous recombination (HR).
In addition, two alternative, less well-understood pathways for DSB repair have been defined. Alternative end joining (aEJ) may take place between ssDNA ends that have been generated following 5′-end resection (Figure 1b). Resection may reveal microhomologous regions (usually in the range 4 to 20 bp) between the processed DNA ends, which can bind to each other, and after removal of 3′-DNA flaps, the ends are ligated. Single-strand annealing (SSA) involves the annealing of more extensively processed DNA ends that carry wider homologous sequences, usually several tens to over a hundred nucleotides long (Figure 1c). Such homologous sequences are typically present if tandem repeats flank the DSB site, which, after resection, can anneal, and following DNA flap excision and limited DNA synthesis, can be ligated to restore the integrity of the double-stranded DNA molecule. SSA and aEJ are inherently mutagenic types of DSB repair since ligation of processed ends is preceded by 3′-flap excision, which generates deletions in DNA. Both SSA and aEJ demand DNA resection of the broken ends, meaning that they are active, like HR, in the S and G2 phases of the cell cycle. There is still a debate if SSA and aEJ are bona fide DSB repair mechanisms or serve as backup mechanisms when the two major pathways, c-NHEJ and HR, are compromised in some way [18,27].
Notably, many widely-used anticancer drugs, such as platinum salts, DNA alkylating agents, and topoisomerase inhibitors, are DNA-damaging agents that suppress cancer cell proliferation by inducing various types of DNA lesions, including DSBs [29], that lead to cell cycle arrest and cell death. Therefore, the proficient repair of DSBs may grant cancer cells with resistance towards these compounds and render chemotherapy largely inefficient [30,31,32]. Enhancing our knowledge of the mechanisms that simultaneously govern DSB repair and regulate the chromatin context in the course of the repair process is pivotal in designing more rational and beneficial chemotherapy regimens for cancer patients.
2.3. Classical Non-Homologous End Joining (c-NHEJ)
c-NHEJ is initiated by the binding of the heterodimer Ku70-Ku80 complex to the broken ends of DNA molecules (Figure 2a). The Ku complex is highly abundant in eukaryotic cells and exhibits high affinity towards blunt DNA ends or ends with short single-strand overhangs [33,34]. The Ku heterodimers bound to both ends of the break act as a platform for the further binding of DNA-dependent protein kinase catalytic subunit (DNA-PKcs), forming the DNA-PK complex [35]. DNA-PKcs is a very large protein belonging to the family of phosphoinositide 3-kinase (PI3K)-related kinases (PIKKs) [36]. DNA-PKcs autophosphorylates itself and, in addition, phosphorylates the chromatin in the vicinity of DSBs and many downstream c-NHEJ factors contributing to their timely recruitment and activation. Blunt DNA ends or ends bearing short homologous overhangs are efficiently and rapidly sealed by the action of the DNA ligase 4–X-ray repair cross-complementing protein 4 (LIG4-XRCC4) complex. XRCC4 is an essential binding partner of LIG4 and stimulates its enzymatic activity [37]. Two more proteins, XRCC4-like factor (XLF, also known as Cernunnos) and Paralogue of XRCC4 and XLF (PAXX), interact with the LIG4-XRCC4 complex and exhibit scaffolding functions facilitating the proper positioning of DNA ends prior to ligation [38,39]. However, broken DNA ends are often not complementary and/or contain modified nucleotides, which necessitates their processing prior to ligation, often in a cyclic fashion. The array of processing enzymes that is used on a particular occasion depends on the nature of the modifications carried by the damaged DNA ends. Artemis is a nuclease recruited to DSBs through its interaction with DNA-PKcs [40]. This interaction stimulates Artemis, which exhibits endonuclease activity and acts upon short overhangs or hairpins. Another c-NHEJ nuclease is aprataxin and PNKP-like factor (APLF), which displays both apurinic-apyrimidinic (AP) endonuclease activity and 3′-5′ exonuclease activity [41,42]. Polynucleotide kinase 3′-phosphatase (PNKP) is a bispecific enzyme, which adds phosphate groups to 5′-ends of DSBs that are lacking one and remove 3′-phosphates which hinder the ligation step [43]. Tyrosyl DNA phosphodiesterase 1 (TDP1) eliminates 3′-phopshoglycolates (3′-PGs) [44] and aprataxin [45] removes adenosine monophosphate (AMP) residues attached to the 5′-ends of DNA breaks owing to abortive DNA ligation reactions. In addition to nucleases and end-processing enzymes, two DNA polymerases belonging to the X-family of DNA polymerases, DNA polymerase λ and DNA polymerase μ, are employed by c-NHEJ. Both polymerases are able to add nucleotides in a template-dependent or template-independent fashion to the 3′-ends of the break until ligatable ends are achieved [34,46]. Typically, as a result of end processing by the abovementioned enzymes, the ends of the breaks drop or gain several nucleotides creating microdeletions or microinsertions, respectively [27]. Therefore, c-NHEJ is considered inherently mutagenic pathway for DSB repair.
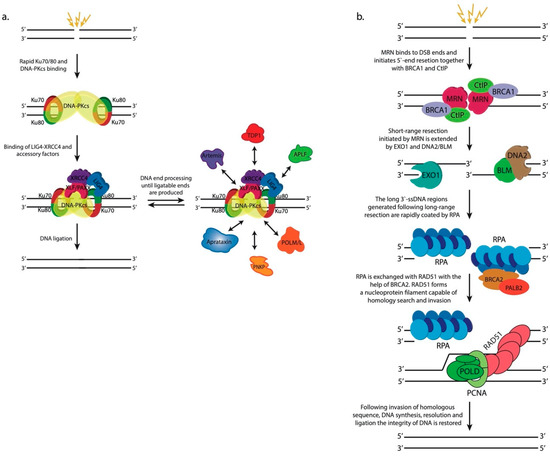
Figure 2.
Schematic representation of the major double-strand DNA breaks (DSB) repair pathways. (a). Classical non-homologous end joining (c-NHEJ). The rapid binding of Ku and DNA-PKcs creates a long-range synapse that stabilizes and protects DNA ends. Binding of LIG4-XRCC4 and accessory factors induce rearrangements leading to the formation of a short-range synapse that is competent for end processing. DNA ends that are not ligatable may be subjected to several cycles of end processing until ligatable ends are generated. (b) Homologous recombination. The MRN complex binds to DSB ends and associates with CtIP and BRCA1 to catalyze short-range 5′-end resection. EXO1 and DNA/BLM2 extend the resected region to several hundred or more than a thousand nucleotides away from the break site. The long 3′-ssDNA tails are rapidly bound by the heterotrimeric RPA complex, which is displaced by BRCA2-PALB2, which is necessary for the formation of a RAD51-ssDNA nucleofilament that is capable of invading a homologous DNA sequence. DNA synthesis performed by DNA polymerases in cooperation with the processivity factor PCNA restores the damaged region, and following resolution and ligation, the integrity of DNA is restored perfectly.
2.4. Homologous Recombination (HR)
The hallmark of homologous recombination compared to c-NHEJ is the extensive 5′-end resection of broken DNA ends that precedes the homology-directed repair of the break (Figure 2b). This processing step is implemented in a highly coordinated and regulated fashion by several nucleases and helicases [47]. HR is initiated by the binding of the heterotrimeric MRE11-RAD50-NBS1 (MRN) complex to DSBs. MRN is a highly dynamic protein complex that interacts with multiple downstream HR factors [48]. Its binding to the CtBP-interacting protein (CtIP, also known as RBBP8) activates the endonuclease activity of the MRE11 subunit, which nicks the strand that ends with a 5′-terminus at the DSB. This nick may be introduced up to several hundred bp away from the break site. MRE11 also exhibits 3′-5′ exonuclease activity, which is responsible for the short-range resection of the nicked strand from the nick as far as to the break site creating a long 3′-ssDNA tail [49,50,51,52].
The MRN complex recruits the prominent DNA damage response (DDR) kinase ataxia-telangiectasia mutated (ATM) through direct interaction between ATM and MRN’s NBS1 subunit [53]. ATM, like DNA-PKcs, is a member of the PIKK family of protein kinases, and following its recruitment to DSB sites, ATM phosphorylates Ser139 of H2A.X (termed γH2A.X after S139 phosphorylation) [54] in the vicinity of the break initiating a cascade of chromatin modifications discussed in detail later in this review. Briefly, γH2A.X serves as a binding site for Mediator of DNA damage checkpoint protein 1 (MDC1), which in turn is modified by ATM [55,56]. Phosphorylated MDC1 recruits the E3 ubiquitin ligase Ring Finger Protein 8 (RNF8) that launches a highly complex ubiquitination cascade of the chromatin [57]. RNF8 polyubiquitinates histone H1 that is bound by Ring Finger Protein 168 (RNF168) [58]. RNF168 ubiquitinates histones H2A/H2A.X at lysine residues K13 and/or K15, which are bound by downstream HR factors [59,60]. The ubiquitination cascade of the chromatin eventually recruits the heterodimeric Breast cancer type 1 susceptibility protein-BRCA1 Associated RING Domain 1 (BRCA1-BARD1) protein complex [61,62,63]. BRCA1-BARD1 is the most important mediator of 5′-end resection that channels DSB repair towards HR. BRCA1-BARD1 interacts with CtIP and MRN in a cell cycle-dependent manner to stimulate MRE11′s activity and initiate resection [64]. The short-range resection completed by the MRN complex is further extended (long-range resection) by exonuclease 1 (EXO1) and the heterodimeric DNA2/Bloom syndrome protein (DNA2/BLM) nuclease-helicase complex [65,66,67,68]. Long-range 5′-end resection generates extensive 3′-ssDNA (at least several hundred bp) that is readily coated by the heterotrimeric replication protein A (RPA) complex [69]. RPA affinity for ssDNA is significant and it must be evicted for the final steps of HR to commence. BRCA1-BARD1 interacts with Partner and localizer of BRCA2 (PALB2) that recruits the Breast cancer type 2 susceptibility protein (BRCA2) to the long 3′-ssDNA regions. BRCA2 is essential for promoting RPA exchange with RAD51 along the single-strand DNA regions [70,71]. RAD51 binds to ssDNA forming helical RAD51-ssDNA nucleoprotein filaments that are capable of homology search and invasion of a homologous DNA sequence [72,73]. BRCA1-BARD1 facilitates homologous pairing and if sufficient base pairing occurs between the invading RAD51-coated strand and the invaded DNA molecule the non-base-paired strand of the invaded molecule is displaced in the form of a loop (D-loop) [74] and the 3′-end of the invasion strand is engaged by DNA polymerase δ or translesion DNA polymerases [75]. DNA polymerases extend the 3′-end of the invasion strand past the break using the invaded homologous strand as a template. This repair DNA synthesis is followed by resolution, annealing, and ligation of the extended invasion strand to the other end of the DSB on the original DNA molecule, effectively resealing the damaged region.
2.5. Alternative End Joining (aEJ)
Contrary to c-NHEJ, alternative end joining requires 5′-DNA end resection of the DSB ends, which usually ranges between 15 and 100 nucleotides from the site of the breaks. This resection step is essential as it exposes short (between 2 bp and 20 bp) microhomology sequences that can anneal and serve as the basis for further rejoining of the break [27]. The resection is accomplished, as in HR initiation, by the MRN complex and CtIP [76]. It has been shown that poly(ADP-ribose) polymerase 1 (PARP1), an abundant DNA damage sensor, promotes aEJ possibly through competition with the Ku heterodimer for DSB binding [77,78]. PARP1 binding to DNA activates its enzymatic activity, and PARP1 starts to form long negatively-charged poly(ADP-ribose) (PAR) chains on itself and on the chromatin proteins surrounding the break (PARylation) that serve as a platform for the recruitment of downstream DNA repair factors [79]. PARP1 is necessary for the recruitment of DNA polymerase Θ, which is the central mediator of aEJ [80]. Pol Θ binds to the 3′-ends of short annealed microhomology sequences stabilizing their association and extends the 3′-ends of the DNA strands to strengthen the association between the ends of the break [81]. If the 3′-ssDNA regions have annealed upstream and their ends are protruding, the association of Pol Θ is preceded by the action of several nucleases such as the xeroderma pigmentosum group F-Excision Repair Cross-Complementation Group 1 (XPF-ERCC1) nuclease, Artemis, and APLF, which remove the 3′-flaps setting the stage for Pol Θ binding [27]. Following Pol Θ-mediated fill-in synthesis at both sides of the annealed region, the break is sealed either by DNA ligase 1 or by the DNA ligase 3-X-ray repair cross-complementing protein 1 (LIG3-XRCC1) complex [82]. aEJ is inherently mutagenic DSB repair pathway since it leads to deletions due to 3′-flaps removal prior to fill-in synthesis and ligation. Currently, it is unclear if aEJ is a bona fide DSB repair mechanism and processes a subset of DSBs that cannot be engaged by c-NHEJ or acts as a backup mechanism when cNHEJ is compromised [18]. The importance of aEJ is apparent in HR-deficient cells, which are heavily dependent on Pol Θ-mediated aEJ for their survival, and this dependency may be exploited in the clinic, but further research is necessary to assess aEJ contribution to DSB repair in normal cells [83,84].
2.6. Single-Strand Annealing (SSA)
Single-strand annealing, compared to aEJ, also requires 5′-end resection of DSB ends, but the extent of the resection is larger since SSA may be accomplished between homologous sequences located along the 3′-ssDNA tails that are significantly longer—in the range between 25 to several hundred nucleotides [85]. Such homologous sequences are most often available due to the presence of tandem repeats flanking both ends of the break [18]. The long-range resection necessary for SSA is initiated by MRN and CtIP and extended by EXO1 and DNA2/BLM [86]. ssDNA is coated by RPA, but the final steps of SSA are RAD51-independent as opposed to HR. The annealing between the complementary ssDNA regions is facilitated by the RAD52 protein, which displaces the RPA molecules coating the ssDNA. If 3′-flaps are present, they are excised by the XPF-ERCC1 endonuclease rendering SSA obligatory mutagenic DSB repair pathway since the intervening sequences between the complementary regions are lost [18,27,87,88].
3. Chromatin Response to Double-Strand DNA Breaks
The accurate pathway choice for DSB repair is fundamental for the efficient repair of the damage. Repair pathway choice depends on many factors, such as the cell cycle phase, the nature of the damage, and the chromatin compartment where the break has occurred. Whether cells choose to execute c-NHEJ or HR on a particular DSB following damage recognition is determined by a remarkably complex array of chromatin modifications which ordain the recruitment of downstream effector proteins that are executing the repair process. The most important protein factors that exert control over DSB repair pathway choice are p53-binding protein 1 (53BP1) and BRCA1, which exhibit opposite functions with respect to initiation of 5′-end DNA resection [18]. 53BP1 opposes 5′-end resection directing DSB repair towards c-NHEJ. 53BP1 achieves this by binding to the chromatin in the vicinity of the breaks and recruiting downstream factors such as Rap1-Interacting Factor 1 (RIF1) and the Shieldin complex, which protect broken DNA ends from nucleolytic degradation [89]. On the other hand, BRCA1 interacts and activates proteins that initiate and execute extensive 5′-end resection, which is a prerequisite for DSB repair by homologous recombination [90]. Both proteins are recruited to DSBs by means of specific chromatin modifications and chromatin remodeling events executed by a multitude of chromatin-modifying DNA repair factors [91]. Next, we discuss in detail the chromatin events that define and orchestrate DSB repair (Figure 3, Table 1 and Table 2).
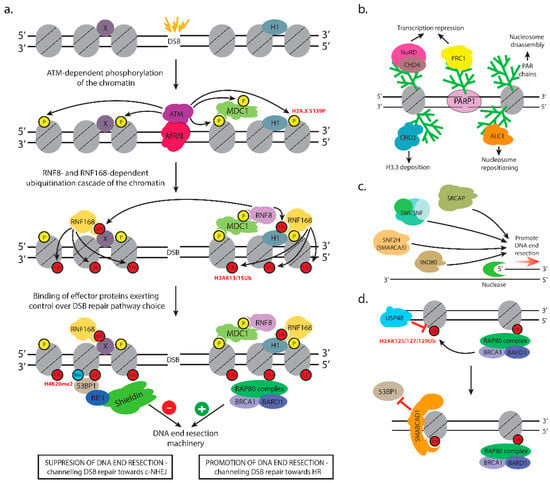
Figure 3.
Chromatin response to double-strand DNA breaks. (a) Major chromatin modifications exerting control over DSB repair pathway choice. (b) PAR-dependent chromatin remodeling following PARP1 binding to DSBs. (c) SRCAP, SWI/SNF, SNF2H, and INO80 lead to chromatin changes facilitating DNA end resection and HR. (d) BRCA1-BARD1 ubiquitinates H2AK125/127/129Ub in the vicinity of DSBs leading to the recruitment of SMARCAD1, which restricts 53BP1 binding to the chromatin surrounding the break. The deubiquitinase USP48 counteracts H2A ubiquitination by BRCA1-BARD1 promoting 53BP1 binding.

Table 1.
Post-translational histone modifications associated with DSB repair.

Table 2.
Chromatin remodelers involved in DSB repair.
3.1. ATM and the Control of Early Events in DSB Repair
ATM is the apical kinase responsible for global orchestration of cellular responses to DSBs, which include DNA repair, checkpoint activation, alterations in chromatin structure, transcription, apoptosis, and senescence [36]. ATM is recruited to chromatin in response to DSBs [92,93] in a process that requires ATM binding to the C terminus of NBS1 [53]. MRN both recruits ATM to DNA lesions and stimulates ATM kinase activity once there [53,94,95] (Figure 3a). In a series of papers, Price et al. have unveiled the chromatin control of ATM activity in response to DSBs. To become activated, the ATM kinase needs to be acetylated by the TAT-interactive protein 60 kDa (TIP60) histone acetyltransferase (HAT). The interaction between ATM and TIP60 is not regulated in response to DNA damage, but the HAT activity is specifically activated by DNA damage [96]. TIP60 is activated by the direct interaction between the chromodomain of TIP60 and histone H3 trimethylated on lysine 9 (H3K9me3) as well as the MRN complex [97]. In turn, H3K9me3 at DSBs is regulated by a complex containing KAP-1, HP1, and the H3K9 methyltransferase Suv39h1. Price et al. demonstrate that a complex containing KRAB-associated protein 1 (KAP-1), heterochromatin protein 1 (HP1), and the H3K9 methyltransferase Suppressor of Variegation 39 Homolog 1 (Suv39h1, KMT1A) is rapidly loaded onto the chromatin at DSBs. Suv39h1 methylates H3K9 facilitating the loading of additional KAP-1/HP1/Suv39h1 through binding of HP1′s chromodomain to the nascent H3K9me3. This process initiates cycles of KAP-1/HP1/Suv39h1 loading and H3K9 methylation that facilitate the spreading of H3K9me3 and KAP-1/HP1/Suv39h1 complexes and transient formation of heterochromatin for tens of kilobases away from the DSB [97]. However, the DSB-specific increase in H3K9me3 has to be reconciled with data showing that H3K9 demethylase KDM4D was recruited to DSBs in a PARylation-dependent manner and controlled recruitment of ATM there. KDM4D deficit affected both HR repair and NHEJ [98,99]. These discrepancies may be explained by temporal differences in the recruitment of activities that increase or decrease H3K9me3.
ATM activation has been shown to depend on histone acetylation. Thus, ATM function in response to ionizing radiation was shown to depend on histone H4K16 acetylation carried out by the histone acetyltransferase Males absent On the First (MOF, KTM8) [100]. The nucleosome-binding protein High-Mobility Group Nucleosome binding 1 (HMGN1) was found to modulate the global organization of ATM throughout the nucleus via promoting DNA damage-induced H3 acetylation. Thus, HMGN1 predetermines the interaction of ATM with chromatin before induction of damage [101].
ATM phosphorylates hundreds of substrates in response to DNA damage [102], although it is unclear what proportion of these are functionally important [36] (Figure 3a). Many ATM substrates are also phosphorylated by ATR in response to replication stress. Multiple crosstalks exist between ATM and ATR as ATM-dependent DNA end resection provides the RPA-ssDNA signal for ATR recruitment and activation [36]. Recent data indicate that the outcomes of the crosstalks depend on the cell cycle phase and the damage load [103,104].
ATM controls accessibility to damaged chromatin. RNF20 and RNF40 (orthologs of the budding yeast protein BREfeldin A sensitivity-Bre1) catalyze the ubiquitination of H2B on lysine 120 [105] (monoubiquitination is mapped on H2B lysine 119 in S. pombe, on lysine 120 in humans and lysine 123 in S. cerevisiae [106]). Throughout the text these numbers are maintained. Upon DSB induction, RNF20-RNF40 is recruited at DSB sites, undergoes ATM-mediated phosphorylation, and ubiquitinates H2B. This is essential for the timely accumulation of NHEJ and HR proteins at DSB sites and subsequent proficient repair via both pathways. The depletion of RNF20-RNF40 led to reduced accumulation of YFP-tagged XRCC4 and Ku80, as well as delayed recruitment to DSBs of RPA, BRCA2, and RAD51. H2BK120ub is thought to initiate chromatin disassembly providing access to both NHEJ and HR DNA repair factors [107]. H2BK120ub did not affect γH2A.X phosphorylation [108]. Another study, however, found that the SAGA complex deubiquitinase activity was required for optimal irradiation-induced γH2A.X formation, and failure to remove H2BK120ub inhibited ATM- and DNA-PKcs-induced γH2A.X formation. These data suggest that deubiquitination is either required at specific regions or that the mark is dynamic at DSBs [109]. Accessibility at DSBs provided by H2BK120ub was dependent on the recruitment of the Imitation Switch Nuclear ATPase 2H (SNF2H)-containing chromatin remodeling activities (Figure 3c). Initially, it was thought to affect RAD51 and BRCA1 recruitment [110], but later it was found to mediate chromatin relaxation and to be necessary for both HR repair and NHEJ [111]. SNF2H recruitment and chromatin relaxation are also dependent on the deacetylation of H3K56ac by Sirtuin 6 (SIRT6), and its deficit impaired both HR and NHEJ [112].
ATM plays a key role in increasing accessibility of heterochromatin. One of ATM substrates, KAP-1, is phosphorylated at the sites of damage, which results in its spread throughout chromatin, thereby promoting relaxation on a global scale and facilitating rapid genome surveillance [113]. Goodarzi et al. [114] have provided evidence that ATM has a specialized role in the repair of DSBs in tightly compacted heterochromatic parts as it loosens the KAP-1 binding affinity for chromatin. Phosphorylated KAP-1 dispersed Chromodomain Helicase DNA binding protein 3 (CHD3) subunit of the Nucleosome Remodeling Deacetylase (NuRD) complex with concomitant chromatin relaxation [115]. ATM silences transcription by two mechanisms—the first one is dependent on RNF8 and RNF168 [116] and the other involves phosphorylation of Polybromo-associated BAF (PBAF) subunit BRG1/BRM-associated factor 180 (BAF180) and mono-ubiquitination of H2A on Lys-119 via polycomb repressive complex 1 and 2 [117]. The downregulation of transcription depends on the distance from the DSB [118].
The primary chromatin target of ATM, ATR, and DNA-PKcs is the histone variant H2A.X, and its phosphorylation is one of the earliest events following break induction [119]. A major role of γH2A.X is to spatially organize repair by the chromatin retention of repair proteins. In both yeast and mammalian cells, phosphorylated H2A.X encompasses rather extensive regions around the break—several tens of kilobases in yeast [120] and megabase-sized domains in mammalian cells [119]. The spread of γH2A.X requires MDC1—a large protein platform that supports the chromatin recruitment of many DSB repair factors (Figure 3a). MDC1 stabilizes the interaction of NBS1 component of MRN with chromatin at DSBs [55], and the MRN complex recruits more ATM [121,122]. This creates a positive feedback loop that propagates H2A.X phosphorylation [123,124]. One way by which ATM can increase MRN binding is by locally increased H3K36me2 after ATM phosphorylation abrogates chromatin binding of KDM2A demethylase [125]. γH2A.X-marked chromatin is transcriptionally inactive [126].
It has been shown that MDC1 binding to γH2A.X is inhibited by phosphorylation of tyrosine 142, which promotes pro-apoptotic factors, thus controlling life-death cell fate decisions [127]. At replication fork stalling lesions, MDC1 and Topoisomerase II Binding Protein 1 (TopBP1) help anchor ATR kinase at chromatin containing damage [128].
MDC1 plays a determining role in the interaction of phosphorylated H2A.X with its downstream partners as it acts as an interaction platform for other DDR components.
3.2. Setting up Chromatin for DSB Repair Pathway Choice
3.2.1. RNF8-RNF168-Mediated Recruitment of Repair Factors
Different sets of events set up the anti- or pro-resection chromatin environment at double-strand breaks. As already noted, one of the MDC1 interactors is the RNF8 E3 ubiquitin ligase [129]. Together with the UBC13 E2 enzyme, RNF8 catalyzes the formation of K63-linked polyubiquitin chains [129,130,131] initially thought to target H2A histones, but now shown to be mainly of H1 linker histones [58]. H1 ubiquitination serves as a recruitment signal for RNF168, which in turn, monoubiquitinates H2A-type histones at K13/K15 in a DSB-dependent manner [59,60,132]. The priming monoubiquitination on H2A by RNF168 gets extended by RNF8 to form K63-linked polyubiquitin chains needed to interact with downstream effectors [60] (Figure 3a). RNF8 also promotes the recruitment of RNF168 via extensive chromatin decondensation by bringing the CHD4 catalytic subunit of the NuRD complex. The chromatin remodeling activity of CHD4 promotes efficient ubiquitin conjugation and assembly of RNF168 at DNA double-strand breaks. Interestingly, RNF8-mediated recruitment of CHD4 and subsequent chromatin remodeling were found to be independent of the ubiquitin-ligase activity of RNF8, suggesting that CHD4 and RNF8 cooperate in creating a chromatin environment that is permissive to repair [133].
53BP1 recognizes H2AK15ub with its ubiquitin-dependent recruitment (UDR) motif [132]. Chromatin retention of 53BP1 requires histone methylation in addition to ubiquitylation. The two methylated lysines associated with retention of 53BP1 to DSB-flanking chromatin are H3K79me and H4K20me, with the latter thought to have a major role in the process [134,135,136]. Both H3K79 and H4K20 methylation are not induced in response to DNA damage. This has led to the suggestion that chromatin changes in response to damage get the chromatin marks exposed to mediate 53BP1 binding. While it is generally accepted that H4K20 methylation is not damage induced, there are data indicating that it may be increased locally upon induction of DSBs by the multiple myeloma SET (MMSET) methyltransferase. Downregulation of MMSET significantly decreased H4K20 dimethylation and subsequent accumulation of 53BP1 at DSBs [137]. Still, mice knocked-out for MMSET show normal recruitment of 53BP1 [138]. H4K20 methylation is catalyzed by several enzymes—monomethylation is carried out by SET Domain Containing 8 (SETD8, PR-SET7, KMT5a) [139,140], while the homologs SUV420H1 and SUV420H2 catalyze the di- and tri-methylation of lysine 20 on histone H4 (H4K20) and require H4K20me1 as substrate [141,142]. No major requirement for the known H4K20 (di-)methylases SUV420H1 and SUV420H2 in 53BP1 recruitment or DSB repair function was found, but the H4K20 monomethylase, PR-SET7 (SET8), was essential [138].
Histone H4K20 methylation is subject to binding by several factors, which can compete with 53BP1 for chromatin recruitment. In response to DNA damage, one of these—Lethal(3)Malignant Brain Tumor-Like Protein 1 (L3MBTL1), which binds H4K20me2 [143]—gets evicted from chromatin [144]. Another competitor is the histone demethylase KDM4A (which binds to H4K20me3) [145], which is targeted for degradation in RNF8- and RNF168-dependent manner after damage induction [146]. The H4K20me2 mark is required for the binding of the TIP60 complex as well, and TIP60 directly competes for binding with 53BP1. TIP60 is able to acetylate H2AK15 on H2A, which is mutually exclusive with K15 ubiquitination (needed for 53BP1). This establishes another switch mechanism for pathway choice [147]. In addition, Tudor interacting repair regulator (TIRR) was shown to bind to the tandem Tudor domains of 53BP1, and both stabilize 53BP1 and impede its binding to H4K20me2 when no damage is present [148]. Following ATM-mediated phosphorylation of 53BP1 after damage induction, TIRR dissociates from 53BP1, allowing its binding to H4K20me2 in the vicinity of the break.
The other histone mark implicated in 53BP1 recruitment, H3K79me [134,149], is deposited by mammalian DOT1 Like protein (DOT1L) [150,151]. Efficient H3K79 methylation was shown to depend on H2BK120 ubiquitination as it directly stimulated DOT1L activity [152,153,154]. In yeast, Dot1 and Rad9 (53BP1 homolog) also contribute to HR via loading of cohesin at sites of damage [155] as well as the repair of other types of damage [156].
The RNF8-RNF168 ubiquitination was reported to recruit BRCA1-BARD1 [61,62,63] (BRCA1-C), the key mediator of 5′-end resection. BRCA1-BARD1 interacts with CtIP and MRN in a cell cycle-dependent manner to stimulate MRE11′s activity and initiate resection [64]. However, recruitment of the Brca1-CtIP-MRN complex was found dependent on ATM and PARylation but not on RNF8/RNF168 mediated ubiquitination [157]. The BRCA1/BARD1 complex was also found to be recruited in the vicinity of the DSBs through BARD1 interaction with K9-dimethylated histone H3 (H3K9me2) mediated by heterochromatin protein 1 (HP1) [157].
At the same time, RNF8/RNF168-mediated K63-linked ubiquitin chains recruit the BRCA1-A complex [62], which consists of BRCA1/BARD1, RAP80, Abraxas, MERIT40, BRCC36, and BRCC45 [62]. Unlike BRCA1/BARD1 (BRCA-C) complex, this one limits DNA end resection [158,159] via BRCC36-mediated cleavage of K63-linked chains [160]. Kinetic data by Aleksandrov et al. obtained by micro-irradiation in living cells clearly show that BARD1 accumulation follows a similar pattern of accumulation as ubiquitin and is recruited much later compared to PARP1 [161]. As of now, the contribution of BRCA1-related pro- vs. anti-resection activities recruited by RNF8-RNF168 H2A ubiquitination remains an open question.
3.2.2. BRCA1-BARD1-Dependent Repositioning of 53BP1 Promotes DNA End Resection
Other factors influence BRCA1 recruitment to DSBs as well. The BRCA1/BARD1 recruitment depends on the recognition of histone H4 unmethylated at lysine 20 (H4K20me0) by the ankyrin repeats domain of BARD1. BARD1 mutations disabling H4K20me0 recognition abrogated accumulation of BRCA1 at DSBs and allowed the anti-resection activity to prevail in S and G2 phases [162]. Methylated histones dilution following DNA replication facilitates BRCA1 recruitment to sister chromatids. Conversely, the recruitment of 53BP1 is dependent on H4K20me2, which guides it to pre-replicative chromatin. [163,164]. The histone variant macroH2A1, together with the H3K9 methyltransferase PRDM2, was required for BRCA1 recruitment [165]. This report, however, needs to be reconciled with an earlier one linking macro H2A1 with 53BP1 recruitment [166].
Super-resolution microscopy performed to examine the spatial distribution of BRCA1 and 53BP1 proteins within a single irradiation-induced focus indicated progressive BRCA1-dependent exclusion of 53BP1 from DNA damage sites during S-phase [167]. Follow-up studies provided insights on a chromatin-based mechanism dependent on BRCA1-BARD1 E3 ubiquitin ligase activity that mediates 53BP1 eviction. BRCA1-BARD1 ubiquitinates [168,169,170] lysines 125, 127, and 129 on H2A [171]. Inactivation of BRCA1-BARD1 E3 ubiquitin ligase activity impaired end resection. The SMARCAD1 remodeler has been shown to bind BRCA1-BARD1-ubiquitinated H2A [172]. SMARCAD1 and its homolog in yeast Fun30 are large, single-subunit nucleosome remodelers, which act in homodimeric form [173]. The role of Fun30 and SMARCAD1 during repair of DNA DSBs by homologous recombination has been reported by several groups [174,175,176] demonstrating a molecular function in counteracting the resection inhibitor 53BP1 [172,174,177] (Figure 3d). In the absence of 53BP1/Rad9, the remodeling activity of SMARCAD1/Fun30 is at least partly dispensable, and phenotypes such as camptothecin sensitivity are suppressed [174,175]. Both the SMARCAD1 ATPase activity and the integrity of its ubiquitin-binding domains are required for the eviction of 53BP1 to stimulate HR repair [172]. These data point to ubiquitin-targeted SMARCAD1 remodeling, rather than competition between 53BP1 and BRCA1 in 53BP1 repositioning and repair choice [172].
SMARCAD1, as well as yeast Fun30, are phosphorylated by cyclin-dependent kinase 1 [177,178] to promote interaction with the N-terminal BRCT repeats of TopBP1 and yeast Dpb11 [177]. The negative regulation to SMARCAD1 involves ubiquitin-specific peptidase 48 (USP48)—a deubiquitinase specific for BRCA1-BARD1 ubiquitination sites (H2AK125/K127/K129) [179] (Figure 3d). Overexpression of USP48 restricted resection, whereas the USP48 deficit resulted in the repositioning of 53BP1 further away from the damage site and extended resection. Cells depleted of USP48 develop a dependence on SSA even though they display normal levels of 53BP1 [179]. The depletion of compacting methyltransferases SETDB1 and Suv39, as well as HP1, prevents changes in irradiation-induced foci and inhibits HR repair [180].
3.3. Recruitment of Effectors
3.3.1. Chromatin Changes that Promote c-NHEJ
Multiple chromatin changes contribute to efficient c-NHEJ, mostly by modulating chromatin accessibility at the sites of damage. DSB induction in human cells caused local nucleosome disassembly in G1. It encompassed a region of 2 kb around the break and was independent of DNA end resection. ATM and the INO80 chromatin remodeler promoted nucleosome disassembly (manifested by removal of histone H3 from the chromatin) during non-homologous end joining [228]. c-NHEJ also appears to be stimulated by the deposition of histone variants that destabilize the nucleosome [242]. It has been reported that the p400 remodeler deposits H2A.Z required for the loading of Ku70/Ku80 [232]. This result is in agreement with an earlier report that yeast SWR1 remodeler participates in yeast c-NHEJ by facilitating the recruitment of the Ku proteins [231]. Another histone variant that stimulates c-NHEJ is histone H3.3. Luijsterburg et al. described a mechanism initiated by PARP1, which recruits the chromatin remodeler CHD2 (Figure 3b). The remodeler, in turn, triggers rapid chromatin expansion and the deposition of histone variant H3.3 at sites of DNA damage. H3.3 deposition promotes efficient assembly of c-NHEJ complexes [235].
SWI/SNF-mediated chromatin remodeling also contributes to mammalian c-NHEJ. The BAF and PBAF complexes include one of two mutually exclusive DNA-dependent ATPases—BRG1/SMARCA4 or BRM/SMARCA2. Together with core and accessory subunits, these ATPases function in mobilizing nucleosomes to regulate transcription, DNA replication, and higher-order chromatin dynamics [243,244]. The BAF complex is needed for efficient c-NHEJ [210], as it controls the accumulation of GFP-tagged Ku70. The amount of Ku70 at laser-irradiated sites was severely reduced in cells after depletion of BRM, BAF47, BAF60a, BAF60c, BAF155, BAF250a, or BAF250b [210]. In turn, SWI/SNF remodeling to recruit Ku70 and Ku80 was dependent on p300 and CREB-binding protein (CBP) acetyltransferases (targeting H3K18, and H4K5, 8, 12, and 16), which were recruited at DSBs. Both RNA interference and HAT inhibitors anacardic acid (inhibits PCAF, p300, and TIP60) and curcumin (which inhibits CBP and p300) produced similar defects in the recruitment of Ku proteins [189].
The SAGA complex was found necessary for c-NHEJ as well. It mediated a switch of H2BK120 from ubiquitinated to acetylated state. The change in the post-translational modifications at this residue promoted histone H1 eviction and 53BP1 accumulation over γH2A.X containing domains [109,187].
Histone acetylation during c-NHEJ appears to be highly dynamic. After break induction, mammalian HDAC1 and HDAC2 are quickly recruited to DSBs and deacetylate H3K56 and H4K16. HDAC1 and HDAC2 knockdown impaired c-NHEJ [245]. The authors suggested that chromatin compaction is needed to keep Ku proteins concentrated at DSB ends and prevent them from sliding away (as they do on naked DNA) [245]. Other HDACs are required for efficient c-NHEJ: knockout of SIRT7 in mice led to an increase in H3K18ac, which impaired c-NHEJ activity [246]. SIRT6 HDAC forms a complex with DNA-PKcs. In response to DSBs, SIRT6 causes an acute decrease in global H3K9ac levels, which stabilize DNA-PKcs at chromatin adjacent to a site-specific DSB [247]. Histone deacetylases control each other. An interplay between SIRT1 and HDAC1 was shown in neurons in which SIRT1 deacetylates and activates HDAC1. HDAC1 targets H4K16ac and promotes c-NHEJ [193]. Other histone marks are reported to modulate c-NHEJ. Metnase-induced H3K36me2 enhanced the recruitment of Ku70 and NHEJ in mammalian cells [204]. Similarly, in yeast Set2-dependent H3K36 (mono-, di-, and tri-) methylation reduced resection and promoted c-NHEJ, while antagonistic Gcn5-dependent H3K36 acetylation increased resection and promoted HR. [248]. These data, however, are in contrast to findings in mammalian cells in which H3K36me3 stimulated HR via Lens Epithelium-Derived Growth Factor (LEDGF), which was found to bind CtIP [206] or TIP60 to promote H4K16ac [249]. However, while these studies suggest H3K36me3 as a DSB repair specific mark, a recent genome-wide study of repair-associated histone modifications found only a modest increase in H3K36me3 at c-NHEJ-repaired break sites [187]. It has been suggested that H3K36me3 might contribute to HDAC1/2 recruitment in order to compact chromatin structure away from the break to serve as a barrier to resection [250].
Phosphorylation of histone H4 at S1 by the casein kinase II has been shown to participate in c-NHEJ in yeast [184]. The mark inhibited NuA4 ability to acetylate H4 [183]. In mammalian cells, phosphorylated H4S1 accumulates at DSBs [187].
PARP1 has been shown to limit the resection process. Inhibition of PARP1 led to hyper-resected DNA DSBs. This phenotype was associated with loss of Ku, 53BP1, and RIF1 from the break site. EXO1-mediated resection was blocked by PARP1 [251]. Contrary to this finding, a recent report showed that the PARP1/2 inhibitor BMN673 (talazoparib) only delays the onset of 5′-end resection in living non-Hoechst-pre-treated cells possibly due to the delay in PCNA-dependent DNA repair pathways in the vicinity of the DNA break. In addition, Ku, 53BP1, BARD1, RPA, and RAD51 were recruited to laser-induced DNA damage sites in the presence of BMN673, albeit some of them with altered kinetics [161]. The recruitment of PARP1 has been shown to be limited by the histone acetyltransferase p400. The deficit of p400 increases the frequency of alternative end joining events, generating large deletions following repair of double-strand breaks [252].
3.3.2. Chromatin Events Promoting the Recruitment of HR Repair Factors
The presence of nucleosomes has been shown to impede resection by both the Exo1- and Sgs1-Dna2-dependent mechanisms [253]. The authors of the study showed that the Sgs1–Dna2 machinery does not require extensive remodeling to initiate resection, which is consistent with previous in vivo data that Remodeling Structure of Chromatin (RSC) remodeling is necessary for the removal of a single nucleosome next to a site-specific DSB [254]. Exo1, on the other hand, was inhibited even when a single nucleosome was surrounded by large tracts of free DNA [217].
ChIP studies in yeast generally indicate nucleosome loss in the immediate vicinity (~500 bp) of a DSB. Histone density is much less affected within 1 kb of the lesion [120,224,231,255]. In a similar manner, data in human cells do not support significant nucleosome loss [187], although resection proceeds to about 3.5 kb from a sequence-specific defined DSB [256]. At the same time, nucleosomes bind resected DNA, and this may even stimulate pro-resection activities. Complexes between ssDNA and recombinant histones have been reconstituted biochemically. Fun30—a key remodeler linked to reversing the 53BP1/Rad9 block to resection, binds preferentially to these complexes over conventional nucleosomes, which activated its ATPase activity [217].
The effect of histone variant H2A.Z on resection is a common theme of several studies. Biochemical and genetic evidence reveal that nucleosomes harboring H2A.Z are more accessible to Exo1 [253]. This might explain the role of the transient incorporation of H2A.Z at DSBs [226,232]. In mammalian cells, the deposition of H2A.Z at damage sites was also linked to increased resection. When H2A.Z removal by Acidic Nuclear Phosphoprotein 32 Family Member E (Anp32e)—an H2A.Z chaperone—was blocked, nucleosomes at DSBs retained elevated levels of H2A.Z. This resulted in increased CtIP-dependent end resection, accumulation of single-stranded DNA, and an increase in repair by aEJ [257]. Other authors found that the deficit of mammalian SRCAP resulted in impaired resection due to defective CtIP recruitment [230] (Figure 3c). Snf2-Related CREBBP Activator Protein (SRCAP) is the mammalian homolog of yeast Swi2/Snf2-related 1 (Swr1), a remodeler that facilitates the exchange of H2A with H2A.Z variant. However, while yeast Swr1 has been shown to stimulate resection [253], an earlier study found defective Ku80 loading but normal end resection in an Swr1 mutant [231]. In addition, in both yeast and mammals, Ino80-deficient cells were found defective in end resection [222,223] (Figure 3c). If the phenotypes were determined by H2A.Z, the opposite would be expected as Ino80 and Swr1 have opposing roles in H2A.Z deposition [258,259].
Fun30 also influences the distribution of H2A.Z genome-wide and particularly in centromeric, pericentromeric, and subtelomeric chromatin [218,219]. However, it is presently unclear if these changes might potentially contribute to resection regulation.
LEDGF protein is constitutively associated with chromatin through its Pro-Trp-Trp-Pro (PWWP) domain, which binds to H3K36me3. LEDGF interacts with CtIP in a DNA damage-dependent manner, thereby enhancing CtIP tethering to the active chromatin and facilitates its access [208]. Several studies have shown that H3K36me3 (associated with active transcription) and the SETD2 methyltransferase are required for HR repair [205,206,207].
In yeast, the absence of functional SWI/SNF activity impaired the recruitment of MRX and significantly delayed the initiation of DNA end resection [216]. The human Helicase, Lymphoid Specific (HELLS) protein—an Snf2-like chromatin remodeler, dysregulated in several cancers—was shown to promote the accumulation of CtIP at IR-induced foci. It contributed to repair within heterochromatic regions during the G2 phase, suggesting that it might be required for efficient repair in specific genomic contexts [260]. CHD1 chromatin remodeler has been shown to be required for the recruitment of CtIP. CHD1 depletion sensitized cells to PARP inhibitors [233].
RAD51 nucleoprotein filament formation is stimulated by several chromatin factors. Asf1 and CAF-1 histone chaperones are needed for RAD51 loading during HR repair. The knockdown of Asf1 or CAF-1 reduced the recruitment of the RAD51 loader MMS22L-TONSL to ssDNA. Deficient cells had persistent RPA foci, extensive DNA end resection, and persistent activation of the ATR-Chk1 pathway [261]. The INO80 chromatin remodeler has also been implicated in stimulating RAD51 binding to resected DNA during HR repair. The authors delineated two distinct functions of the INO80 remodeler during HR—DSB end resection (see above) and presynaptic filament formation. The latter function depended on INO80-mediated removal of H2A.Z from damage sites, which promoted RAD51 loading [227]. A similar function has been reported for the p400 remodeling complex. p400 was found important for the recruitment of RAD51 to DSB sites and for survival after DNA damage. p400 and RAD51 were present in the same complex, and both favored chromatin remodeling around DSBs [262].
Data in yeast indicated that the SWI/SNF chromatin remodeler is recruited to HO endonuclease-induced DNA break site and participates in HR repair by remodeling nucleosomes at the donor locus, while another remodeler, RSC, is required following synapsis to complete the repair event [214].
Depletion of BRG1 (the catalytic subunit of mammalian SWI/SNF complexes) impaired HR repair as measured using reporter constructs specific to this repair pathway. The defect was attributed to defective RAD51 filament assembly and increased RPA retention, suggesting that BAF and/or PBAF may specifically promote the exchange of RPA with RAD51 on ssDNA [263].
Chromatin factors also modulate HR repair machinery via transcriptional effects. The histone lysine demethylase PHD finger protein 2 (PHF2) is deregulated in various cancers. PHF2 knockdown decreased CtIP and BRCA1 protein and mRNA levels. The authors also suggested that it might interfere with H4K20me2 and 53BP1 recruitment to DNA lesions [264]. In addition to its direct roles at DSBs, the NuRD complex affects RAD51 abundance transcriptionally. CHD4, a component of the complex, has recently been shown to be required for proper RAD51 gene transcription [265].
3.3.3. Chromatin Compaction in HR Repair
A number of chromatin changes that promote compaction are necessary for efficient HR repair.
The NuRD remodeling complex has been implicated in promoting HR repair by repressing transcription at DSBs [237,238] (Figure 3b). The complex has also been linked to the recruitment of a number of repair proteins, including BRCA1 [239]. The depletion of the NuRD subunit CHD4 affects the phosphorylation of the ssDNA-binding complex RPA, which is indicative of a defect in DNA end resection [266]. The association of NuRD itself to DSBs was found to depend on PARP activity [238,239,267]. More recent studies have established that NuRD is recruited to DNA breaks by Zinc Finger MYND Domain-Containing Protein 8 (ZMYND8) [268]. ZMYND8 contains a BRD domain, which binds to N-terminally acetylated histone H4 (mediated by TIP60), as well as a PHD and PWWP domains [269]. The N-terminal PHD-BRD-PWWP domains of ZMYND8 engage chromatin through multivalent interactions [270]. It has been reported that H3K4me3 demethylation near DSB sites carried out by the KDM5A demethylase is required for ZMYND8-NuRD binding to the lesions [198]. ZMYND8 binding is spatially controlled by another bromodomain protein—BRD2. BRD2 was found to be spatially restricted to a chromatin domain extending only 2 kb on either side of the DSB, and ZMYND8 spreads along the flanking chromatin but is excluded from the BRD2 region [271].
Another factor repressing transcription to promote HR repair is H2AK119 ubiquitination [195]. B Lymphoma Mo-MLV Insertion Region 1 protein (BMI1) and RNF2, as components of the Polycomb repressive complex 1 (PRC1), accumulate at DSB sites and carry out DNA damage-dependent H2AK119 monoubiquitination [272]. The major role of H2AK119 monoubiquitination is to transcriptionally silence genes. A reporter system visualizing transcription at DSB sites has shown that it was silenced locally following DSB induction [116]. Inhibition of both RNA polymerase I and II at DSB sites depended on RNF2/BMI-dependent monoubiquitination of H2AK119 [272] (as well as on ATM [273]). H2AK119ub was also dependent on PBAF (but not BAF complex) [117]. Surprisingly, the reversal of H2AK119ub by the BAP1 deubiquitinase was reported to promote homologous recombination. The authors suggested that the mark might be dynamic at damage sites [196]. The NuRD complex might also be involved in reversing H2AK119 ubiquitination. NuRD has been reported to interact with the deubiquitinase USP11 and to perform coordinated deubiquitination (by USP11) and deacetylation (by NuRD) at a DSB site [274].
Surprisingly, PRC1-mediated transcriptional repression also appears to stimulate c-NHEJ. Both BMI1 and its interactor eleven-nineteen leukemia protein (ENL)—a transcription elongation factor that is phosphorylated by ATM following DNA damage [202]—were required for the accumulation of Ku70 at DSBs; however, it is unclear if this is linked to PRC1-mediated transcription repression there. It has been suggested that silencing might be needed for retention of c-NHEJ factors, as transcription machinery could evict them [275]
It has been found that the PRC2 complex is also required for DSB-induced silencing of transcription and stimulation of repair [117]. Enhancer of Zeste Homolog 2 (EZH2) methyltransferase, a component of PRC2, is brought to sites of damage via the chromodomain Y-like protein 1 (CDYL1). CDYL1 recruitment to DSBs was reported to be PARP1-dependent, and its deficiency impaired HR repair [203]. Localization of EZH2 at DSBs by this mechanism increased the H3K27me3 repressive mark at DSBs.
H2A.Z, which, as discussed above, might promote resection, also represses transcription when it becomes incorporated into a gene body (but not when it is incorporated at a promoter [276]), suggesting that it leads to transcription silencing if a DSB arises in gene bodies.
It should be noted, however, that while chromatin factors silence transcription at DSBs, multiple recent reports suggest accumulation of RNA polymerase II at DSBs and initiation of transcription there. These DNA damage response RNAs regulate the early recruitment of crucial DSB signaling factors, including MDC1, RNF168, 53BP1, and RIF1, as well as the formation of K63-linked ubiquitin moieties, and future studies will have to reconcile the various aspects of transcriptional regulation in DSB repair [275].
3.4. Dynamics of DSB Repair in Living Cells
Although highly informative regarding the functions of individual DNA repair proteins and the interactions between them, in vitro studies cannot provide information about the exact time scale of DSB repair factor binding to damage sites. The temporal and spatial coordination between different DNA repair pathways can only be obtained by live-cell methods, such as live-cell imaging of the recruitment of fluorescently-labeled DNA repair proteins at laser-induced DNA damage sites [277,278,279]. A recent study reported the kinetics of recruitment and removal of 70 DNA repair proteins at laser-induced DNA damage sites in living HeLa Kyoto cells, including numerous proteins involved in DSB repair [161]. PARP1 and PARP2 are among the first proteins to be recruited to DNA damage sites (with halftimes of recruitment in the range between 2.5–5 s), and their binding to DNA lesions is required for the efficient and timely recruitment of downstream repair factors. HDAC1 and HDAC2 were also amid the fastest proteins recruited, with halftimes between 3.5 s and 5 s, indicating the need for rapid histone deacetylation following DNA damage induction. The chromatin remodeler proteins SPT16 (a subunit of the FACT complex), SNF2H (SMARCA5), and SMARCAD1 are recruited shortly after PARP1/2 and HDAC1/2 with halftimes of recruitment between 8 s and 23 s. These proteins, as discussed, have been implicated in the early chromatin remodeling events around DSBs. Early responders to DSBs, such as Ku70, LIG4, RAD50 (MRN complex), ATM, MDC1, and notably the histone chaperone CAF-1, were recruited immediately after the chromatin remodelers with halftimes of recruitment between 23 s and 60 s. These were followed by the binding of the RNF168 ubiquitin ligase (halftime of 78 s), which, as already discussed, is part of a complex ubiquitination cascade of the chromatin in the vicinity of the break that is responsible for the recruitment of downstream effector proteins. Interestingly, it was found that ubiquitin, which is recruited with a halftime of about 180 s, is the most abundant protein at laser-induced damage sites, highlighting once more the important role of chromatin ubiquitination in the organization and orchestration of DNA repair. Proteins implicated in the DSB repair pathway choice, such as 53BP1, PTIP (PAXIP1), RNF169, and BARD1, were recruited to DNA damage sites following the extensive ubiquitination of the chromatin with halftimes of recruitment between 200 s and 300 s. RPA1, s subunit of the RPA complex, RAD17, a specific subunit of the RAD17-RFC2-5 clamp loader complex, and RAD1, a subunit of the RAD9-RAD1-HUS1 DNA clamp that is loaded onto ssDNA by the RAD17-RFC2-5 complex and regulates resection, are recruited simultaneously (halftimes of recruitment between 600 s and 700 s) to laser-induced DNA damage sites with their presence indicating the implementation of 5′-end resection [280,281]. The latest proteins to be recruited (halftimes of over 900 s) are RAD51 and its associated factors, such as RAD51AP1 and RAD54B, which participate in the late steps of DSB repair by HR. Such type of studies can answer many pressing questions regarding the chronology and coordination of the numerous chromatin events that govern DSB repair. Importantly, such a systematic live-cell imaging approach may be employed to gain an in-depth knowledge of the mechanisms of action of DSB repair-targeting drugs (as has been shown for the PARP1/2 inhibitor BMN673) [161] and assess their potency in specific genetic context (e.g., BRCA1/2-deficiency).
4. Targeting the Chromatin Response to DSB DNA Damage
4.1. A New Hope: Synthetic Lethality and PARP1/2 Inhibitors
Overcoming the current flaws of anticancer chemotherapy necessitates the development of new therapeutic paradigms that take into account the specific genetic makeup of cancer cells leading to more personalized approaches to treating cancer patients. Such a novel therapeutic approach is the notion of synthetic lethality. It combines the consequences of specific genetic mutations in cancer cells with the pharmacological inhibition of a particular cellular protein, which results in a synergistic effect between these two events leading to cancer cell death [282,283]. Synthetic lethal interactions between pairs of proteins and/or pairs of pathways are especially prominent in DDR, which offers multiple redundant and/or interdependent pathways that, if restrained simultaneously, lead to profound cancer cell death [284].
The first drugs that act by the principle of synthetic lethality, and currently still the only approved synthetically-lethal drugs for clinical use in cancer patients, are the PARP1/2 inhibitors (PARPis) [284]. These drugs encompass small molecules that inhibit the enzymatic activity of PARP1 and PARP2. The PARP1 protein is an essential DNA repair protein that acts as a DNA damage sensor that recognizes single- and double-strand breaks in DNA (Figure 4a) [285,286]. Following its binding to the lesion, which is very rapid in living cells [161,287], PARP1 activates its enzymatic activity, which leads to the synthesis of long, negatively charged poly(ADP-ribose) chains on the nearby chromatin proteins. These PAR chains serve as binding sites for the recruitment of downstream DNA repair factors, which actually repair the lesion [79,286,288,289]. Importantly, PARP1 and PARylation exert significant remodeling effect on the chromatin in the vicinity of DSBs that is necessary for proper DNA repair [80]. PARP1 modifies nucleosomal histones causing nucleosome disassembly, which in turn leads to chromatin relaxation [290] (Figure 3b). In addition, several chromatin remodeling complexes possess PAR-binding motifs (PBMs) and bind to DNA damage sites in a PAR-dependent manner. Such remodelers are amplified in liver cancer protein 1 (ALC1) [234], chromodomain helicase DNA-binding protein 2 (CHD2) [235], CHD4—a subunit of the nucleosome remodeling and deacetylase complex (NuRD)—and Polycomb repressive complex 1 (PRC1) [237]. Interestingly, these chromatin remodelers exhibit opposing functions with respect to the chromatin changes they induce. ALC1 repositions nucleosomes in the chromatin surrounding the break, which increases the accessibility of the damaged site to downstream factors [234], while CHD2 deposits the histone variant H3.3 in the vicinity of DSBs resulting in chromatin relaxation that promotes c-NHEJ [235] (Figure 3b).
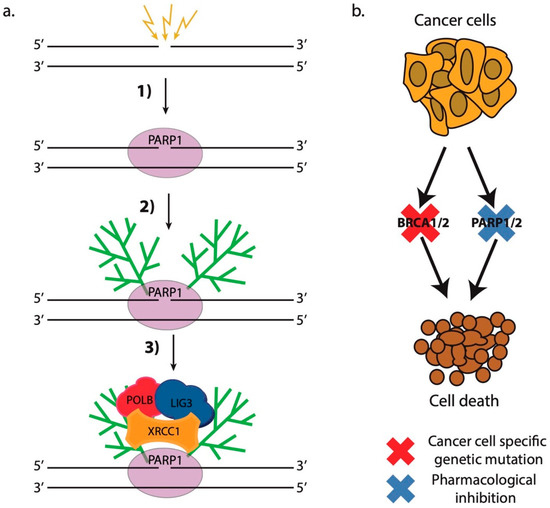
Figure 4.
PARP1 and synthetic lethality. (a) (1) PARP1 binds to single- and double-strand breaks in DNA; (2) Binding to DNA lesions stimulates PARP1 activity which results in the synthesis of long negatively charged chains of poly(ADP-ribose) (PAR) on the chromatin surrounding the break and on itself; (3) Multiple PAR-binding proteins, such as XRCC1, bind to the PAR chains and repair the lesion; (b) Cancer cell-specific genetic defects, which are not present in normal non-cancerous cells, such as BRCA1/2 mutations, may be exploited in combination with specific pharmacological inhibitors, such as PARP1/2 inhibitors, to specifically kill cancer cells.
In contrast, the NuRD and PRC1 complexes have well-defined functions as transcriptional repressors, and their recruitment to DSB sites may lead to shut down of transcription near DNA breaks, thereby preventing possible conflicts between transcription and DNA repair machinery [80,237] (Figure 3b).
PARPi have been shown to effectively kill cancer cells harboring gene mutations in BRCA1 and BRCA2 (Figure 4b) [291,292,293,294]. A significant number of breast and ovarian tumors, but also prostate and brain tumors, bear such mutations that make them prone to PARPi treatment [295]. PARP1/2 inhibitors not only inhibit PARP1 enzymatic activity, but they also change PARP1 conformation so that it becomes trapped onto the lesion following damage recognition hindering the faithful repair of the damaged region. It has been shown that PARPi cytotoxicity against BRCA1/2-deficient cancer cells correlates with their trapping potency and not with their inhibitor properties [296,297,298,299]. PARPi treatment leads to a significant rearrangement in the order of recruitment and dissociation of DNA repair proteins at complex DNA damage sites, altering the coordination of the DNA damage response and possibly the outcome of the repair process [161]. In addition, it has been shown that tumors that are defective with respect to other HR factors such as ATM, RAD51, PALB2, etc., are also prone to PARPi treatment [283,300]. Moreover, PARP1 has been implicated in the protection and stabilization of stalled replication forks due to replication stress—a function that gains extreme importance in BRCA1/2-deficient context and rationalizes once more the use of PARPi in treating BRCA-deficient cancer patients [80,301].
PARP1/2 inhibitors being superior to existing cancer treatment regimens for HR-deficient cancer patients, as clearly manifested in clinical trials, led to their approval for clinical use for the first time in 2014. Currently, there are four clinically-approved PARP1/2 inhibitors—olaparib (Lynparza®, Astra Zeneca), rucaparib (Rubraca®, Clovis), niraparib (Zejula®, Tesaro), and talazoparib (Talzenna®, Pfizer)—which, at least for now, are almost exclusively used for treating HR-deficient types of cancer or cancers that have responded well to previous therapy with platinum compounds [282,302]. As of the end of 2019, there are almost 200 ongoing clinical trials that assess the potency of the abovementioned PARPi, and many others are still not yet approved for clinical use (https://clinicaltrials.gov/). PARP1 facilitates not only the repair of single- and double-strand breaks but interacts with TDP1, which removes DNA topoisomerase I covalent complexes from DNA that are generated in the presence of topoisomerase inhibitors such as camptothecin and its derivatives (TOPccs, topoisomerase cleavage complexes) [300,303]. PARP1 associates with and PARylates TDP1, stimulating its enzymatic activity to eliminate transcription- and replication-blocking TOPccs. This justifies the most frequently used combinations used in the clinic–PARPi + temozolomide (a DNA alkylating agents) and PARPi + DNA topoisomerase I inhibitors [300].
Unfortunately, as has been shown for virtually all classes of anticancer drugs, cancer cells are able to evolve resistance. Currently, multiple mechanisms that could lead to PARPi resistance have been described, including BRCA1/2 reversal mutations [304,305], loss of non-homologous end joining factors such as 53BP1 [306], RIF1 [307,308] and subunits of the Shieldin complex [309,310,311,312], mutations in the PARP1 protein itself [313], loss of DYNLL1–an end resection inhibitor protein [314], and probably many more yet to be discovered [315]. All these resistance mechanisms heavily involve the chromatin landscape in the vicinity of the breaks, and understanding the dynamics of chromatin changes during DSB repair may provide novel avenues for combating PARPi resistance in the clinic.
PARP1 and BRCA1/2 is not the only synthetically lethal pair of genes linked to DSB repair that has been described. Another such pair is formed between Pol Θ and BRCA1/2. As described above, Pol Θ is a DNA polymerase that is essential for alternative end joining [316,317]. aEJ becomes a critical pathway for DSB repair in HR-deficient cells harboring mutations in BRCA1/2 (and possibly other HR genes); therefore, Pol Θ inhibition or knockdown significantly reduces cell survival in a BRCA1/2-deficient context [83,84]. The SWI/SNF chromatin remodelers are huge protein complexes that are vital for reorganizing chromatin in processes such as transcription, replication, and DNA repair [243]. Tumors harboring mutations in SNF5—a subunit of the SWI/SNF complex that exhibits transcriptional co-activator properties—are profoundly sensitive to the inhibition of EZH2—the enzymatic methyltransferase subunit of the PRC2 complex that represses active chromatin and drives epigenetic silencing. EZH2 leads to suppression of p16—a prominent tumor suppressor gene—in SNF5-deficient cancer cells [318]. Moreover, EZH2 levels are increased in SNF5-deficient cells. Hence, EZH2 inhibitors (EZH2i), such as EPZ-6438, have been shown to abrogate EZH2 activity and reactivate p16 expression in SNF5-deficient context both in vivo and in vitro significantly inhibiting their proliferation [318,319,320]. These results prompted the design and implementation of clinical trial studies to investigate the efficiency of EZH2i in treating SNF5-deficient cancer patients (https://clinicaltrials.gov). Undoubtedly, with the advancement of more elaborate functional genomics approaches such as CRISPRi screens, more synthetically lethal pairs of genes will be uncovered, driving the development of a new generation of anticancer drugs that are more effective and tailored to the specific mutational context of the cancer patients [284].
4.2. Targeting Chromatin Regulators
The complicated nature of the chromatin response to DSBs offers multiple possibilities for drug targets. In the following paragraphs, we provide examples of targeting the chromatin regulators that participate in DSB repair.
Early studies of ATM and ATR have shown that caffeine sensitizes cells to genotoxic agents [321,322] and those with p53 defects were particularly sensitive [323,324,325]. However, the doses required for radiosensitization cannot be achieved in patients [326]. Similarly, wortmannin, a pan-PI3K inhibitor [327], is a potent radiosensitizer [328], but lacks selectivity and has too high in vivo toxicity [329] to be useful as a drug.
The first potent and selective ATM inhibitor was KU-55933. KU-55933 sensitized cancer cells to DSB-inducing chemotherapeutics, such as the topoisomerase II inhibitors etoposide and doxorubicin as well as ionizing radiation. In cells without functional ATM, radiosensitization was not observed [330]. A similar inhibitor, KU-60019, sensitized to irradiation highly resistant glioblastoma cell lines [331,332]. KU59403 was the first compound to achieve tissue distribution with concentrations exceeding those required for in vitro activity. It caused significant chemosensitization in in vivo models of human cancer [333].
ATM is also increasingly being recognized as a target for synthetic lethality with PARP inhibitors. The ATM inhibition combined with PARP1 depletion or inhibition caused a massive synergistic increase of DNA damage suggesting the effectiveness of combined treatments [334].
Very few tumors harbor ATR mutations. Increasing evidence indicates that high levels of replication stress in tumors make them particularly vulnerable to the loss of ATR [335].
In spite of concerns that ATR inhibition could cause severe side effects in highly proliferative normal tissues, studies suggest that ATR inhibition might be cytotoxic specifically to cancer cells, especially in p53-defective backgrounds [336,337]. The reduction of ATR expression to 10% of normal levels was synthetically lethal in Ras-driven tumors, while normal bone marrow and intestinal cells were minimally affected [338]. Schisandrin B, a naturally-occurring compound and the active ingredient of Fructus schisandrae, was reported as a selective inhibitor of ATR [339]. A compound called VE-821 was shown to be a potent inhibitor of ATR with minimal cross-reactivity (>600-fold selectivity over ATM or DNA-PKcs) [340]. It showed strong synergy with genotoxic agents and ionizing radiation in HCT116 cancer cells. Not surprisingly, synergy was most pronounced with the cross-linking drugs cisplatin and carboplatin [341]. A related compound, VE-822, markedly increased the sensitivity of ATM-deficient lung adenocarcinoma cells leading the authors to propose that ATM deficiency could be useful as a marker for ATR and PARP sensitivity [342]. In a cellular model of oxaliplatin resistance, VE-822 successfully overcame the drug resistance [343].
AZD6738, an orally available inhibitor of both ATR and Chk1 [344], showed synthetic lethality in various cancer cell lines [345,346], including in combination with the WEE1 inhibitor AZD1775, causing mitotic catastrophe in triple-negative breast cancer cell lines [347].
An ATR inhibitor, AY1895344, developed by Bayer, showed strong monotherapy efficacy in cancer xenograft models that carry DNA damage repair deficiencies and a strong synergistic anti-proliferative activity in vitro in combination with olaparib [348].
NU5455, a novel highly selective oral inhibitor of DNA-PKcs, increased the efficacy of a parenterally administered topoisomerase inhibitor in liver tumor xenografts without inducing any adverse effects. It augmented the effect of targeted radiotherapy of lung tumors and helped to avoid inflicting acute DNA damage and toxicity to normal surrounding tissues [349]. For a detailed review of PI3K inhibitors, including natural compounds, the reader is referred to [350].
Histone deacetylase inhibitors (HDACi) are also regarded as promising anticancer drugs. They were initially described as compounds that induced tumor cell differentiation [351,352]. The effects of HDAC inhibition depend on altered histone acetylation or acetylation of DSB repair factors, but mostly on expression changes. Trichostatin A (TSA) and suberoylanilide hydroxamic acid (SAHA) belong to the same group of pan-HDAC inhibitors and have been shown to downregulate DSB repair factors [353,354,355,356] with concomitant radiosensitization. Other HDAC inhibitors—valproic acid [357], panobinostat [358], MS-275 [359], and entinostat [360]—inhibited the expression of DSB repair and signaling proteins sensitizing cellular models of various tumors.
HDAC inhibitors also affect the DNA damage response in non-transcription-related ways. TSA treatment significantly increases PARP1 binding to DSBs, resembling PARP “trapping.” The knockdown of PARP1 mitigated the inhibitory effect of HDACi on c-NHEJ. These results indicate that a combination of HDACi and PARP1 inhibition might augment the cytotoxicity in leukemia cells by impairing c-NHEJ [361]. Sirtuin-type HDACs affect HR repair by directly deacetylating resection machinery—SIRT1 deacetylates NBS1, which stimulates HR repair [362]. As already noted, TSA and sodium butyrate led to longer persistence of GFP-tagged Ku70 and Artemis at micro-irrigation sites [245].
HDAC3 inhibition using a first-in-class selective inhibitor, RGFP966, resulted in increased apoptosis in cutaneous T cell lymphoma cell lines. These effects were associated with DNA damage and impaired S-phase progression. Isolation of proteins on nascent DNA (iPOND) indicated that HDAC3 associated with chromatin around replication forks and its inhibition resulted in a significant reduction in DNA replication fork velocity [363].
HDACi are used to treat other diseases as well [364,365] and their application should go along with the understanding that they disrupt genome integrity and may promote cellular transformation [366,367,368,369].
Several known compounds of plant origin inhibit histone acetyltransferases. Anacardic acid is a bioactive phytochemical, structurally related to salicylic acid, isolated from the shell of Anacardium occidentale nuts. It is a noncompetitive inhibitor of p300, PCAF, and TIP60 [370,371], which are involved in both HR and c-NHEJ [189,192,372]. As expected, anacardic acid sensitizes cells to ionizing radiation [373]; however, its low cell permeability limits its practical applications [374]. Garcinol targets p300 and PCAF [375] causing radiosensitization [376], and curcumin, found in the spice turmeric, is a specific inhibitor of CBP and p300 [377]. All three inhibited c-NHEJ when assessed by a chromosomally integrated reporter construct [189,378]. Curcumin also reduced the expression of BRCA1 by changes to the BRCA1 promoter and was also shown to inhibit the ATR kinase [378]. Due to poor bio-absorption, several analogs of curcumin have been developed. One of these analogs—H-4073, enhanced the therapeutic efficacy of cisplatin [379]. A novel low molecular weight compound, C646, is a selective HAT inhibitor that binds p300 and induces apoptosis in prostate [380] and gastric [381] cancer cell lines.
Targeting enzymes that mediate methylation in the course of DSB repair is significantly less explored. In mammalian cells, DOT1L (KMT4) methylates H3K79. The interest to develop inhibitors of DOT1L is fueled by the fact that this enzyme is a key factor in MLL-rearranged leukemia, where the MLL gene is rearranged to fuse with several different genes [382], and these fusions aberrantly recruit DOT1L enhancing the expression of genes required for pathogenesis [383]. PZ004777—a selective inhibitor of DOT1L—blocked the expression of MLL-fusion target genes, and displayed anti-proliferative effects [384]. Although the effects of this drug on DNA repair have not been directly studied, existing data suggest synergistic anti-proliferative activity in combination with chemotherapy compounds, including topoisomerase inhibitors [385]. The interest in developing novel DOT1L inhibitors is likely to increase [386], given that they might be effective in diseases other than MLL [387,388,389].
H4K20 monomethylation by SETD8 could be inhibited by nahuoic acid A—a polyketide produced in culture by Streptomyces sp. obtained from tropical marine sediment and the first known selective inhibitor of SETD8 [390]. It has also been reported the discovery of UNC0379, the first substrate-competitive inhibitor of SETD8, selective for SETD8 over 15 other methyltransferases [391].
Recently, A-196, a potent and selective inhibitor of SUV420H1 and SUV420H2, has been identified as a substrate-competitive inhibitor of both enzymes. In cells, A-196 induced a global decrease in H4K20me2 and H4K20me3 and a concomitant increase in H4K20me1. A-196 inhibited 53BP1 foci formation upon ionizing radiation and reduced NHEJ-mediated DNA-break repair [392]. The development of two other compounds that could markedly reduce H4K20me2 in a dose-dependent manner has been reported [393].
Another histone methyltransferase actively targeted for inhibitor development is EZH2, a subunit of the PRC2 complex, responsible for the majority of genome-wide H3K27me3 trimethylation [394]. EZH2 is overexpressed in prostate, breast, kidney, and lung cancers in which EZH2 up-regulation induces cell migration, colony formation, and genomic instability [395]. Gain-of-function mutations were also described in follicular and diffuse large B-cell lymphomas. These have led to the development of several inhibitors, such as GSK126, EPZ005687, and EPZ-6438 [395]. Although there are no data if these affect DSB repair, as already discussed, PRC2 is recruited to DSBs in a PARP-dependent way, and loss of its components sensitized cells to ionizing radiation [237,396] suggesting that EZH2 inhibitors may be effective in combination with DSB-inducing chemotherapeutics and ionizing radiation.
BMI1 (a subunit of both PRC1 and PRC2 complexes) is overexpressed in pancreatic cancer, glioblastoma multiforme, pediatric diffuse intrinsic pontine glioma, colorectal cancer, epithelial ovarian cancer, and acute myeloid leukemia [397]. It has been found that BMI1 is necessary for the maintenance of chemoresistant stem cells capable of re-establishing disease. The BMI1 inhibitor PTC596 induced apoptosis in a p53-independent manner [398]. It is unknown if it would synergize with treatments that induce DSB breaks or stall replication forks. Similarly, it would be of interest to study the combined effects of genotoxic treatments and the recently described small-molecule orally-available inhibitors of the SWI/SNF proteins Brm1 and Brg1, given the various effects of the latter in DSB repair and replication fork stability [399].
While the list of inhibitors and targets is by no means complete, it does give a sense of the vast spectrum of opportunities for therapeutic modulation of the DNA damage response. There is no doubt that the future will see more promising and, above all, clinically applicable approaches harnessing the knowledge of the chromatin response to DNA damage.
5. Conclusions and Future Perspectives
Double-strand DNA breaks are among the most perilous events that cells and organisms have to cope with. Accordingly, DSB repair mechanisms, which counteract the grievous consequences of DSBs, are vastly complex cellular pathways that recognize and repair this type of lesion. The DNA damage response operates in the context of chromatin, and the ever-changing chromatin environment exerts significant control over it. Proficient DSB repair is dependent on hundreds of different DNA repair proteins that act in a concerted fashion, most of which are recruited and regulated by specific chromatin changes such as histone modifications and chromatin remodeling events. Although much is known about the activities and the interactions between these proteins and the chromatin, the precise dynamics and coordination of the events during DSB repair is largely an open question. Moreover, different cells and tissues in the body or cells with specific genetic background, such as BRCA1/2-deficient cancer cells, may accomplish DSB repair in different manners that are adapted to their specific needs [400,401,402]. The advent and implication of novel techniques, such as laser micro-irradiation followed by live-cell imaging [161] or CRISPR-guided DSB induction in living cells at specific loci complemented with live-cell imaging of DNA repair protein recruitment [403], may provide crucial new knowledge and shed light on unanticipated events during DSB repair. Functional genomics approaches such as CRISPR screens may uncover novel DSB repair factors and expose interdependent sets of events in the course of DSB repair [32,312].
In the past decade, targeting of specific players in the DNA damage response has turned into a promising avenue for the introduction of novel therapies for cancer patients. The first PARP1/2 inhibitors—the first class of anticancer drugs that employ the tactics of synthetic lethality—have already hit the clinic. Many more DDR-targeting drugs are in various phases of clinical trials, such as ATM, ATR, and DNA-PKcs inhibitors. Molecules that suppress certain histone-modifying enzymes such as HDAC inhibitors have proven efficient in the treatment of various types of cancer in the clinic and are pursued as therapy in other non-cancer-related conditions. Although the specific biochemical effects of these molecules are well investigated, much less is known about their systematic impact on the dynamics and coordination of DSB repair pathways. Moreover, the efficient reorganization and implementation of DDR in cancer patients following chemotherapy with DNA damage-inducing anticancer drugs is one of the most important reasons behind the evolution of resistance, which is often seen in the clinic. Given the complexity of the DDR, many resistance mechanisms that rely on it have been described. To deal with this prominent problem and offer cancer patients more beneficial and sparing chemotherapy regimens, novel drug targets must be uncovered. Recent years have seen tremendous progress in the discovery of novel DSB protein factors and their functions, largely due to the implementation of cutting-edge high-throughput methods. CRISPR screens have identified numerous synthetically lethal pairs of genes that can be targeted in various cancer-specific contexts [284]. Tens, if not hundreds, of DDR-targeting molecules are pursued for clinical use, and there can be no doubt that at least some of them will reach the clinic carrying benefit for cancer patients.
Funding
R.A. was supported by the National Scientific Program “Young Scientists and Postdoctoral Fellows” by the Ministry of Education. R.A., R.H., S.S., and A.G. acknowledge the support of the Bulgarian National Science Fund grant# K06-N21-9 to S.S. and DN11/17 to A.G. The research work of R.A., R.H., S.S., and A.G. is also supported by the National Scientific Program BioActivMed (D01-217) by the Bulgarian Ministry of Education.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Friedberg, E.C. A brief history of the DNA repair field. Cell Res. 2008, 18, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef]
- Lindahl, T.; Barnes, D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Unsal-Kacmaz, K.; Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004, 73, 39–85. [Google Scholar] [CrossRef] [PubMed]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef]
- Peterson, C.L.; Almouzni, G. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb. Perspect. Biol. 2013, 5. [Google Scholar] [CrossRef]
- Price, B.D.; D’Andrea, A.D. Chromatin remodeling at DNA double-strand breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e236. [Google Scholar] [CrossRef]
- Madhusudan, S.; Hickson, I.D. DNA repair inhibition: A selective tumour targeting strategy. Trends Mol. Med. 2005, 11, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Raghavan, S.C. DNA double-strand break repair inhibitors as cancer therapeutics. Chem. Biol. 2015, 22, 17–29. [Google Scholar] [CrossRef]
- Cadet, J.; Douki, T.; Gasparutto, D.; Ravanat, J.L. Oxidative damage to DNA: Formation, measurement and biochemical features. Mutat. Res. 2003, 531, 5–23. [Google Scholar] [CrossRef]
- Stingele, J.; Jentsch, S. DNA-protein crosslink repair. Nat. Rev. Mol. Cell Biol. 2015, 16, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Poulsen, H. Measurement of oxidatively generated base damage in cellular DNA and urine. Free Radic. Biol. Med. 2010, 48, 1457–1459. [Google Scholar] [CrossRef] [PubMed]
- Winterbourn, C.C. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008, 4, 278–286. [Google Scholar] [CrossRef]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef]
- Scully, R.; Panday, A.; Elango, R.; Willis, N.A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019, 20, 698–714. [Google Scholar] [CrossRef]
- Sankar, T.S.; Wastuwidyaningtyas, B.D.; Dong, Y.; Lewis, S.A.; Wang, J.D. The nature of mutations induced by replication–transcription collisions. Nature 2016, 535, 178–181. [Google Scholar] [CrossRef]
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef]
- Lippert, M.J.; Kim, N.; Cho, J.E.; Larson, R.P.; Schoenly, N.E.; O’Shea, S.H.; Jinks-Robertson, S. Role for topoisomerase 1 in transcription-associated mutagenesis in yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 698–703. [Google Scholar] [CrossRef]
- Soulas-Sprauel, P.; Rivera-Munoz, P.; Malivert, L.; Le Guyader, G.; Abramowski, V.; Revy, P.; de Villartay, J.P. V(D)J and immunoglobulin class switch recombinations: A paradigm to study the regulation of DNA end-joining. Oncogene 2007, 26, 7780–7791. [Google Scholar] [CrossRef] [PubMed]
- de Massy, B. Initiation of meiotic recombination: How and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013, 47, 563–599. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Mahato, D.K.; Kamle, M.; Mohanta, T.K.; Kang, S.G. Aflatoxins: A Global Concern for Food Safety, Human Health and Their Management. Front. Microbiol. 2016, 7, 2170. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Denissenko, M.F.; Olivier, M.; Tretyakova, N.; Hecht, S.S.; Hainaut, P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene 2002, 21, 7435–7451. [Google Scholar] [CrossRef] [PubMed]
- Hodskinson, M.R.; Bolner, A.; Sato, K.; Kamimae-Lanning, A.N.; Rooijers, K.; Witte, M.; Mahesh, M.; Silhan, J.; Petek, M.; Williams, D.M.; et al. Alcohol-derived DNA crosslinks are repaired by two distinct mechanisms. Nature 2020, 579, 603–608. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016, 19, 1–9. [Google Scholar] [CrossRef]
- Helleday, T.; Petermann, E.; Lundin, C.; Hodgson, B.; Sharma, R.A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. [Google Scholar] [CrossRef]
- Ali, S.; Coombes, R.C. Endocrine-responsive breast cancer and strategies for combating resistance. Nat. Rev. Cancer 2002, 2, 101–112. [Google Scholar] [CrossRef]
- Maginn, E.N.; de Sousa, C.H.; Wasan, H.S.; Stronach, E.A. Opportunities for translation: Targeting DNA repair pathways in pancreatic cancer. Biochim. Biophys. Acta 2014, 1846, 45–54. [Google Scholar] [CrossRef]
- Pettitt, S.J.; Lord, C.J. Dissecting PARP inhibitor resistance with functional genomics. Curr. Opin. Genet. Dev. 2019, 54, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Britton, S.; Coates, J.; Jackson, S.P. A new method for high-resolution imaging of Ku foci to decipher mechanisms of DNA double-strand break repair. J. Cell Biol. 2013, 202, 579–595. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Lu, H.; Tippin, B.; Shimazaki, N.; Goodman, M.F.; Lieber, M.R. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007, 26, 1010–1023. [Google Scholar] [CrossRef] [PubMed]
- Meek, K.; Dang, V.; Lees-Miller, S.P. DNA-PK: The means to justify the ends? Adv. Immunol. 2008, 99, 33–58. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Grawunder, U.; Wilm, M.; Wu, X.; Kulesza, P.; Wilson, T.E.; Mann, M.; Lieber, M.R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 1997, 388, 492–495. [Google Scholar] [CrossRef]
- Brouwer, I.; Sitters, G.; Candelli, A.; Heerema, S.J.; Heller, I.; de Melo, A.J.; Zhang, H.; Normanno, D.; Modesti, M.; Peterman, E.J.; et al. Sliding sleeves of XRCC4-XLF bridge DNA and connect fragments of broken DNA. Nature 2016, 535, 566–569. [Google Scholar] [CrossRef]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef]
- Goodarzi, A.A.; Yu, Y.; Riballo, E.; Douglas, P.; Walker, S.A.; Ye, R.; Härer, C.; Marchetti, C.; Morrice, N.; Jeggo, P.A.; et al. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. EMBO J. 2006, 25, 3880–3889. [Google Scholar] [CrossRef]
- Grundy, G.J.; Rulten, S.L.; Zeng, Z.; Arribas-Bosacoma, R.; Iles, N.; Manley, K.; Oliver, A.; Caldecott, K.W. APLF promotes the assembly and activity of non-homologous end joining protein complexes. EMBO J. 2013, 32, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Iles, N.; Rulten, S.; El-Khamisy, S.F.; Caldecott, K.W. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 2007, 27, 3793–3803. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, N.K.; Williams, R.S.; Rakovszky, M.L.; Cui, D.; Green, R.; Karimi-Busheri, F.; Mani, R.S.; Galicia, S.; Koch, C.A.; Cass, C.E.; et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell 2005, 17, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Flett, F.J.; Ruksenaite, E.; Armstrong, L.A.; Bharati, S.; Carloni, R.; Morris, E.R.; Mackay, C.L.; Interthal, H.; Richardson, J.M. Structural basis for DNA 3’-end processing by human tyrosyl-DNA phosphodiesterase 1. Nat. Commun. 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Ahel, I.; Rass, U.; El-Khamisy, S.F.; Katyal, S.; Clements, P.M.; McKinnon, P.J.; Caldecott, K.W.; West, S.C. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 2006, 443, 713–716. [Google Scholar] [CrossRef]
- Nick McElhinny, S.A.; Havener, J.M.; Garcia-Diaz, M.; Juárez, R.; Bebenek, K.; Kee, B.L.; Blanco, L.; Kunkel, T.A.; Ramsden, D.A. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell 2005, 19, 357–366. [Google Scholar] [CrossRef]
- Symington, L.S.; Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011, 45, 247–271. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.; Tainer, J.A. The MRE11-RAD50-NBS1 Complex Conducts the Orchestration of Damage Signaling and Outcomes to Stress in DNA Replication and Repair. Annu. Rev. Biochem 2018, 87, 263–294. [Google Scholar] [CrossRef]
- Cannavo, E.; Cejka, P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 2014, 514, 122–125. [Google Scholar] [CrossRef]
- Garcia, V.; Phelps, S.E.; Gray, S.; Neale, M.J. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 2011, 479, 241–244. [Google Scholar] [CrossRef]
- Limbo, O.; Chahwan, C.; Yamada, Y.; de Bruin, R.A.; Wittenberg, C.; Russell, P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol. Cell 2007, 28, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Uziel, T.; Lerenthal, Y.; Moyal, L.; Andegeko, Y.; Mittelman, L.; Shiloh, Y. Requirement of the MRN complex for ATM activation by DNA damage. EMBO J. 2003, 22, 5612–5621. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Pilch, D.R.; Orr, A.H.; Ivanova, V.S.; Bonner, W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998, 273, 5858–5868. [Google Scholar] [CrossRef] [PubMed]
- Lukas, C.; Melander, F.; Stucki, M.; Falck, J.; Bekker-Jensen, S.; Goldberg, M.; Lerenthal, Y.; Jackson, S.P.; Bartek, J.; Lukas, J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004, 23, 2674–2683. [Google Scholar] [CrossRef] [PubMed]
- Stucki, M.; Clapperton, J.A.; Mohammad, D.; Yaffe, M.B.; Smerdon, S.J.; Jackson, S.P. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 2005, 123, 1213–1226. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Durocher, D. Push back to respond better: Regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 2013, 14, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Thorslund, T.; Ripplinger, A.; Hoffmann, S.; Wild, T.; Uckelmann, M.; Villumsen, B.; Narita, T.; Sixma, T.K.; Choudhary, C.; Bekker-Jensen, S.; et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 2015, 527, 389–393. [Google Scholar] [CrossRef]
- Doil, C.; Mailand, N.; Bekker-Jensen, S.; Menard, P.; Larsen, D.H.; Pepperkok, R.; Ellenberg, J.; Panier, S.; Durocher, D.; Bartek, J.; et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009, 136, 435–446. [Google Scholar] [CrossRef]
- Mattiroli, F.; Vissers, J.H.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef]
- Kim, H.; Chen, J.; Yu, X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 2007, 316, 1202–1205. [Google Scholar] [CrossRef] [PubMed]
- Sobhian, B.; Shao, G.; Lilli, D.R.; Culhane, A.C.; Moreau, L.A.; Xia, B.; Livingston, D.M.; Greenberg, R.A. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 2007, 316, 1198–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Matsuoka, S.; Ballif, B.A.; Zhang, D.; Smogorzewska, A.; Gygi, S.P.; Elledge, S.J. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 2007, 316, 1194–1198. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.H.; Hiom, K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature 2009, 459, 460–463. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 2008, 455, 770–774. [Google Scholar] [CrossRef]
- Nimonkar, A.V.; Ozsoy, A.Z.; Genschel, J.; Modrich, P.; Kowalczykowski, S.C. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. USA 2008, 105, 16906–16911. [Google Scholar] [CrossRef]
- Nimonkar, A.V.; Genschel, J.; Kinoshita, E.; Polaczek, P.; Campbell, J.L.; Wyman, C.; Modrich, P.; Kowalczykowski, S.C. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011, 25, 350–362. [Google Scholar] [CrossRef]
- Zhu, Z.; Chung, W.H.; Shim, E.Y.; Lee, S.E.; Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 2008, 134, 981–994. [Google Scholar] [CrossRef]
- Chen, H.; Lisby, M.; Symington, L.S. RPA coordinates DNA end resection and prevents formation of DNA hairpins. Mol. Cell 2013, 50, 589–600. [Google Scholar] [CrossRef]
- Carreira, A.; Kowalczykowski, S.C. Two classes of BRC repeats in BRCA2 promote RAD51 nucleoprotein filament function by distinct mechanisms. Proc. Natl. Acad. Sci. USA 2011, 108, 10448–10453. [Google Scholar] [CrossRef]
- Esashi, F.; Galkin, V.E.; Yu, X.; Egelman, E.H.; West, S.C. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007, 14, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Renkawitz, J.; Lademann, C.A.; Jentsch, S. Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 2014, 15, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Zhao, L.; Xu, Y.; Zhao, W.; Sung, P.; Wang, H.W. Cryo-EM structures of human RAD51 recombinase filaments during catalysis of DNA-strand exchange. Nat. Struct. Mol. Biol. 2017, 24, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. BRCA1-BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef]
- Kane, D.P.; Shusterman, M.; Rong, Y.; McVey, M. Competition between replicative and translesion polymerases during homologous recombination repair in Drosophila. PLoS Genet. 2012, 8, e1002659. [Google Scholar] [CrossRef] [PubMed]
- Myler, L.R.; Gallardo, I.F.; Soniat, M.M.; Deshpande, R.A.; Gonzalez, X.B.; Kim, Y.; Paull, T.T.; Finkelstein, I.J. Single-Molecule Imaging Reveals How Mre11-Rad50-Nbs1 Initiates DNA Break Repair. Mol. Cell 2017, 67, 891–898.e894. [Google Scholar] [CrossRef]
- Robert, I.; Dantzer, F.; Reina-San-Martin, B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 2009, 206, 1047–1056. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wu, W.; Wu, W.; Rosidi, B.; Zhang, L.; Wang, H.; Iliakis, G. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006, 34, 6170–6182. [Google Scholar] [CrossRef]
- Gibson, B.A.; Kraus, W.L. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012, 13, 411–424. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Wyatt, D.W.; Feng, W.; Conlin, M.P.; Yousefzadeh, M.J.; Roberts, S.A.; Mieczkowski, P.; Wood, R.D.; Gupta, G.P.; Ramsden, D.A. Essential Roles for Polymerase θ-Mediated End Joining in the Repair of Chromosome Breaks. Mol. Cell 2016, 63, 662–673. [Google Scholar] [CrossRef] [PubMed]
- Masani, S.; Han, L.; Meek, K.; Yu, K. Redundant function of DNA ligase 1 and 3 in alternative end-joining during immunoglobulin class switch recombination. Proc. Natl. Acad. Sci. USA 2016, 113, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Ceccaldi, R.; Liu, J.C.; Amunugama, R.; Hajdu, I.; Primack, B.; Petalcorin, M.I.; O’Connor, K.W.; Konstantinopoulos, P.A.; Elledge, S.J.; Boulton, S.J.; et al. Homologous-recombination-deficient tumours are dependent on Polθ-mediated repair. Nature 2015, 518, 258–262. [Google Scholar] [CrossRef] [PubMed]
- Mateos-Gomez, P.A.; Gong, F.; Nair, N.; Miller, K.M.; Lazzerini-Denchi, E.; Sfeir, A. Mammalian polymerase θ promotes alternative NHEJ and suppresses recombination. Nature 2015, 518, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Symington, L.S. Mechanism and regulation of DNA end resection in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Mimitou, E.P.; Symington, L.S. DNA end resection--unraveling the tail. DNA Repair 2011, 10, 344–348. [Google Scholar] [CrossRef]
- Symington, L.S. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002, 66, 630–670. [Google Scholar] [CrossRef]
- Mendez-Dorantes, C.; Bhargava, R.; Stark, J.M. Repeat-mediated deletions can be induced by a chromosomal break far from a repeat, but multiple pathways suppress such rearrangements. Genes Dev. 2018, 32, 524–536. [Google Scholar] [CrossRef]
- Setiaputra, D.; Durocher, D. Shieldin-the protector of DNA ends. EMBO Rep. 2019, 20. [Google Scholar] [CrossRef]
- Tarsounas, M.; Sung, P. The antitumorigenic roles of BRCA1-BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 2020, 21, 284–299. [Google Scholar] [CrossRef]
- Daley, J.M.; Sung, P. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014, 34, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Andegeko, Y.; Moyal, L.; Mittelman, L.; Tsarfaty, I.; Shiloh, Y.; Rotman, G. Nuclear retention of ATM at sites of DNA double strand breaks. J. Biol. Chem. 2001, 276, 38224–38230. [Google Scholar] [CrossRef] [PubMed]
- Bekker-Jensen, S.; Lukas, C.; Kitagawa, R.; Melander, F.; Kastan, M.B.; Bartek, J.; Lukas, J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006, 173, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Paull, T.T. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007, 26, 7741–7748. [Google Scholar] [CrossRef] [PubMed]
- Carson, C.T.; Schwartz, R.A.; Stracker, T.H.; Lilley, C.E.; Lee, D.V.; Weitzman, M.D. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003, 22, 6610–6620. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Chen, S.; Fernandes, N.; Price, B.D. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. USA 2005, 102, 13182–13187. [Google Scholar] [CrossRef]
- Ayrapetov, M.K.; Gursoy-Yuzugullu, O.; Xu, C.; Xu, Y.; Price, B.D. DNA double-strand breaks promote methylation of histone H3 on lysine 9 and transient formation of repressive chromatin. Proc. Natl. Acad. Sci. USA 2014, 111, 9169–9174. [Google Scholar] [CrossRef]
- Young, L.C.; McDonald, D.W.; Hendzel, M.J. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 2013, 288, 21376–21388. [Google Scholar] [CrossRef]
- Khoury-Haddad, H.; Guttmann-Raviv, N.; Ipenberg, I.; Huggins, D.; Jeyasekharan, A.D.; Ayoub, N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc. Natl. Acad. Sci. USA 2014, 111, E728–E737. [Google Scholar] [CrossRef]
- Gupta, A.; Sharma, G.G.; Young, C.S.; Agarwal, M.; Smith, E.R.; Paull, T.T.; Lucchesi, J.C.; Khanna, K.K.; Ludwig, T.; Pandita, T.K. Involvement of human MOF in ATM function. Mol. Cell. Biol. 2005, 25, 5292–5305. [Google Scholar] [CrossRef]
- Kim, Y.C.; Gerlitz, G.; Furusawa, T.; Catez, F.; Nussenzweig, A.; Oh, K.S.; Kraemer, K.H.; Shiloh, Y.; Bustin, M. Activation of ATM depends on chromatin interactions occurring before induction of DNA damage. Nat. Cell Biol. 2009, 11, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Matsuoka, S.; Ballif, B.A.; Smogorzewska, A.; McDonald, E.R., 3rd; Hurov, K.E.; Luo, J.; Bakalarski, C.E.; Zhao, Z.; Solimini, N.; Lerenthal, Y.; et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007, 316, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Fan, X.; Paul-Konietzko, K.; Soni, A.; Iliakis, G. DNA-PKcs and ATM epistatically suppress DNA end resection and hyperactivation of ATR-dependent G(2)-checkpoint in S-phase irradiated cells. Sci. Rep. 2019, 9, 14597. [Google Scholar] [CrossRef] [PubMed]
- Mladenov, E.; Fan, X.; Dueva, R.; Soni, A.; Iliakis, G. Radiation-dose-dependent functional synergisms between ATM, ATR and DNA-PKcs in checkpoint control and resection in G(2)-phase. Sci. Rep. 2019, 9, 8255. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y.; Shema, E.; Moyal, L.; Oren, M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS Lett. 2011, 585, 2795–2802. [Google Scholar] [CrossRef]
- Feng, J.; Shen, W.H. Dynamic regulation and function of histone monoubiquitination in plants. Front. Plant Sci. 2014, 5, 83. [Google Scholar] [CrossRef]
- Moyal, L.; Lerenthal, Y.; Gana-Weisz, M.; Mass, G.; So, S.; Wang, S.Y.; Eppink, B.; Chung, Y.M.; Shalev, G.; Shema, E.; et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell 2011, 41, 529–542. [Google Scholar] [CrossRef]
- So, C.C.; Ramachandran, S.; Martin, A. E3 Ubiquitin Ligases RNF20 and RNF40 Are Required for Double-Stranded Break (DSB) Repair: Evidence for Monoubiquitination of Histone H2B Lysine 120 as a Novel Axis of DSB Signaling and Repair. Mol. Cell. Biol. 2019, 39. [Google Scholar] [CrossRef]
- Ramachandran, S.; Haddad, D.; Li, C.; Le, M.X.; Ling, A.K.; So, C.C.; Nepal, R.M.; Gommerman, J.L.; Yu, K.; Ketela, T.; et al. The SAGA Deubiquitination Module Promotes DNA Repair and Class Switch Recombination through ATM and DNAPK-Mediated γH2AX Formation. Cell Rep. 2016, 15, 1554–1565. [Google Scholar] [CrossRef]
- Nakamura, K.; Kato, A.; Kobayashi, J.; Yanagihara, H.; Sakamoto, S.; Oliveira, D.V.; Shimada, M.; Tauchi, H.; Suzuki, H.; Tashiro, S.; et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 2011, 41, 515–528. [Google Scholar] [CrossRef]
- Kato, A.; Komatsu, K. RNF20-SNF2H Pathway of Chromatin Relaxation in DNA Double-Strand Break Repair. Genes 2015, 6, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Toiber, D.; Erdel, F.; Bouazoune, K.; Silberman, D.M.; Zhong, L.; Mulligan, P.; Sebastian, C.; Cosentino, C.; Martinez-Pastor, B.; Giacosa, S.; et al. SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Mol. Cell 2013, 51, 454–468. [Google Scholar] [CrossRef] [PubMed]
- Ziv, Y.; Bielopolski, D.; Galanty, Y.; Lukas, C.; Taya, Y.; Schultz, D.C.; Lukas, J.; Bekker-Jensen, S.; Bartek, J.; Shiloh, Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 2006, 8, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Noon, A.T.; Deckbar, D.; Ziv, Y.; Shiloh, Y.; Löbrich, M.; Jeggo, P.A. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol. Cell 2008, 31, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Goodarzi, A.A.; Kurka, T.; Jeggo, P.A. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 2011, 18, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010, 141, 970–981. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Ismail, A.; Chambers, A.L.; Riballo, E.; Herbert, A.D.; Künzel, J.; Löbrich, M.; Jeggo, P.A.; Downs, J.A. Requirement for PBAF in transcriptional repression and repair at DNA breaks in actively transcribed regions of chromatin. Mol. Cell 2014, 55, 723–732. [Google Scholar] [CrossRef]
- Iannelli, F.; Galbiati, A.; Capozzo, I.; Nguyen, Q.; Magnuson, B.; Michelini, F.; D’Alessandro, G.; Cabrini, M.; Roncador, M.; Francia, S.; et al. A damaged genome’s transcriptional landscape through multilayered expression profiling around in situ-mapped DNA double-strand breaks. Nat. Commun. 2017, 8, 15656. [Google Scholar] [CrossRef]
- Rogakou, E.P.; Boon, C.; Redon, C.; Bonner, W.M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. [Google Scholar] [CrossRef]
- Shroff, R.; Arbel-Eden, A.; Pilch, D.; Ira, G.; Bonner, W.M.; Petrini, J.H.; Haber, J.E.; Lichten, M. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 2004, 14, 1703–1711. [Google Scholar] [CrossRef]
- Falck, J.; Coates, J.; Jackson, S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005, 434, 605–611. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Chahwan, C.; Bailis, J.; Hunter, T.; Russell, P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell. Biol. 2005, 25, 5363–5379. [Google Scholar] [CrossRef] [PubMed]
- Kinner, A.; Wu, W.; Staudt, C.; Iliakis, G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008, 36, 5678–5694. [Google Scholar] [CrossRef] [PubMed]
- Soria, G.; Polo, S.E.; Almouzni, G. Prime, repair, restore: The active role of chromatin in the DNA damage response. Mol. Cell 2012, 46, 722–734. [Google Scholar] [CrossRef]
- Cao, L.L.; Wei, F.; Du, Y.; Song, B.; Wang, D.; Shen, C.; Lu, X.; Cao, Z.; Yang, Q.; Gao, Y.; et al. ATM-mediated KDM2A phosphorylation is required for the DNA damage repair. Oncogene 2016, 35, 402. [Google Scholar] [CrossRef]
- Polo, S.E.; Blackford, A.N.; Chapman, J.R.; Baskcomb, L.; Gravel, S.; Rusch, A.; Thomas, A.; Blundred, R.; Smith, P.; Kzhyshkowska, J.; et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell 2012, 45, 505–516. [Google Scholar] [CrossRef]
- Cook, P.J.; Ju, B.G.; Telese, F.; Wang, X.; Glass, C.K.; Rosenfeld, M.G. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 2009, 458, 591–596. [Google Scholar] [CrossRef]
- Wang, J.; Gong, Z.; Chen, J. MDC1 collaborates with TopBP1 in DNA replication checkpoint control. J. Cell Biol. 2011, 193, 267–273. [Google Scholar] [CrossRef]
- Mailand, N.; Bekker-Jensen, S.; Faustrup, H.; Melander, F.; Bartek, J.; Lukas, C.; Lukas, J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007, 131, 887–900. [Google Scholar] [CrossRef]
- Huen, M.S.; Grant, R.; Manke, I.; Minn, K.; Yu, X.; Yaffe, M.B.; Chen, J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007, 131, 901–914. [Google Scholar] [CrossRef]
- Kolas, N.K.; Chapman, J.R.; Nakada, S.; Ylanko, J.; Chahwan, R.; Sweeney, F.D.; Panier, S.; Mendez, M.; Wildenhain, J.; Thomson, T.M.; et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007, 318, 1637–1640. [Google Scholar] [CrossRef] [PubMed]
- Fradet-Turcotte, A.; Canny, M.D.; Escribano-Diaz, C.; Orthwein, A.; Leung, C.C.; Huang, H.; Landry, M.C.; Kitevski-LeBlanc, J.; Noordermeer, S.M.; Sicheri, F.; et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 2013, 499, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Luijsterburg, M.S.; Acs, K.; Ackermann, L.; Wiegant, W.W.; Bekker-Jensen, S.; Larsen, D.H.; Khanna, K.K.; van Attikum, H.; Mailand, N.; Dantuma, N.P. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012, 31, 2511–2527. [Google Scholar] [CrossRef] [PubMed]
- Huyen, Y.; Zgheib, O.; Ditullio, R.A., Jr.; Gorgoulis, V.G.; Zacharatos, P.; Petty, T.J.; Sheston, E.A.; Mellert, H.S.; Stavridi, E.S.; Halazonetis, T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004, 432, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Portoso, M.; Mata, J.; Bahler, J.; Allshire, R.C.; Kouzarides, T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004, 119, 603–614. [Google Scholar] [CrossRef]
- Schotta, G.; Sengupta, R.; Kubicek, S.; Malin, S.; Kauer, M.; Callen, E.; Celeste, A.; Pagani, M.; Opravil, S.; De La Rosa-Velazquez, I.A.; et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008, 22, 2048–2061. [Google Scholar] [CrossRef]
- Pei, H.; Zhang, L.; Luo, K.; Qin, Y.; Chesi, M.; Fei, F.; Bergsagel, P.L.; Wang, L.; You, Z.; Lou, Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 2011, 470, 124–128. [Google Scholar] [CrossRef]
- Hartlerode, A.J.; Guan, Y.; Rajendran, A.; Ura, K.; Schotta, G.; Xie, A.; Shah, J.V.; Scully, R. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS ONE 2012, 7, e49211. [Google Scholar] [CrossRef]
- Nishioka, K.; Rice, J.C.; Sarma, K.; Erdjument-Bromage, H.; Werner, J.; Wang, Y.; Chuikov, S.; Valenzuela, P.; Tempst, P.; Steward, R.; et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol. Cell 2002, 9, 1201–1213. [Google Scholar] [CrossRef]
- Beck, D.B.; Oda, H.; Shen, S.S.; Reinberg, D. PR-Set7 and H4K20me1: At the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012, 26, 325–337. [Google Scholar] [CrossRef]
- Yang, H.; Pesavento, J.J.; Starnes, T.W.; Cryderman, D.E.; Wallrath, L.L.; Kelleher, N.L.; Mizzen, C.A. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J. Biol. Chem. 2008, 283, 12085–12092. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, S.; Schotta, G.; Sørensen, C.S. Histone H4 lysine 20 methylation: Key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013, 41, 2797–2806. [Google Scholar] [CrossRef]
- Min, J.; Allali-Hassani, A.; Nady, N.; Qi, C.; Ouyang, H.; Liu, Y.; MacKenzie, F.; Vedadi, M.; Arrowsmith, C.H. L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol. 2007, 14, 1229–1230. [Google Scholar] [CrossRef] [PubMed]
- Acs, K.; Luijsterburg, M.S.; Ackermann, L.; Salomons, F.A.; Hoppe, T.; Dantuma, N.P. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 2011, 18, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Thompson, J.R.; Botuyan, M.V.; Mer, G. Distinct binding modes specify the recognition of methylated histones H3K4 and H4K20 by JMJD2A-tudor. Nat. Struct. Mol. Biol. 2008, 15, 109–111. [Google Scholar] [CrossRef]
- Mallette, F.A.; Mattiroli, F.; Cui, G.; Young, L.C.; Hendzel, M.J.; Mer, G.; Sixma, T.K.; Richard, S. RNF8- and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 2012, 31, 1865–1878. [Google Scholar] [CrossRef]
- Jacquet, K.; Fradet-Turcotte, A.; Avvakumov, N.; Lambert, J.P.; Roques, C.; Pandita, R.K.; Paquet, E.; Herst, P.; Gingras, A.C.; Pandita, T.K.; et al. The TIP60 Complex Regulates Bivalent Chromatin Recognition by 53BP1 through Direct H4K20me Binding and H2AK15 Acetylation. Mol. Cell 2016, 62, 409–421. [Google Scholar] [CrossRef]
- Drane, P.; Brault, M.E.; Cui, G.; Meghani, K.; Chaubey, S.; Detappe, A.; Parnandi, N.; He, Y.; Zheng, X.F.; Botuyan, M.V.; et al. TIRR regulates 53BP1 by masking its histone methyl-lysine binding function. Nature 2017, 543, 211–216. [Google Scholar] [CrossRef]
- Wakeman, T.P.; Wang, Q.; Feng, J.; Wang, X.F. Bat3 facilitates H3K79 dimethylation by DOT1L and promotes DNA damage-induced 53BP1 foci at G1/G2 cell-cycle phases. EMBO J. 2012, 31, 2169–2181. [Google Scholar] [CrossRef]
- Feng, Q.; Wang, H.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Struhl, K.; Zhang, Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002, 12, 1052–1058. [Google Scholar] [CrossRef]
- Nguyen, A.T.; Zhang, Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Köhn, M.; Chatterjee, C.; Chiang, K.P.; Pratt, M.R.; Muir, T.W. Structure-activity analysis of semisynthetic nucleosomes: Mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. Acs Chem. Biol. 2009, 4, 958–968. [Google Scholar] [CrossRef] [PubMed]
- McGinty, R.K.; Kim, J.; Chatterjee, C.; Roeder, R.G.; Muir, T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 2008, 453, 812–816. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Holt, M.T.; Ohashi, N.; Zhao, A.; Müller, M.M.; Wang, B.; Muir, T.W. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat. Commun. 2016, 7, 10589. [Google Scholar] [CrossRef] [PubMed]
- Conde, F.; Refolio, E.; Cordón-Preciado, V.; Cortés-Ledesma, F.; Aragón, L.; Aguilera, A.; San-Segundo, P.A. The Dot1 histone methyltransferase and the Rad9 checkpoint adaptor contribute to cohesin-dependent double-strand break repair by sister chromatid recombination in Saccharomyces cerevisiae. Genetics 2009, 182, 437–446. [Google Scholar] [CrossRef]
- Wood, K.; Tellier, M.; Murphy, S. DOT1L and H3K79 Methylation in Transcription and Genomic Stability. Biomolecules 2018, 8, 11. [Google Scholar] [CrossRef]
- Wu, W.; Nishikawa, H.; Fukuda, T.; Vittal, V.; Asano, M.; Miyoshi, Y.; Klevit, R.E.; Ohta, T. Interaction of BARD1 and HP1 Is Required for BRCA1 Retention at Sites of DNA Damage. Cancer Res. 2015, 75, 1311–1321. [Google Scholar] [CrossRef]
- Hu, Y.; Scully, R.; Sobhian, B.; Xie, A.; Shestakova, E.; Livingston, D.M. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011, 25, 685–700. [Google Scholar] [CrossRef]
- Savage, K.I.; Harkin, D.P. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J. 2015, 282, 630–646. [Google Scholar] [CrossRef]
- Ng, H.M.; Wei, L.; Lan, L.; Huen, M.S. The Lys63-deubiquitylating Enzyme BRCC36 Limits DNA Break Processing and Repair. J. Biol. Chem. 2016, 291, 16197–16207. [Google Scholar] [CrossRef]
- Aleksandrov, R.; Dotchev, A.; Poser, I.; Krastev, D.; Georgiev, G.; Panova, G.; Babukov, Y.; Danovski, G.; Dyankova, T.; Hubatsch, L.; et al. Protein Dynamics in Complex DNA Lesions. Mol. Cell 2018, 69, 1046–1061.e1045. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Saredi, G.; Becker, J.R.; Foster, B.M.; Nguyen, N.V.; Beyer, T.E.; Cesa, L.C.; Faull, P.A.; Lukauskas, S.; Frimurer, T.; et al. H4K20me0 recognition by BRCA1-BARD1 directs homologous recombination to sister chromatids. Nat. Cell Biol. 2019, 21, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Pellegrino, S.; Michelena, J.; Teloni, F.; Imhof, R.; Altmeyer, M. Replication-Coupled Dilution of H4K20me2 Guides 53BP1 to Pre-replicative Chromatin. Cell Rep. 2017, 19, 1819–1831. [Google Scholar] [CrossRef] [PubMed]
- Simonetta, M.; de Krijger, I.; Serrat, J.; Moatti, N.; Fortunato, D.; Hoekman, L.; Bleijerveld, O.B.; Altelaar, A.F.M.; Jacobs, J.J.L. H4K20me2 distinguishes pre-replicative from post-replicative chromatin to appropriately direct DNA repair pathway choice by 53BP1-RIF1-MAD2L2. Cell Cycle 2018, 17, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Khurana, S.; Kruhlak, M.J.; Kim, J.; Tran, A.D.; Liu, J.; Nyswaner, K.; Shi, L.; Jailwala, P.; Sung, M.H.; Hakim, O.; et al. A macrohistone variant links dynamic chromatin compaction to BRCA1-dependent genome maintenance. Cell Rep. 2014, 8, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Xu, Y.; Gursoy-Yuzugullu, O.; Price, B.D. The histone variant macroH2A1.1 is recruited to DSBs through a mechanism involving PARP1. FEBS Lett. 2012, 586, 3920–3925. [Google Scholar] [CrossRef]
- Chapman, J.R.; Sossick, A.J.; Boulton, S.J.; Jackson, S.P. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J. Cell Sci 2012, 125, 3529–3534. [Google Scholar] [CrossRef]
- Lorick, K.L.; Jensen, J.P.; Fang, S.; Ong, A.M.; Hatakeyama, S.; Weissman, A.M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 1999, 96, 11364–11369. [Google Scholar] [CrossRef]
- Ruffner, H.; Joazeiro, C.A.; Hemmati, D.; Hunter, T.; Verma, I.M. Cancer-predisposing mutations within the RING domain of BRCA1: Loss of ubiquitin protein ligase activity and protection from radiation hypersensitivity. Proc. Natl. Acad. Sci. USA 2001, 98, 5134–5139. [Google Scholar] [CrossRef]
- Xia, Y.; Pao, G.M.; Chen, H.W.; Verma, I.M.; Hunter, T. Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J. Biol. Chem. 2003, 278, 5255–5263. [Google Scholar] [CrossRef]
- Kalb, R.; Mallery, D.L.; Larkin, C.; Huang, J.T.; Hiom, K. BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 2014, 8, 999–1005. [Google Scholar] [CrossRef] [PubMed]
- Densham, R.M.; Garvin, A.J.; Stone, H.R.; Strachan, J.; Baldock, R.A.; Daza-Martin, M.; Fletcher, A.; Blair-Reid, S.; Beesley, J.; Johal, B.; et al. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 2016, 23, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Awad, S.; Ryan, D.; Prochasson, P.; Owen-Hughes, T.; Hassan, A.H. The Snf2 homolog Fun30 acts as a homodimeric ATP-dependent chromatin-remodeling enzyme. J. Biol. Chem. 2010, 285, 9477–9484. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Cui, D.; Papusha, A.; Zhang, X.; Chu, C.D.; Tang, J.; Chen, K.; Pan, X.; Ira, G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 2012, 489, 576–580. [Google Scholar] [CrossRef]
- Costelloe, T.; Louge, R.; Tomimatsu, N.; Mukherjee, B.; Martini, E.; Khadaroo, B.; Dubois, K.; Wiegant, W.W.; Thierry, A.; Burma, S.; et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature 2012, 489, 581–584. [Google Scholar] [CrossRef]
- Eapen, V.V.; Sugawara, N.; Tsabar, M.; Wu, W.H.; Haber, J.E. The Saccharomyces cerevisiae chromatin remodeler Fun30 regulates DNA end resection and checkpoint deactivation. Mol. Cell. Biol. 2012, 32, 4727–4740. [Google Scholar] [CrossRef]
- Bantele, S.C.; Ferreira, P.; Gritenaite, D.; Boos, D.; Pfander, B. Targeting of the Fun30 nucleosome remodeller by the Dpb11 scaffold facilitates cell cycle-regulated DNA end resection. eLife 2017, 6. [Google Scholar] [CrossRef]
- Chen, X.; Niu, H.; Yu, Y.; Wang, J.; Zhu, S.; Zhou, J.; Papusha, A.; Cui, D.; Pan, X.; Kwon, Y.; et al. Enrichment of Cdk1-cyclins at DNA double-strand breaks stimulates Fun30 phosphorylation and DNA end resection. Nucleic Acids Res. 2016, 44, 2742–2753. [Google Scholar] [CrossRef]
- Uckelmann, M.; Densham, R.M.; Baas, R.; Winterwerp, H.H.K.; Fish, A.; Sixma, T.K.; Morris, J.R. USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat. Commun. 2018, 9, 229. [Google Scholar] [CrossRef]
- Alagoz, M.; Katsuki, Y.; Ogiwara, H.; Ogi, T.; Shibata, A.; Kakarougkas, A.; Jeggo, P. SETDB1, HP1 and SUV39 promote repositioning of 53BP1 to extend resection during homologous recombination in G2 cells. Nucleic Acids Res. 2015, 43, 7931–7944. [Google Scholar] [CrossRef]
- Keogh, M.C.; Kim, J.A.; Downey, M.; Fillingham, J.; Chowdhury, D.; Harrison, J.C.; Onishi, M.; Datta, N.; Galicia, S.; Emili, A.; et al. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 2006, 439, 497–501. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Capetillo, O.; Allis, C.D.; Nussenzweig, A. Phosphorylation of histone H2B at DNA double-strand breaks. J. Exp. Med. 2004, 199, 1671–1677. [Google Scholar] [CrossRef]
- Utley, R.T.; Lacoste, N.; Jobin-Robitaille, O.; Allard, S.; Côté, J. Regulation of NuA4 histone acetyltransferase activity in transcription and DNA repair by phosphorylation of histone H4. Mol. Cell. Biol. 2005, 25, 8179–8190. [Google Scholar] [CrossRef] [PubMed]
- Cheung, W.L.; Turner, F.B.; Krishnamoorthy, T.; Wolner, B.; Ahn, S.H.; Foley, M.; Dorsey, J.A.; Peterson, C.L.; Berger, S.L.; Allis, C.D. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 2005, 15, 656–660. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Dong, L.; Tang, M.; Zhang, P.; Zhang, C.; Cao, Z.; Zhu, Q.; Chen, Y.; Wang, H.; et al. Histone H1 acetylation at lysine 85 regulates chromatin condensation and genome stability upon DNA damage. Nucleic Acids Res. 2018, 46, 7716–7730. [Google Scholar] [CrossRef] [PubMed]
- Ikura, T.; Tashiro, S.; Kakino, A.; Shima, H.; Jacob, N.; Amunugama, R.; Yoder, K.; Izumi, S.; Kuraoka, I.; Tanaka, K.; et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 2007, 27, 7028–7040. [Google Scholar] [CrossRef] [PubMed]
- Clouaire, T.; Rocher, V.; Lashgari, A.; Arnould, C.; Aguirrebengoa, M.; Biernacka, A.; Skrzypczak, M.; Aymard, F.; Fongang, B.; Dojer, N.; et al. Comprehensive Mapping of Histone Modifications at DNA Double-Strand Breaks Deciphers Repair Pathway Chromatin Signatures. Mol. Cell 2018, 72, 250–262.e256. [Google Scholar] [CrossRef]
- Tamburini, B.A.; Tyler, J.K. Localized histone acetylation and deacetylation triggered by the homologous recombination pathway of double-strand DNA repair. Mol. Cell. Biol. 2005, 25, 4903–4913. [Google Scholar] [CrossRef]
- Ogiwara, H.; Ui, A.; Otsuka, A.; Satoh, H.; Yokomi, I.; Nakajima, S.; Yasui, A.; Yokota, J.; Kohno, T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011, 30, 2135–2146. [Google Scholar] [CrossRef]
- Das, C.; Lucia, M.S.; Hansen, K.C.; Tyler, J.K. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459, 113–117. [Google Scholar] [CrossRef]
- Chen, C.C.; Carson, J.J.; Feser, J.; Tamburini, B.; Zabaronick, S.; Linger, J.; Tyler, J.K. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell 2008, 134, 231–243. [Google Scholar] [CrossRef]
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Dobbin, M.M.; Madabhushi, R.; Pan, L.; Chen, Y.; Kim, D.; Gao, J.; Ahanonu, B.; Pao, P.C.; Qiu, Y.; Zhao, Y.; et al. SIRT1 collaborates with ATM and HDAC1 to maintain genomic stability in neurons. Nat. Neurosci. 2013, 16, 1008–1015. [Google Scholar] [CrossRef] [PubMed]
- Stewart, G.S.; Panier, S.; Townsend, K.; Al-Hakim, A.K.; Kolas, N.K.; Miller, E.S.; Nakada, S.; Ylanko, J.; Olivarius, S.; Mendez, M.; et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 2009, 136, 420–434. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Pak, H.; Hammond-Martel, I.; Ghram, M.; Rodrigue, A.; Daou, S.; Barbour, H.; Corbeil, L.; Hébert, J.; Drobetsky, E.; et al. Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2014, 111, 285–290. [Google Scholar] [CrossRef]
- Yan, Q.; Dutt, S.; Xu, R.; Graves, K.; Juszczynski, P.; Manis, J.P.; Shipp, M.A. BBAP monoubiquitylates histone H4 at lysine 91 and selectively modulates the DNA damage response. Mol. Cell 2009, 36, 110–120. [Google Scholar] [CrossRef]
- Gong, F.; Clouaire, T.; Aguirrebengoa, M.; Legube, G.; Miller, K.M. Histone demethylase KDM5A regulates the ZMYND8-NuRD chromatin remodeler to promote DNA repair. J. Cell Biol. 2017, 216, 1959–1974. [Google Scholar] [CrossRef]
- Mosammaparast, N.; Kim, H.; Laurent, B.; Zhao, Y.; Lim, H.J.; Majid, M.C.; Dango, S.; Luo, Y.; Hempel, K.; Sowa, M.E.; et al. The histone demethylase LSD1/KDM1A promotes the DNA damage response. J. Cell Biol. 2013, 203, 457–470. [Google Scholar] [CrossRef]
- Ayoub, N.; Jeyasekharan, A.D.; Bernal, J.A.; Venkitaraman, A.R. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 2008, 453, 682–686. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, X.; Xu, Y.; Ayrapetov, M.K.; Moreau, L.A.; Whetstine, J.R.; Price, B.D. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 2009, 11, 1376–1382. [Google Scholar] [CrossRef] [PubMed]
- Ui, A.; Nagaura, Y.; Yasui, A. Transcriptional elongation factor ENL phosphorylated by ATM recruits polycomb and switches off transcription for DSB repair. Mol. Cell 2015, 58, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Abu-Zhayia, E.R.; Awwad, S.W.; Ben-Oz, B.M.; Khoury-Haddad, H.; Ayoub, N. CDYL1 fosters double-strand break-induced transcription silencing and promotes homology-directed repair. J. Mol. Cell Biol. 2018, 10, 341–357. [Google Scholar] [CrossRef] [PubMed]
- Fnu, S.; Williamson, E.A.; De Haro, L.P.; Brenneman, M.; Wray, J.; Shaheen, M.; Radhakrishnan, K.; Lee, S.H.; Nickoloff, J.A.; Hromas, R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA 2011, 108, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Aymard, F.; Bugler, B.; Schmidt, C.K.; Guillou, E.; Caron, P.; Briois, S.; Iacovoni, J.S.; Daburon, V.; Miller, K.M.; Jackson, S.P.; et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014, 21, 366–374. [Google Scholar] [CrossRef]
- Pfister, S.X.; Ahrabi, S.; Zalmas, L.P.; Sarkar, S.; Aymard, F.; Bachrati, C.Z.; Helleday, T.; Legube, G.; La Thangue, N.B.; Porter, A.C.; et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014, 7, 2006–2018. [Google Scholar] [CrossRef]
- Carvalho, S.; Vítor, A.C.; Sridhara, S.C.; Martins, F.B.; Raposo, A.C.; Desterro, J.M.; Ferreira, J.; de Almeida, S.F. SETD2 is required for DNA double-strand break repair and activation of the p53-mediated checkpoint. eLife 2014, 3, e02482. [Google Scholar] [CrossRef]
- Daugaard, M.; Baude, A.; Fugger, K.; Povlsen, L.K.; Beck, H.; Sørensen, C.S.; Petersen, N.H.; Sorensen, P.H.; Lukas, C.; Bartek, J.; et al. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012, 19, 803–810. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, J.H.; Kim, S.J.; Kwon, S.J.; Kwon, J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010, 29, 1434–1445. [Google Scholar] [CrossRef]
- Watanabe, R.; Ui, A.; Kanno, S.; Ogiwara, H.; Nagase, T.; Kohno, T.; Yasui, A. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014, 74, 2465–2475. [Google Scholar] [CrossRef]
- Kwon, J.; Morshead, K.B.; Guyon, J.R.; Kingston, R.E.; Oettinger, M.A. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol. Cell 2000, 6, 1037–1048. [Google Scholar] [CrossRef]
- Morshead, K.B.; Ciccone, D.N.; Taverna, S.D.; Allis, C.D.; Oettinger, M.A. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA 2003, 100, 11577–11582. [Google Scholar] [CrossRef] [PubMed]
- Patenge, N.; Elkin, S.K.; Oettinger, M.A. ATP-dependent remodeling by SWI/SNF and ISWI proteins stimulates V(D)J cleavage of 5 S arrays. J. Biol. Chem. 2004, 279, 35360–35367. [Google Scholar] [CrossRef]
- Chai, B.; Huang, J.; Cairns, B.R.; Laurent, B.C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 2005, 19, 1656–1661. [Google Scholar] [CrossRef]
- Sinha, M.; Watanabe, S.; Johnson, A.; Moazed, D.; Peterson, C.L. Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 2009, 138, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Wiest, N.E.; Houghtaling, S.; Sanchez, J.C.; Tomkinson, A.E.; Osley, M.A. The SWI/SNF ATP-dependent nucleosome remodeler promotes resection initiation at a DNA double-strand break in yeast. Nucleic Acids Res. 2017, 45, 5887–5900. [Google Scholar] [CrossRef]
- Adkins, N.L.; Swygert, S.G.; Kaur, P.; Niu, H.; Grigoryev, S.A.; Sung, P.; Wang, H.; Peterson, C.L. Nucleosome-like, Single-stranded DNA (ssDNA)-Histone Octamer Complexes and the Implication for DNA Double Strand Break Repair. J. Biol. Chem. 2017, 292, 5271–5281. [Google Scholar] [CrossRef]
- Strålfors, A.; Walfridsson, J.; Bhuiyan, H.; Ekwall, K. The FUN30 chromatin remodeler, Fft3, protects centromeric and subtelomeric domains from euchromatin formation. PLoS Genet. 2011, 7, e1001334. [Google Scholar] [CrossRef]
- Durand-Dubief, M.; Will, W.R.; Petrini, E.; Theodorou, D.; Harris, R.R.; Crawford, M.R.; Paszkiewicz, K.; Krueger, F.; Correra, R.M.; Vetter, A.T.; et al. SWI/SNF-like chromatin remodeling factor Fun30 supports point centromere function in S. cerevisiae. PLoS Genet. 2012, 8, e1002974. [Google Scholar] [CrossRef]
- Shen, X.; Mizuguchi, G.; Hamiche, A.; Wu, C. A chromatin remodelling complex involved in transcription and DNA processing. Nature 2000, 406, 541–544. [Google Scholar] [CrossRef]
- Shen, X.; Ranallo, R.; Choi, E.; Wu, C. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 2003, 12, 147–155. [Google Scholar] [CrossRef]
- Gospodinov, A.; Vaissiere, T.; Krastev, D.B.; Legube, G.; Anachkova, B.; Herceg, Z. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Mol. Cell. Biol. 2011, 31, 4735–4745. [Google Scholar] [CrossRef] [PubMed]
- van Attikum, H.; Fritsch, O.; Hohn, B.; Gasser, S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004, 119, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Tsukuda, T.; Fleming, A.B.; Nickoloff, J.A.; Osley, M.A. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 2005, 438, 379–383. [Google Scholar] [CrossRef] [PubMed]
- Park, E.J.; Hur, S.K.; Kwon, J. Human INO80 chromatin-remodelling complex contributes to DNA double-strand break repair via the expression of Rad54B and XRCC3 genes. Biochem. J. 2010, 431, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Papamichos-Chronakis, M.; Krebs, J.E.; Peterson, C.L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006, 20, 2437–2449. [Google Scholar] [CrossRef] [PubMed]
- Lademann, C.A.; Renkawitz, J.; Pfander, B.; Jentsch, S. The INO80 Complex Removes H2A.Z to Promote Presynaptic Filament Formation during Homologous Recombination. Cell Rep. 2017, 19, 1294–1303. [Google Scholar] [CrossRef]
- Li, X.; Tyler, J.K. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. eLife 2016, 5. [Google Scholar] [CrossRef]
- Poli, J.; Gerhold, C.B.; Tosi, A.; Hustedt, N.; Seeber, A.; Sack, R.; Herzog, F.; Pasero, P.; Shimada, K.; Hopfner, K.P.; et al. Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 2016, 30, 337–354. [Google Scholar] [CrossRef]
- Dong, S.; Han, J.; Chen, H.; Liu, T.; Huen, M.S.Y.; Yang, Y.; Guo, C.; Huang, J. The human SRCAP chromatin remodeling complex promotes DNA-end resection. Curr. Biol. 2014, 24, 2097–2110. [Google Scholar] [CrossRef]
- van Attikum, H.; Fritsch, O.; Gasser, S.M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007, 26, 4113–4125. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ayrapetov, M.K.; Xu, C.; Gursoy-Yuzugullu, O.; Hu, Y.; Price, B.D. Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair. Mol. Cell 2012, 48, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Kari, V.; Mansour, W.Y.; Raul, S.K.; Baumgart, S.J.; Mund, A.; Grade, M.; Sirma, H.; Simon, R.; Will, H.; Dobbelstein, M.; et al. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016, 17, 1609–1623. [Google Scholar] [CrossRef] [PubMed]
- Ahel, D.; Horejsí, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef]
- Luijsterburg, M.S.; de Krijger, I.; Wiegant, W.W.; Shah, R.G.; Smeenk, G.; de Groot, A.J.L.; Pines, A.; Vertegaal, A.C.O.; Jacobs, J.J.L.; Shah, G.M.; et al. PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining. Mol. Cell 2016, 61, 547–562. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Callen, E.; Ding, X.; Gogola, E.; Duarte, A.A.; Lee, J.E.; Wong, N.; Lafarga, V.; Calvo, J.A.; Panzarino, N.J.; et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016, 535, 382–387. [Google Scholar] [CrossRef]
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiácovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. USA 2010, 107, 18475–18480. [Google Scholar] [CrossRef]
- Polo, S.E.; Kaidi, A.; Baskcomb, L.; Galanty, Y.; Jackson, S.P. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010, 29, 3130–3139. [Google Scholar] [CrossRef]
- Smeenk, G.; Wiegant, W.W.; Vrolijk, H.; Solari, A.P.; Pastink, A.; van Attikum, H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010, 190, 741–749. [Google Scholar] [CrossRef]
- Lan, L.; Ui, A.; Nakajima, S.; Hatakeyama, K.; Hoshi, M.; Watanabe, R.; Janicki, S.M.; Ogiwara, H.; Kohno, T.; Kanno, S.; et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol. Cell 2010, 40, 976–987. [Google Scholar] [CrossRef]
- Mehrotra, P.V.; Ahel, D.; Ryan, D.P.; Weston, R.; Wiechens, N.; Kraehenbuehl, R.; Owen-Hughes, T.; Ahel, I. DNA repair factor APLF is a histone chaperone. Mol. Cell 2011, 41, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007, 21, 1519–1529. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.G.; Roberts, C.W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 2011, 11, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Dykhuizen, E.C.; Hargreaves, D.C.; Miller, E.L.; Cui, K.; Korshunov, A.; Kool, M.; Pfister, S.; Cho, Y.J.; Zhao, K.; Crabtree, G.R. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature 2013, 497, 624–627. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.M.; Tjeertes, J.V.; Coates, J.; Legube, G.; Polo, S.E.; Britton, S.; Jackson, S.P. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 2010, 17, 1144–1151. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, B.N.; Thackray, J.K.; Simonet, N.G.; Kane-Goldsmith, N.; Martinez-Redondo, P.; Nguyen, T.; Bunting, S.; Vaquero, A.; Tischfield, J.A.; Serrano, L. SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J. 2016, 35, 1488–1503. [Google Scholar] [CrossRef]
- McCord, R.A.; Michishita, E.; Hong, T.; Berber, E.; Boxer, L.D.; Kusumoto, R.; Guan, S.; Shi, X.; Gozani, O.; Burlingame, A.L.; et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging 2009, 1, 109–121. [Google Scholar] [CrossRef]
- Pai, C.C.; Deegan, R.S.; Subramanian, L.; Gal, C.; Sarkar, S.; Blaikley, E.J.; Walker, C.; Hulme, L.; Bernhard, E.; Codlin, S.; et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 2014, 5, 4091. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J. Biol. Chem. 2017, 292, 11951–11959. [Google Scholar] [CrossRef]
- Clouaire, T.; Legube, G. A Snapshot on the Cis Chromatin Response to DNA Double-Strand Breaks. Trends Genet. 2019, 35, 330–345. [Google Scholar] [CrossRef]
- Caron, M.C.; Sharma, A.K.; O’Sullivan, J.; Myler, L.R.; Ferreira, M.T.; Rodrigue, A.; Coulombe, Y.; Ethier, C.; Gagné, J.P.; Langelier, M.F.; et al. Poly(ADP-ribose) polymerase-1 antagonizes DNA resection at double-strand breaks. Nat. Commun. 2019, 10, 2954. [Google Scholar] [CrossRef] [PubMed]
- Taty-Taty, G.C.; Chailleux, C.; Quaranta, M.; So, A.; Guirouilh-Barbat, J.; Lopez, B.S.; Bertrand, P.; Trouche, D.; Canitrot, Y. Control of alternative end joining by the chromatin remodeler p400 ATPase. Nucleic Acids Res. 2016, 44, 1657–1668. [Google Scholar] [CrossRef] [PubMed]
- Adkins, N.L.; Niu, H.; Sung, P.; Peterson, C.L. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013, 20, 836–842. [Google Scholar] [CrossRef] [PubMed]
- Shim, E.Y.; Hong, S.J.; Oum, J.H.; Yanez, Y.; Zhang, Y.; Lee, S.E. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 2007, 27, 1602–1613. [Google Scholar] [CrossRef] [PubMed]
- Bennett, G.; Papamichos-Chronakis, M.; Peterson, C.L. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 2013, 4, 2084. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Caron, P.; Legube, G.; Paull, T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014, 42, e19. [Google Scholar] [CrossRef]
- Gursoy-Yuzugullu, O.; Ayrapetov, M.K.; Price, B.D. Histone chaperone Anp32e removes H2A.Z from DNA double-strand breaks and promotes nucleosome reorganization and DNA repair. Proc. Natl. Acad. Sci. USA 2015, 112, 7507–7512. [Google Scholar] [CrossRef]
- Mizuguchi, G.; Shen, X.; Landry, J.; Wu, W.H.; Sen, S.; Wu, C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 2004, 303, 343–348. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Watanabe, S.; Rando, O.J.; Peterson, C.L. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 2011, 144, 200–213. [Google Scholar] [CrossRef]
- Kollárovič, G.; Topping, C.E.; Shaw, E.P.; Chambers, A.L. The human HELLS chromatin remodelling protein promotes end resection to facilitate homologous recombination and contributes to DSB repair within heterochromatin. Nucleic Acids Res. 2020, 48, 1872–1885. [Google Scholar] [CrossRef]
- Huang, T.H.; Fowler, F.; Chen, C.C.; Shen, Z.J.; Sleckman, B.; Tyler, J.K. The Histone Chaperones ASF1 and CAF-1 Promote MMS22L-TONSL-Mediated Rad51 Loading onto ssDNA during Homologous Recombination in Human Cells. Mol. Cell 2018, 69, 879–892.e875. [Google Scholar] [CrossRef] [PubMed]
- Courilleau, C.; Chailleux, C.; Jauneau, A.; Grimal, F.; Briois, S.; Boutet-Robinet, E.; Boudsocq, F.; Trouche, D.; Canitrot, Y. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. J. Cell Biol. 2012, 199, 1067–1081. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Wang, R.; Chen, H.; Wang, X.; Xiao, T.; Boldogh, I.; Ba, X.; Han, L.; Zeng, X. BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci 2015, 128, 317–330. [Google Scholar] [CrossRef]
- Alonso-de Vega, I.; Paz-Cabrera, M.C.; Rother, M.B.; Wiegant, W.W.; Checa-Rodríguez, C.; Hernández-Fernaud, J.R.; Huertas, P.; Freire, R.; van Attikum, H.; Smits, V.A.J. PHF2 regulates homology-directed DNA repair by controlling the resection of DNA double strand breaks. Nucleic Acids Res. 2020, 48, 4915–4927. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, L.D.; LeClair, J.W.; Miller, K.N.; Strong, A.D.; Chan, H.L.; Oates, E.L.; Ligon, K.L.; Brennan, C.W.; Chheda, M.G. CHD4 regulates the DNA damage response and RAD51 expression in glioblastoma. Sci. Rep. 2019, 9, 4444. [Google Scholar] [CrossRef]
- Pan, M.R.; Hsieh, H.J.; Dai, H.; Hung, W.C.; Li, K.; Peng, G.; Lin, S.Y. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J. Biol. Chem. 2012, 287, 6764–6772. [Google Scholar] [CrossRef]
- Larsen, D.H.; Poinsignon, C.; Gudjonsson, T.; Dinant, C.; Payne, M.R.; Hari, F.J.; Rendtlew Danielsen, J.M.; Menard, P.; Sand, J.C.; Stucki, M.; et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J. Cell Biol. 2010, 190, 731–740. [Google Scholar] [CrossRef]
- Gong, F.; Chiu, L.Y.; Cox, B.; Aymard, F.; Clouaire, T.; Leung, J.W.; Cammarata, M.; Perez, M.; Agarwal, P.; Brodbelt, J.S.; et al. Screen identifies bromodomain protein ZMYND8 in chromatin recognition of transcription-associated DNA damage that promotes homologous recombination. Genes Dev. 2015, 29, 197–211. [Google Scholar] [CrossRef]
- Gong, F.; Miller, K.M. Double duty: ZMYND8 in the DNA damage response and cancer. Cell Cycle 2018, 17, 414–420. [Google Scholar] [CrossRef]
- Savitsky, P.; Krojer, T.; Fujisawa, T.; Lambert, J.P.; Picaud, S.; Wang, C.Y.; Shanle, E.K.; Krajewski, K.; Friedrichsen, H.; Kanapin, A.; et al. Multivalent Histone and DNA Engagement by a PHD/BRD/PWWP Triple Reader Cassette Recruits ZMYND8 to K14ac-Rich Chromatin. Cell Rep. 2016, 17, 2724–2737. [Google Scholar] [CrossRef]
- Gursoy-Yuzugullu, O.; Carman, C.; Price, B.D. Spatially restricted loading of BRD2 at DNA double-strand breaks protects H4 acetylation domains and promotes DNA repair. Sci. Rep. 2017, 7, 12921. [Google Scholar] [CrossRef] [PubMed]
- Ginjala, V.; Nacerddine, K.; Kulkarni, A.; Oza, J.; Hill, S.J.; Yao, M.; Citterio, E.; van Lohuizen, M.; Ganesan, S. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 2011, 31, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Kruhlak, M.; Crouch, E.E.; Orlov, M.; Montaño, C.; Gorski, S.A.; Nussenzweig, A.; Misteli, T.; Phair, R.D.; Casellas, R. The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 2007, 447, 730–734. [Google Scholar] [CrossRef]
- Ting, X.; Xia, L.; Yang, J.; He, L.; Si, W.; Shang, Y.; Sun, L. USP11 acts as a histone deubiquitinase functioning in chromatin reorganization during DNA repair. Nucleic Acids Res. 2019, 47, 9721–9740. [Google Scholar] [CrossRef]
- Caron, P.; van der Linden, J.; van Attikum, H. Bon voyage: A transcriptional journey around DNA breaks. DNA Repair 2019, 82, 102686. [Google Scholar] [CrossRef] [PubMed]
- Lashgari, A.; Millau, J.F.; Jacques, P.; Gaudreau, L. Global inhibition of transcription causes an increase in histone H2A.Z incorporation within gene bodies. Nucleic Acids Res. 2017, 45, 12715–12722. [Google Scholar] [CrossRef]
- Vermeulen, W. Dynamics of mammalian NER proteins. DNA Repair 2011, 10, 760–771. [Google Scholar] [CrossRef]
- Feuerhahn, S.; Egly, J.M. Tools to study DNA repair: What’s in the box? Trends Genet. 2008, 24, 467–474. [Google Scholar] [CrossRef]
- Kong, X.; Mohanty, S.K.; Stephens, J.; Heale, J.T.; Gomez-Godinez, V.; Shi, L.Z.; Kim, J.S.; Yokomori, K.; Berns, M.W. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009, 37, e68. [Google Scholar] [CrossRef]
- Zou, L.; Liu, D.; Elledge, S.J. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 2003, 100, 13827–13832. [Google Scholar] [CrossRef]
- Ngo, G.H.; Lydall, D. The 9-1-1 checkpoint clamp coordinates resection at DNA double strand breaks. Nucleic Acids Res. 2015, 43, 5017–5032. [Google Scholar] [CrossRef]
- Ashworth, A.; Lord, C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018, 15, 564–576. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020, 19, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Beck, C.; Robert, I.; Reina-San-Martin, B.; Schreiber, V.; Dantzer, F. Poly(ADP-ribose) polymerases in double-strand break repair: Focus on PARP1, PARP2 and PARP3. Exp. Cell Res. 2014, 329, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, V.; Dantzer, F.; Ame, J.C.; de Murcia, G. Poly(ADP-ribose): Novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006, 7, 517–528. [Google Scholar] [CrossRef]
- Gassman, N.R.; Wilson, S.H. Micro-irradiation tools to visualize base excision repair and single-strand break repair. DNA Repair 2015, 31, 52–63. [Google Scholar] [CrossRef]
- Beneke, S. Regulation of chromatin structure by poly(ADP-ribosyl)ation. Front. Genet. 2012, 3, 169. [Google Scholar] [CrossRef]
- Ko, H.L.; Ren, E.C. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules 2012, 2, 524–548. [Google Scholar] [CrossRef]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; Hottiger, M.O. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar] [CrossRef]
- Bryant, H.E.; Schultz, N.; Thomas, H.D.; Parker, K.M.; Flower, D.; Lopez, E.; Kyle, S.; Meuth, M.; Curtin, N.J.; Helleday, T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 2005, 434, 913–917. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.N.; Lord, C.J.; McCabe, N.; Farmer, H.; Turner, N.; Martin, N.M.; Jackson, S.P.; Smith, G.C.; Ashworth, A. Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb. Symp. Quant. Biol. 2005, 70, 139–148. [Google Scholar] [CrossRef]
- Hosoya, N.; Miyagawa, K. Targeting DNA damage response in cancer therapy. Cancer Sci. 2014, 105, 370–388. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Bouberhan, S.; Philp, L.; Hill, S.; Al-Alem, L.F.; Rueda, B. Exploiting the Prevalence of Homologous Recombination Deficiencies in High-Grade Serous Ovarian Cancer. Cancers 2020, 12, 1206. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef]
- Murai, J.; Huang, S.Y.; Renaud, A.; Zhang, Y.; Ji, J.; Takeda, S.; Morris, J.; Teicher, B.; Doroshow, J.H.; Pommier, Y. Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol. Cancer Ther. 2014, 13, 433–443. [Google Scholar] [CrossRef]
- Shen, Y.; Rehman, F.L.; Feng, Y.; Boshuizen, J.; Bajrami, I.; Elliott, R.; Wang, B.; Lord, C.J.; Post, L.E.; Ashworth, A. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin. Cancer Res. 2013, 19, 5003–5015. [Google Scholar] [CrossRef]
- Zandarashvili, L.; Langelier, M.F.; Velagapudi, U.K.; Hancock, M.A.; Steffen, J.D.; Billur, R.; Hannan, Z.M.; Wicks, A.J.; Krastev, D.B.; Pettitt, S.J.; et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020, 368. [Google Scholar] [CrossRef]
- Pommier, Y.; O’Connor, M.J.; de Bono, J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci. Transl. Med. 2016, 8, 362ps317. [Google Scholar] [CrossRef]
- Maya-Mendoza, A.; Moudry, P.; Merchut-Maya, J.M.; Lee, M.; Strauss, R.; Bartek, J. High speed of fork progression induces DNA replication stress and genomic instability. Nature 2018, 559, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Nowsheen, S.; Maraboyina, S.; Xia, F. The role of poly(ADP-ribose) polymerase inhibitors in the treatment of cancer and methods to overcome resistance: A review. Cell Biosci. 2020, 10, 35. [Google Scholar] [CrossRef]
- Pommier, Y. Topoisomerase I inhibitors: Camptothecins and beyond. Nat. Rev. Cancer 2006, 6, 789–802. [Google Scholar] [CrossRef]
- Norquist, B.; Wurz, K.A.; Pennil, C.C.; Garcia, R.; Gross, J.; Sakai, W.; Karlan, B.Y.; Taniguchi, T.; Swisher, E.M. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 2011, 29, 3008–3015. [Google Scholar] [CrossRef] [PubMed]
- Swisher, E.M.; Sakai, W.; Karlan, B.Y.; Wurz, K.; Urban, N.; Taniguchi, T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008, 68, 2581–2586. [Google Scholar] [CrossRef] [PubMed]
- Bunting, S.F.; Callen, E.; Wong, N.; Chen, H.T.; Polato, F.; Gunn, A.; Bothmer, A.; Feldhahn, N.; Fernandez-Capetillo, O.; Cao, L.; et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 2010, 141, 243–254. [Google Scholar] [CrossRef]
- Daley, J.M.; Sung, P. RIF1 in DNA break repair pathway choice. Mol. Cell 2013, 49, 840–841. [Google Scholar] [CrossRef]
- Zimmermann, M.; Lottersberger, F.; Buonomo, S.B.; Sfeir, A.; de Lange, T. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science 2013, 339, 700–704. [Google Scholar] [CrossRef]
- Dev, H.; Chiang, T.W.; Lescale, C.; de Krijger, I.; Martin, A.G.; Pilger, D.; Coates, J.; Sczaniecka-Clift, M.; Wei, W.; Ostermaier, M.; et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 2018, 20, 954–965. [Google Scholar] [CrossRef]
- Gupta, R.; Somyajit, K.; Narita, T.; Maskey, E.; Stanlie, A.; Kremer, M.; Typas, D.; Lammers, M.; Mailand, N.; Nussenzweig, A.; et al. DNA Repair Network Analysis Reveals Shieldin as a Key Regulator of NHEJ and PARP Inhibitor Sensitivity. Cell 2018, 173, 972–988.e923. [Google Scholar] [CrossRef]
- Mirman, Z.; Lottersberger, F.; Takai, H.; Kibe, T.; Gong, Y.; Takai, K.; Bianchi, A.; Zimmermann, M.; Durocher, D.; de Lange, T. 53BP1-RIF1-shieldin counteracts DSB resection through CST- and Polalpha-dependent fill-in. Nature 2018, 560, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Noordermeer, S.M.; Adam, S.; Setiaputra, D.; Barazas, M.; Pettitt, S.J.; Ling, A.K.; Olivieri, M.; Alvarez-Quilon, A.; Moatti, N.; Zimmermann, M.; et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature 2018, 560, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Pettitt, S.J.; Krastev, D.B.; Brandsma, I.; Dréan, A.; Song, F.; Aleksandrov, R.; Harrell, M.I.; Menon, M.; Brough, R.; Campbell, J.; et al. Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 2018, 9, 1849. [Google Scholar] [CrossRef]
- He, Y.J.; Meghani, K.; Caron, M.C.; Yang, C.; Ronato, D.A.; Bian, J.; Sharma, A.; Moore, J.; Niraj, J.; Detappe, A.; et al. DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 2018, 563, 522–526. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.D. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Arana, M.E.; Seki, M.; Wood, R.D.; Rogozin, I.B.; Kunkel, T.A. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008, 36, 3847–3856. [Google Scholar] [CrossRef]
- Black, S.J.; Ozdemir, A.Y.; Kashkina, E.; Kent, T.; Rusanov, T.; Ristic, D.; Shin, Y.; Suma, A.; Hoang, T.; Chandramouly, G.; et al. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 2019, 10, 4423. [Google Scholar] [CrossRef]
- Knutson, S.K.; Warholic, N.M.; Wigle, T.J.; Klaus, C.R.; Allain, C.J.; Raimondi, A.; Porter Scott, M.; Chesworth, R.; Moyer, M.P.; Copeland, R.A.; et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc. Natl. Acad. Sci. USA 2013, 110, 7922–7927. [Google Scholar] [CrossRef]
- Kia, S.K.; Gorski, M.M.; Giannakopoulos, S.; Verrijzer, C.P. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell. Biol. 2008, 28, 3457–3464. [Google Scholar] [CrossRef]
- Wilson, B.G.; Wang, X.; Shen, X.; McKenna, E.S.; Lemieux, M.E.; Cho, Y.J.; Koellhoffer, E.C.; Pomeroy, S.L.; Orkin, S.H.; Roberts, C.W. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010, 18, 316–328. [Google Scholar] [CrossRef]
- Blasina, A.; Price, B.D.; Turenne, G.A.; McGowan, C.H. Caffeine inhibits the checkpoint kinase ATM. Curr. Biol. 1999, 9, 1135–1138. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Busby, E.C.; Tibbetts, R.S.; Roos, P.; Taya, Y.; Karnitz, L.M.; Abraham, R.T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999, 59, 4375–4382. [Google Scholar] [PubMed]
- Powell, S.N.; DeFrank, J.S.; Connell, P.; Eogan, M.; Preffer, F.; Dombkowski, D.; Tang, W.; Friend, S. Differential sensitivity of p53(-) and p53(+) cells to caffeine-induced radiosensitization and override of G2 delay. Cancer Res. 1995, 55, 1643–1648. [Google Scholar] [CrossRef]
- Yao, S.L.; Akhtar, A.J.; McKenna, K.A.; Bedi, G.C.; Sidransky, D.; Mabry, M.; Ravi, R.; Collector, M.I.; Jones, R.J.; Sharkis, S.J.; et al. Selective radiosensitization of p53-deficient cells by caffeine-mediated activation of p34cdc2 kinase. Nat. Med. 1996, 2, 1140–1143. [Google Scholar] [CrossRef]
- Bracey, T.S.; Williams, A.C.; Paraskeva, C. Inhibition of radiation-induced G2 delay potentiates cell death by apoptosis and/or the induction of giant cells in colorectal tumor cells with disrupted p53 function. Clin. Cancer Res. 1997, 3, 1371–1381. [Google Scholar]
- Newton, R.; Broughton, L.J.; Lind, M.J.; Morrison, P.J.; Rogers, H.J.; Bradbrook, I.D. Plasma and salivary pharmacokinetics of caffeine in man. Eur. J. Clin. Pharmacol. 1981, 21, 45–52. [Google Scholar] [CrossRef]
- Sarkaria, J.N.; Tibbetts, R.S.; Busby, E.C.; Kennedy, A.P.; Hill, D.E.; Abraham, R.T. Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res. 1998, 58, 4375–4382. [Google Scholar]
- Price, B.D.; Youmell, M.B. The phosphatidylinositol 3-kinase inhibitor wortmannin sensitizes murine fibroblasts and human tumor cells to radiation and blocks induction of p53 following DNA damage. Cancer Res. 1996, 56, 246–250. [Google Scholar]
- Karve, S.; Werner, M.E.; Sukumar, R.; Cummings, N.D.; Copp, J.A.; Wang, E.C.; Li, C.; Sethi, M.; Chen, R.C.; Pacold, M.E.; et al. Revival of the abandoned therapeutic wortmannin by nanoparticle drug delivery. Proc. Natl. Acad. Sci. USA 2012, 109, 8230–8235. [Google Scholar] [CrossRef]
- Hickson, I.; Zhao, Y.; Richardson, C.J.; Green, S.J.; Martin, N.M.; Orr, A.I.; Reaper, P.M.; Jackson, S.P.; Curtin, N.J.; Smith, G.C. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004, 64, 9152–9159. [Google Scholar] [CrossRef]
- Golding, S.E.; Rosenberg, E.; Adams, B.R.; Wignarajah, S.; Beckta, J.M.; O’Connor, M.J.; Valerie, K. Dynamic inhibition of ATM kinase provides a strategy for glioblastoma multiforme radiosensitization and growth control. Cell Cycle 2012, 11, 1167–1173. [Google Scholar] [CrossRef] [PubMed]
- Vecchio, D.; Daga, A.; Carra, E.; Marubbi, D.; Raso, A.; Mascelli, S.; Nozza, P.; Garrè, M.L.; Pitto, F.; Ravetti, J.L.; et al. Pharmacokinetics, pharmacodynamics and efficacy on pediatric tumors of the glioma radiosensitizer KU60019. Int. J. Cancer 2015, 136, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Batey, M.A.; Zhao, Y.; Kyle, S.; Richardson, C.; Slade, A.; Martin, N.M.; Lau, A.; Newell, D.R.; Curtin, N.J. Preclinical evaluation of a novel ATM inhibitor, KU59403, in vitro and in vivo in p53 functional and dysfunctional models of human cancer. Mol. Cancer Ther. 2013, 12, 959–967. [Google Scholar] [CrossRef]
- Mak, J.P.Y.; Ma, H.T.; Poon, R.Y.C. Synergism between ATM and PARP1 Inhibition Involves DNA Damage and Abrogating the G(2) DNA Damage Checkpoint. Mol. Cancer Ther. 2020, 19, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 2015, 15, 276–289. [Google Scholar] [CrossRef]
- Murga, M.; Campaner, S.; Lopez-Contreras, A.J.; Toledo, L.I.; Soria, R.; Montaña, M.F.; Artista, L.; Schleker, T.; Guerra, C.; Garcia, E.; et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat. Struct. Mol. Biol. 2011, 18, 1331–1335. [Google Scholar] [CrossRef]
- Ruzankina, Y.; Schoppy, D.W.; Asare, A.; Clark, C.E.; Vonderheide, R.H.; Brown, E.J. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat. Genet. 2009, 41, 1144–1149. [Google Scholar] [CrossRef]
- Schoppy, D.W.; Ragland, R.L.; Gilad, O.; Shastri, N.; Peters, A.A.; Murga, M.; Fernandez-Capetillo, O.; Diehl, J.A.; Brown, E.J. Oncogenic stress sensitizes murine cancers to hypomorphic suppression of ATR. J. Clin. Investig. 2012, 122, 241–252. [Google Scholar] [CrossRef]
- Nishida, H.; Tatewaki, N.; Nakajima, Y.; Magara, T.; Ko, K.M.; Hamamori, Y.; Konishi, T. Inhibition of ATR protein kinase activity by schisandrin B in DNA damage response. Nucleic Acids Res. 2009, 37, 5678–5689. [Google Scholar] [CrossRef]
- Charrier, J.D.; Durrant, S.J.; Golec, J.M.; Kay, D.P.; Knegtel, R.M.; MacCormick, S.; Mortimore, M.; O’Donnell, M.E.; Pinder, J.L.; Reaper, P.M.; et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J. Med. Chem. 2011, 54, 2320–2330. [Google Scholar] [CrossRef]
- Reaper, P.M.; Griffiths, M.R.; Long, J.M.; Charrier, J.D.; Maccormick, S.; Charlton, P.A.; Golec, J.M.; Pollard, J.R. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat. Chem. Biol. 2011, 7, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, A.; Knittel, G.; Welcker, D.; Yang, T.P.; George, J.; Nowak, M.; Leeser, U.; Büttner, R.; Perner, S.; Peifer, M.; et al. ATM Deficiency Is Associated with Sensitivity to PARP1- and ATR Inhibitors in Lung Adenocarcinoma. Cancer Res. 2017, 77, 3040–3056. [Google Scholar] [CrossRef] [PubMed]
- Combès, E.; Andrade, A.F.; Tosi, D.; Michaud, H.A.; Coquel, F.; Garambois, V.; Desigaud, D.; Jarlier, M.; Coquelle, A.; Pasero, P.; et al. Inhibition of Ataxia-Telangiectasia Mutated and RAD3-Related (ATR) Overcomes Oxaliplatin Resistance and Promotes Antitumor Immunity in Colorectal Cancer. Cancer Res. 2019, 79, 2933–2946. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Zhang, J.; He, K.; Zhang, J. Ataxia telangiectasia and Rad3-related inhibitors and cancer therapy: Where we stand. J. Hematol. Oncol. 2019, 12, 43. [Google Scholar] [CrossRef]
- Vendetti, F.P.; Lau, A.; Schamus, S.; Conrads, T.P.; O’Connor, M.J.; Bakkenist, C.J. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget 2015, 6, 44289–44305. [Google Scholar] [CrossRef]
- Kwok, M.; Davies, N.; Agathanggelou, A.; Smith, E.; Oldreive, C.; Petermann, E.; Stewart, G.; Brown, J.; Lau, A.; Pratt, G.; et al. ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 2016, 127, 582–595. [Google Scholar] [CrossRef]
- Jin, J.; Fang, H.; Yang, F.; Ji, W.; Guan, N.; Sun, Z.; Shi, Y.; Zhou, G.; Guan, X. Combined Inhibition of ATR and WEE1 as a Novel Therapeutic Strategy in Triple-Negative Breast Cancer. Neoplasia 2018, 20, 478–488. [Google Scholar] [CrossRef]
- Wengner, A.M.; Siemeister, G.; Lücking, U.; Lefranc, J.; Wortmann, L.; Lienau, P.; Bader, B.; Bömer, U.; Moosmayer, D.; Eberspächer, U.; et al. The Novel ATR Inhibitor BAY 1895344 Is Efficacious as Monotherapy and Combined with DNA Damage-Inducing or Repair-Compromising Therapies in Preclinical Cancer Models. Mol. Cancer Ther. 2020, 19, 26–38. [Google Scholar] [CrossRef]
- Willoughby, C.E.; Jiang, Y.; Thomas, H.D.; Willmore, E.; Kyle, S.; Wittner, A.; Phillips, N.; Zhao, Y.; Tudhope, S.J.; Prendergast, L.; et al. Selective DNA-PKcs inhibition extends the therapeutic index of localized radiotherapy and chemotherapy. J. Clin. Investig. 2020, 130, 258–271. [Google Scholar] [CrossRef]
- Liu, Y.; Wan, W.Z.; Li, Y.; Zhou, G.L.; Liu, X.G. Recent development of ATP-competitive small molecule phosphatidylinostitol-3-kinase inhibitors as anticancer agents. Oncotarget 2017, 8, 7181–7200. [Google Scholar] [CrossRef] [PubMed]
- Leder, A.; Leder, P. Butyric acid, a potent inducer of erythroid differentiation in cultured erythroleukemic cells. Cell 1975, 5, 319–322. [Google Scholar] [CrossRef]
- Riggs, M.G.; Whittaker, R.G.; Neumann, J.R.; Ingram, V.M. n-Butyrate causes histone modification in HeLa and Friend erythroleukaemia cells. Nature 1977, 268, 462–464. [Google Scholar] [CrossRef] [PubMed]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, T.; Teng, Z.H.; Zhang, R.; Wang, J.B.; Mei, Q.B. Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in non-small cell lung cancer (NSCLC) cells. Cancer Biol. Ther. 2009, 8, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Blattmann, C.; Oertel, S.; Ehemann, V.; Thiemann, M.; Huber, P.E.; Bischof, M.; Witt, O.; Deubzer, H.E.; Kulozik, A.E.; Debus, J.; et al. Enhancement of radiation response in osteosarcoma and rhabdomyosarcoma cell lines by histone deacetylase inhibition. Int. J. Radiat. Oncol. Biol. Phys. 2010, 78, 237–245. [Google Scholar] [CrossRef]
- Lee, J.H.; Choy, M.L.; Ngo, L.; Foster, S.S.; Marks, P.A. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc. Natl. Acad. Sci. USA 2010, 107, 14639–14644. [Google Scholar] [CrossRef]
- Kachhap, S.K.; Rosmus, N.; Collis, S.J.; Kortenhorst, M.S.; Wissing, M.D.; Hedayati, M.; Shabbeer, S.; Mendonca, J.; Deangelis, J.; Marchionni, L.; et al. Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor. PLoS ONE 2010, 5, e11208. [Google Scholar] [CrossRef]
- Groselj, B.; Kerr, M.; Kiltie, A.E. Radiosensitisation of bladder cancer cells by panobinostat is modulated by Ku80 expression. Radiother. Oncol. 2013, 108, 429–433. [Google Scholar] [CrossRef]
- Krumm, A.; Barckhausen, C.; Kücük, P.; Tomaszowski, K.H.; Loquai, C.; Fahrer, J.; Krämer, O.H.; Kaina, B.; Roos, W.P. Enhanced Histone Deacetylase Activity in Malignant Melanoma Provokes RAD51 and FANCD2-Triggered Drug Resistance. Cancer Res. 2016, 76, 3067–3077. [Google Scholar] [CrossRef]
- Maertens, O.; Kuzmickas, R.; Manchester, H.E.; Emerson, C.E.; Gavin, A.G.; Guild, C.J.; Wong, T.C.; De Raedt, T.; Bowman-Colin, C.; Hatchi, E.; et al. MAPK Pathway Suppression Unmasks Latent DNA Repair Defects and Confers a Chemical Synthetic Vulnerability in BRAF-, NRAS-, and NF1-Mutant Melanomas. Cancer Discov. 2019, 9, 526–545. [Google Scholar] [CrossRef]
- Robert, C.; Nagaria, P.K.; Pawar, N.; Adewuyi, A.; Gojo, I.; Meyers, D.J.; Cole, P.A.; Rassool, F.V. Histone deacetylase inhibitors decrease NHEJ both by acetylation of repair factors and trapping of PARP1 at DNA double-strand breaks in chromatin. Leuk. Res. 2016, 45, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Uhl, M.; Csernok, A.; Aydin, S.; Kreienberg, R.; Wiesmüller, L.; Gatz, S.A. Role of SIRT1 in homologous recombination. DNA Repair 2010, 9, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Wells, C.E.; Bhaskara, S.; Stengel, K.R.; Zhao, Y.; Sirbu, B.; Chagot, B.; Cortez, D.; Khabele, D.; Chazin, W.J.; Cooper, A.; et al. Inhibition of histone deacetylase 3 causes replication stress in cutaneous T cell lymphoma. PLoS ONE 2013, 8, e68915. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dai, X.; Qi, Y.; He, Y.; Du, W.; Pang, J.J. Histone Deacetylases Inhibitors in the Treatment of Retinal Degenerative Diseases: Overview and Perspectives. J. Ophthalmol. 2015, 2015, 250812. [Google Scholar] [CrossRef]
- Das Gupta, K.; Shakespear, M.R.; Iyer, A.; Fairlie, D.P.; Sweet, M.J. Histone deacetylases in monocyte/macrophage development, activation and metabolism: Refining HDAC targets for inflammatory and infectious diseases. Clin. Transl. Immunol. 2016, 5, e62. [Google Scholar] [CrossRef]
- Bhaskara, S.; Knutson, S.K.; Jiang, G.; Chandrasekharan, M.B.; Wilson, A.J.; Zheng, S.; Yenamandra, A.; Locke, K.; Yuan, J.L.; Bonine-Summers, A.R.; et al. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell 2010, 18, 436–447. [Google Scholar] [CrossRef]
- Dovey, O.M.; Foster, C.T.; Conte, N.; Edwards, S.A.; Edwards, J.M.; Singh, R.; Vassiliou, G.; Bradley, A.; Cowley, S.M. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood 2013, 121, 1335–1344. [Google Scholar] [CrossRef]
- Heideman, M.R.; Wilting, R.H.; Yanover, E.; Velds, A.; de Jong, J.; Kerkhoven, R.M.; Jacobs, H.; Wessels, L.F.; Dannenberg, J.H. Dosage-dependent tumor suppression by histone deacetylases 1 and 2 through regulation of c-Myc collaborating genes and p53 function. Blood 2013, 121, 2038–2050. [Google Scholar] [CrossRef]
- Santoro, F.; Botrugno, O.A.; Dal Zuffo, R.; Pallavicini, I.; Matthews, G.M.; Cluse, L.; Barozzi, I.; Senese, S.; Fornasari, L.; Moretti, S.; et al. A dual role for Hdac1: Oncosuppressor in tumorigenesis, oncogene in tumor maintenance. Blood 2013, 121, 3459–3468. [Google Scholar] [CrossRef]
- Hemshekhar, M.; Sebastin Santhosh, M.; Kemparaju, K.; Girish, K.S. Emerging roles of anacardic acid and its derivatives: A pharmacological overview. Basic Clin. Pharmacol. Toxicol. 2012, 110, 122–132. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Swaminathan, V.; Ranganathan, A.; Kundu, T.K. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 2003, 278, 19134–19140. [Google Scholar] [CrossRef] [PubMed]
- Ogiwara, H.; Kohno, T. CBP and p300 histone acetyltransferases contribute to homologous recombination by transcriptionally activating the BRCA1 and RAD51 genes. PLoS ONE 2012, 7, e52810. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Jiang, X.; Chen, S.; Price, B.D. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006, 580, 4353–4356. [Google Scholar] [CrossRef] [PubMed]
- Eliseeva, E.D.; Valkov, V.; Jung, M.; Jung, M.O. Characterization of novel inhibitors of histone acetyltransferases. Mol. Cancer Ther. 2007, 6, 2391–2398. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Altaf, M.; Varier, R.A.; Swaminathan, V.; Ravindran, A.; Sadhale, P.P.; Kundu, T.K. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J. Biol. Chem. 2004, 279, 33716–33726. [Google Scholar] [CrossRef]
- Oike, T.; Ogiwara, H.; Torikai, K.; Nakano, T.; Yokota, J.; Kohno, T. Garcinol, a histone acetyltransferase inhibitor, radiosensitizes cancer cells by inhibiting non-homologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 2012, 84, 815–821. [Google Scholar] [CrossRef]
- Balasubramanyam, K.; Varier, R.A.; Altaf, M.; Swaminathan, V.; Siddappa, N.B.; Ranga, U.; Kundu, T.K. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004, 279, 51163–51171. [Google Scholar] [CrossRef]
- Ogiwara, H.; Ui, A.; Shiotani, B.; Zou, L.; Yasui, A.; Kohno, T. Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor. Carcinogenesis 2013, 34, 2486–2497. [Google Scholar] [CrossRef]
- Kumar, B.; Yadav, A.; Hideg, K.; Kuppusamy, P.; Teknos, T.N.; Kumar, P. A novel curcumin analog (H-4073) enhances the therapeutic efficacy of cisplatin treatment in head and neck cancer. PLoS ONE 2014, 9, e93208. [Google Scholar] [CrossRef]
- Santer, F.R.; Höschele, P.P.; Oh, S.J.; Erb, H.H.; Bouchal, J.; Cavarretta, I.T.; Parson, W.; Meyers, D.J.; Cole, P.A.; Culig, Z. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol. Cancer Ther. 2011, 10, 1644–1655. [Google Scholar] [CrossRef]
- Wang, Y.M.; Gu, M.L.; Meng, F.S.; Jiao, W.R.; Zhou, X.X.; Yao, H.P.; Ji, F. Histone acetyltransferase p300/CBP inhibitor C646 blocks the survival and invasion pathways of gastric cancer cell lines. Int. J. Oncol. 2017, 51, 1860–1868. [Google Scholar] [CrossRef] [PubMed]
- de Boer, J.; Walf-Vorderwülbecke, V.; Williams, O. In focus: MLL-rearranged leukemia. Leukemia 2013, 27, 1224–1228. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y.; Feng, Q.; Lin, Y.; Jiang, Q.; Li, Y.; Coffield, V.M.; Su, L.; Xu, G.; Zhang, Y. hDOT1L links histone methylation to leukemogenesis. Cell 2005, 121, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Daigle, S.R.; Olhava, E.J.; Therkelsen, C.A.; Majer, C.R.; Sneeringer, C.J.; Song, J.; Johnston, L.D.; Scott, M.P.; Smith, J.J.; Xiao, Y.; et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 2011, 20, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Klaus, C.R.; Iwanowicz, D.; Johnston, D.; Campbell, C.A.; Smith, J.J.; Moyer, M.P.; Copeland, R.A.; Olhava, E.J.; Scott, M.P.; Pollock, R.M.; et al. DOT1L inhibitor EPZ-5676 displays synergistic antiproliferative activity in combination with standard of care drugs and hypomethylating agents in MLL-rearranged leukemia cells. J. Pharmacol. Exp. Ther. 2014, 350, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, G.S.; Chakraborty, A.; Grigsby, S.M.; Umeano, A.C.; Liao, C.; Moukha-Chafiq, O.; Pathak, V.; Mathew, B.; Lee, Y.T.; Dou, Y.; et al. Identification of DOT1L inhibitors by structure-based virtual screening adapted from a nucleoside-focused library. Eur. J. Med. Chem. 2020, 189, 112023. [Google Scholar] [CrossRef]
- Byun, W.S.; Kim, W.K.; Yoon, J.S.; Jarhad, D.B.; Jeong, L.S.; Lee, S.K. Antiproliferative and Antimigration Activities of Fluoro-Neplanocin A via Inhibition of Histone H3 Methylation in Triple-Negative Breast Cancer. Biomolecules 2020, 10, 530. [Google Scholar] [CrossRef]
- Yang, L.; Lei, Q.; Li, L.; Yang, J.; Dong, Z.; Cui, H. Silencing or inhibition of H3K79 methyltransferase DOT1L induces cell cycle arrest by epigenetically modulating c-Myc expression in colorectal cancer. Clin. Epigenetics 2019, 11, 199. [Google Scholar] [CrossRef]
- Yang, C.; Chen, Z.; Yu, H.; Liu, X. Inhibition of Disruptor of Telomeric Silencing 1-Like Alleviated Renal Ischemia and Reperfusion Injury-Induced Fibrosis by Blocking PI3K/AKT-Mediated Oxidative Stress. Drug Des. Dev. Ther. 2019, 13, 4375–4387. [Google Scholar] [CrossRef]
- Williams, D.E.; Dalisay, D.S.; Li, F.; Amphlett, J.; Maneerat, W.; Chavez, M.A.; Wang, Y.A.; Matainaho, T.; Yu, W.; Brown, P.J.; et al. Nahuoic acid A produced by a Streptomyces sp. isolated from a marine sediment is a selective SAM-competitive inhibitor of the histone methyltransferase SETD8. Org. Lett. 2013, 15, 414–417. [Google Scholar] [CrossRef]
- Ma, A.; Yu, W.; Li, F.; Bleich, R.M.; Herold, J.M.; Butler, K.V.; Norris, J.L.; Korboukh, V.; Tripathy, A.; Janzen, W.P.; et al. Discovery of a selective, substrate-competitive inhibitor of the lysine methyltransferase SETD8. J. Med. Chem. 2014, 57, 6822–6833. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, K.D.; Mitchell, T.R.; Upadhyay, A.K.; Jakob, C.G.; Jhala, M.A.; Comess, K.M.; Lasko, L.M.; Li, C.; Tuzon, C.T.; Dai, Y.; et al. The SUV4-20 inhibitor A-196 verifies a role for epigenetics in genomic integrity. Nat. Chem. Biol. 2017, 13, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Liang, D.; Jiang, N.; Zhou, J.; Long, X.; Wang, X.; Wu, M.; Wu, C.; Bao, J. Computer-aided screening for suppressor of variegation 4-20 homolog 1 inhibitors and their preliminary activity validation in human osteosarcoma. J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gieni, R.S.; Ismail, I.H.; Campbell, S.; Hendzel, M.J. Polycomb group proteins in the DNA damage response: A link between radiation resistance and “stemness”. Cell Cycle 2011, 10, 883–894. [Google Scholar] [CrossRef]
- Simó-Riudalbas, L.; Esteller, M. Targeting the histone orthography of cancer: Drugs for writers, erasers and readers. Br. J. Pharmacol. 2015, 172, 2716–2732. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Ismail, I.H.; Young, L.C.; Poirier, G.G.; Hendzel, M.J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell Cycle 2013, 12, 2675–2683. [Google Scholar] [CrossRef]
- Jeusset, L.M.; McManus, K.J. Developing Targeted Therapies That Exploit Aberrant Histone Ubiquitination in Cancer. Cells 2019, 8, 165. [Google Scholar] [CrossRef]
- Nishida, Y.; Maeda, A.; Kim, M.J.; Cao, L.; Kubota, Y.; Ishizawa, J.; AlRawi, A.; Kato, Y.; Iwama, A.; Fujisawa, M.; et al. The novel BMI-1 inhibitor PTC596 downregulates MCL-1 and induces p53-independent mitochondrial apoptosis in acute myeloid leukemia progenitor cells. Blood Cancer J. 2017, 7, e527. [Google Scholar] [CrossRef] [PubMed]
- Papillon, J.P.N.; Nakajima, K.; Adair, C.D.; Hempel, J.; Jouk, A.O.; Karki, R.G.; Mathieu, S.; Möbitz, H.; Ntaganda, R.; Smith, T.; et al. Discovery of Orally Active Inhibitors of Brahma Homolog (BRM)/SMARCA2 ATPase Activity for the Treatment of Brahma Related Gene 1 (BRG1)/SMARCA4-Mutant Cancers. J. Med. Chem. 2018, 61, 10155–10172. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef]
- Mandal, P.K.; Blanpain, C.; Rossi, D.J. DNA damage response in adult stem cells: Pathways and consequences. Nat. Rev. Mol. Cell Biol. 2011, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wiese, C.; Kwon, Y.; Hromas, R.; Sung, P. The BRCA Tumor Suppressor Network in Chromosome Damage Repair by Homologous Recombination. Annu. Rev. Biochem. 2019, 88, 221–245. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zou, R.S.; He, S.; Nihongaki, Y.; Li, X.; Razavi, S.; Wu, B.; Ha, T. Very fast CRISPR on demand. Science 2020, 368, 1265–1269. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).