Expression of miR-135b in Psoriatic Skin and Its Association with Disease Improvement
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects and Sample Collection
2.2. Small RNA NGS
2.3. Tissue miRNA Isolation, Reverse Transcription and RT-PCR
2.4. miRNA Target Profiling
2.5. Statistics
3. Results
3.1. miRNA Profiling of Psoriatic Skin by NGS
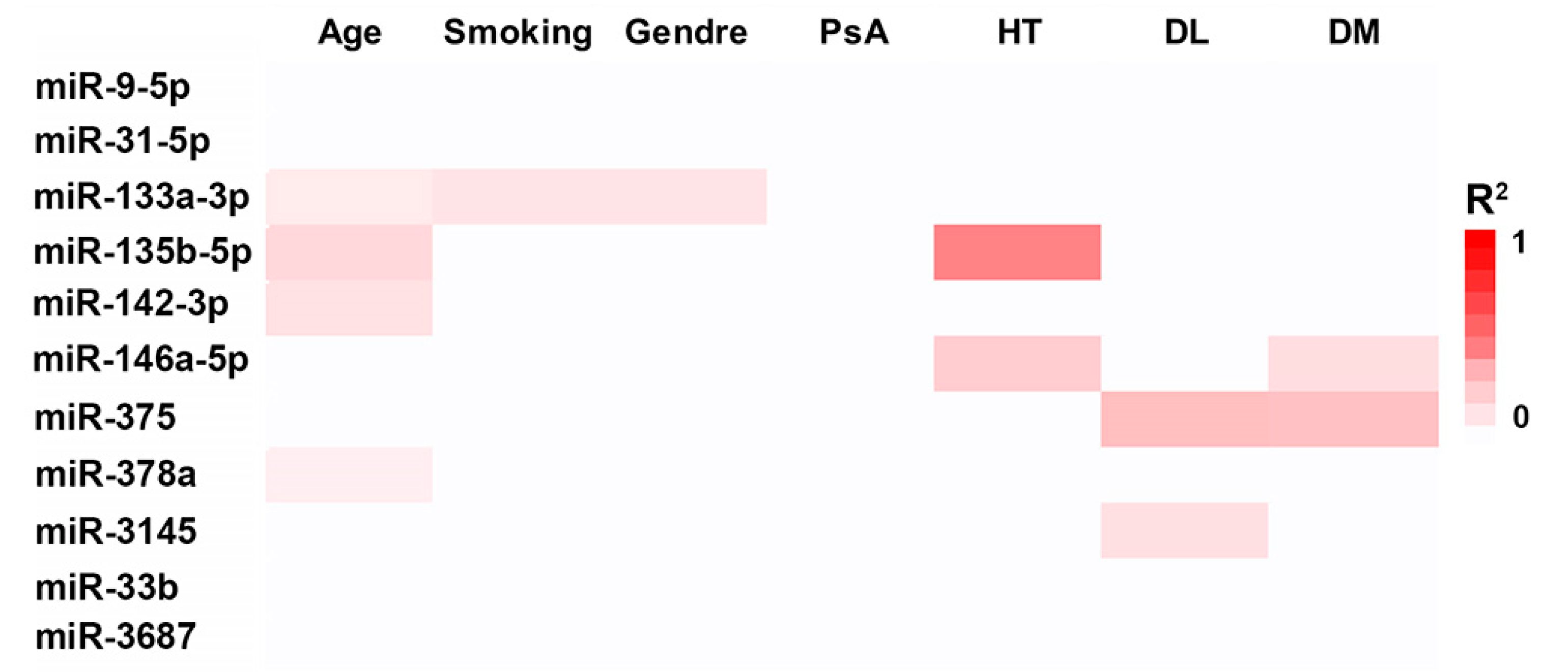
3.2. Effect of Demographic and Clinical Variables on miRNA Expression
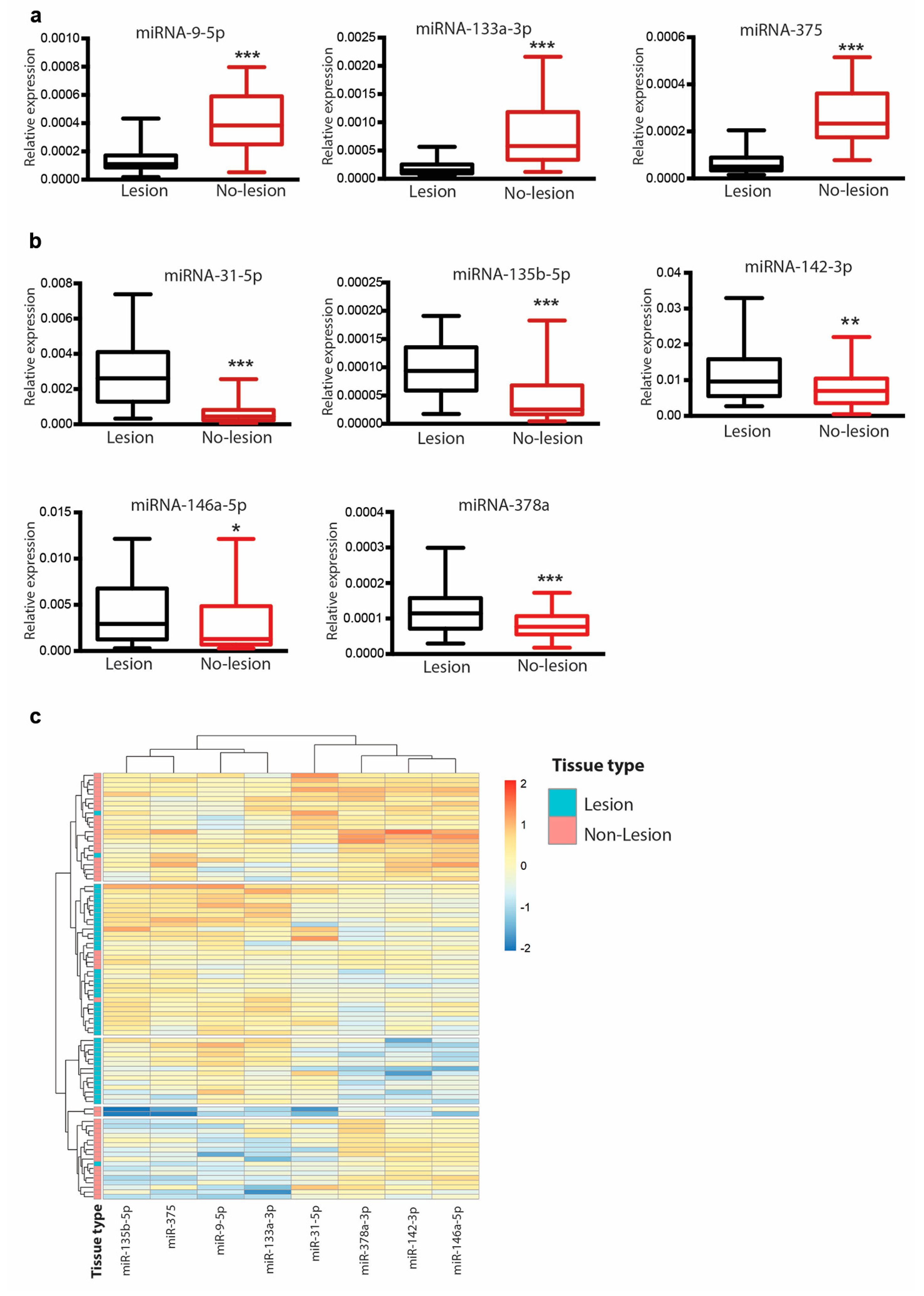
3.3. Differential Expression of miR-9-5p, miR-375, miR-133a and Their Modulation Following Treatment
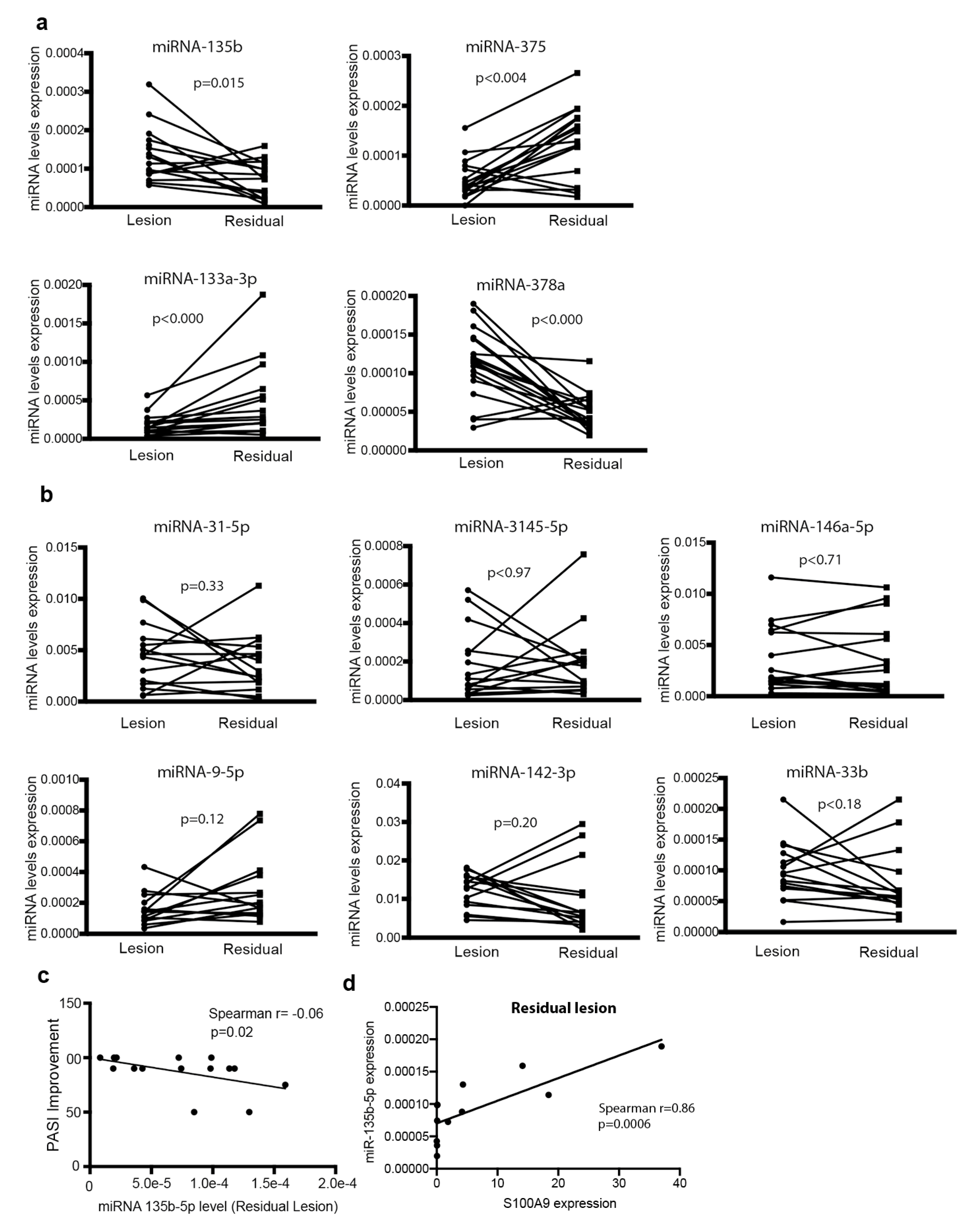
3.4. Relationship between miRNA Expression and Psoriasis Activity
3.5. Basal Expression of miR-146a and miR-135b Is Associated with Disease Improvement
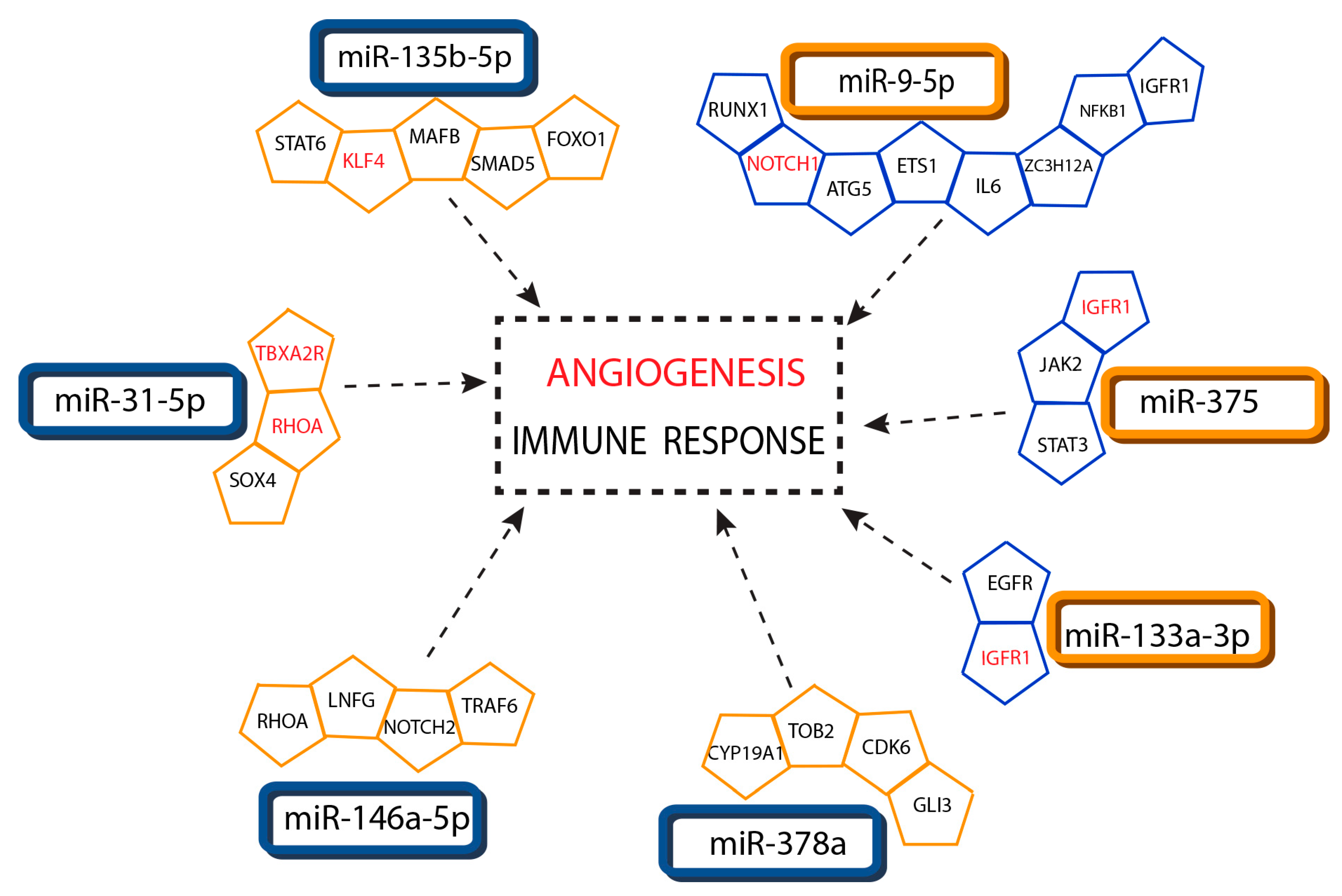
3.6. Association of Interacting miRNA-Targets with Psoriasis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perera, G.; Di Meglio, P.; Nestle, F.O. Psoriasis. Annu. Rev. Pathol. 2012, 7, 385–422. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2014, 25, 137–147. [Google Scholar] [CrossRef]
- Miska, E.A.; Alvarez-Saavedra, E.; Abbott, A.L.; Lau, N.C.; Hellman, A.B.; McGonagle, S.M.; Bartel, D.P.; Ambros, V.R.; Horvitz, H.R. Most Caenorhabditis elegans microRNAs are Individually Not Essential for Development or Viability. PLoS Genet. 2007, 3, e215. [Google Scholar] [CrossRef]
- Park, C.Y.; Jeker, L.T.; Carver-Moore, K.; Oh, A.; Liu, H.J.; Cameron, R.; Richards, H.; Li, Z.; Adler, D.; Yoshinaga, Y.; et al. A Resource for the Conditional Ablation of microRNAs in the Mouse. Cell Rep. 2012, 1, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.; Farh, K.K.-H.; Burge, C.B.; Bartel, B. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2008, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.I.; Yamagata, K.; Sugimoto, K.; Iwamoto, T.; Kato, S.; Miyazono, K. Modulation of microRNA processing by p53. Nature 2009, 460, 529–533. [Google Scholar] [CrossRef]
- Wiesen, J.L.; Tomasi, T.B. Dicer is regulated by cellular stresses and interferons. Mol. Immunol. 2009, 46, 1222–1228. [Google Scholar] [CrossRef]
- Simon, L.M.; Edelstein, L.C.; Nagalla, S.; Woodley, A.B.; Chen, E.S.; Kong, X.; Ma, L.; Fortina, P.; Kunapuli, S.; Holinstat, M.; et al. Human platelet microRNA-mRNA networks associated with age and gender revealed by integrated plateletomics. Blood 2014, 123, e37–e45. [Google Scholar] [CrossRef]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-biased MicroRNAs. Genom. Proteom. Bioinform. 2018, 16, 200–211. [Google Scholar] [CrossRef]
- Srivastava, A.; Karlsson, M.; Marionnet, C.; Bernerd, F.; Gueniche, A.; Rawadi, C.E.L.; Ståhle, M.; Sonkoly, E.; Breton, L.; Pivarcsi, A. Identification of chronological and photoageing-associated microRNAs in human skin. Sci. Rep. 2018, 8, 12990. [Google Scholar] [CrossRef] [PubMed]
- Momi, N.; Kaur, S.; Rachagani, S.; Ganti, A.K.; Batra, S.K.; Rachgani, S. Smoking and microRNA dysregulation: A cancerous combination. Trends Mol. Med. 2013, 20, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, J.E.; Nguyen, G.H.; Fujita, M.; Florell, S.R.; Duffin, K.C.; Krueger, G.G.; O’Connell, R.M. microRNAs in Psoriasis. J. Investig. Dermatol. 2016, 136, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Joyce, C.E.; Zhou, X.; Xia, J.; Ryan, C.; Thrash, B.; Menter, A.; Zhang, W.; Bowcock, A.M. Deep sequencing of small RNAs from human skin reveals major alterations in the psoriasis miRNAome. Hum. Mol. Genet. 2011, 20, 4025–4040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yi, X.; Guo, S.; Shi, Q.; Wei, C.; Li, X.; Gao, L.; Wang, G.; Gao, T.; Wang, L.; et al. A single-nucleotide polymorphism of miR-146a and psoriasis: An association and functional study. J. Cell. Mol. Med. 2014, 18, 2225–2234. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.-B.; Zhang, S.-L.; Wu, X.-J.; Pu, X.-M.; Kang, X.-J. Association of rs2910164 polymorphism in MiR-146a gene with psoriasis susceptibility. Medicine 2019, 98, e14401. [Google Scholar] [CrossRef]
- Swindell, W.R.; Remmer, H.A.; Sarkar, M.K.; Xing, X.; Barnes, D.H.; Wolterink, L.; Voorhees, J.J.; Nair, R.P.; Johnston, A.; Elder, J.T.; et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015, 7, 86. [Google Scholar] [CrossRef]
- Masalha, M.; Sidi, Y.; Avni, D. The contribution of feedback loops between miRNAs, cytokines and growth factors to the pathogenesis of psoriasis. Exp. Dermatol. 2018, 27, 603–610. [Google Scholar] [CrossRef]
- Srivastava, A.; Nikamo, P.; Lohcharoenkal, W.; Li, D.; Meisgen, F.; Landén, N.X.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. MicroRNA-146a suppresses IL-17–mediated skin inflammation and is genetically associated with psoriasis. J. Allergy Clin. Immunol. 2017, 139, 550–561. [Google Scholar] [CrossRef]
- Hermann, H.; Runnel, T.; Aab, A.; Baurecht, H.; Rodriguez, E.; Magilnick, N.; Urgard, E.; Šahmatova, L.; Prans, E.; Maslovskaja, J.; et al. miR-146b Probably Assists miRNA-146a in the Suppression of Keratinocyte Proliferation and Inflammatory Responses in Psoriasis. J. Investig. Dermatol. 2017, 137, 1945–1954. [Google Scholar] [CrossRef]
- Torri, A.; Carpi, N.; Bulgheroni, E.; Crosti, M.-C.; Moro, M.; Gruarin, P.; Rossi, R.L.; Rossetti, G.; Di Vizio, L.; Hoxha, M.; et al. Extracellular MicroRNA Signature of Human Helper T Cell Subsets in Health and Autoimmunity. J. Biol. Chem. 2017, 292, 2903–2915. [Google Scholar] [CrossRef] [PubMed]
- Landén, N.X.; Meisgen, F.; Butler, L.M.; Han, G.; Wang, X.; Söderberg-Nauclér, C.; Ståhle, M.; Pivarcsi, A.; Sonkoly, E. MicroRNA-31 Is Overexpressed in Psoriasis and Modulates Inflammatory Cytokine and Chemokine Production in Keratinocytes via Targeting Serine/Threonine Kinase 40. J. Immunol. 2012, 190, 678–688. [Google Scholar] [CrossRef]
- Yan, S.; Xu, Z.; Lou, F.; Zhang, L.; Ke, F.; Bai, J.; Liu, Z.; Liu, J.; Wang, H.; Zhu, H.; et al. NF-κB-induced microRNA-31 promotes epidermal hyperplasia by repressing protein phosphatase 6 in psoriasis. Nat. Commun. 2015, 6, 7652. [Google Scholar] [CrossRef] [PubMed]
- Pivarcsi, A.; Meisgen, F.; Xu, N.; Stahle, M.; Sonkoly, E. Changes in the Level of Serum Micrornas in Patients with Psoriasis after Antitumour Necrosis Factor-Alpha Therapy. Br. J. Dermatol. 2013, 169, 563–570. [Google Scholar] [CrossRef]
- Mensà, E.; Recchioni, R.; Marcheselli, F.; Giuliodori, K.; Consales, V.; Molinelli, E.; Prattichizzo, F.; Rippo, M.R.; Campanati, A.; Procopio, A.D.; et al. MiR-146a-5p correlates with clinical efficacy in patients with psoriasis treated with the tumour necrosis factor-alpha inhibitor adalimumab. Br. J. Dermatol. 2018, 179, 787–789. [Google Scholar] [CrossRef]
- Lu, L.-F.; Boldin, M.P.; Chaudhry, A.; Lin, L.-L.; Taganov, K.D.; Hanada, T.; Yoshimura, A.; Baltimore, D.; Rudensky, A.Y. Function of miR-146a in Controlling Treg Cell-Mediated Regulation of Th1 Responses. Cell 2010, 142, 914–929. [Google Scholar] [CrossRef]
- Ou, M.; Zhang, Y.; Cui, S.; Zhao, S.; Tu, J. Upregulated Mir-9–5p Protects against Inflammatory Response in Rats with Deep Vein Thrombosis Via Inhibition of Nf-Kappab P50. Inflammation 2019, 42, 1925–1938. [Google Scholar] [CrossRef]
- Yue, P.; Jing, L.; Zhao, X.; Zhu, H.; Teng, J. Down-Regulation of Taurine-up-Regulated Gene 1 Attenuates Inflammation by Sponging Mir-9–5p Via Targeting Nf-Kappab1/P50 in Multiple Sclerosis. Life Sci. 2019, 233, 116731. [Google Scholar] [CrossRef]
- Gundara, J.S.; Zhao, J.; Gill, A.J.; Lee, J.C.; Delbridge, L.; Robinson, B.G.; McLean, C.; Serpell, J.; Sidhu, S.B. Noncoding RNA blockade of autophagy is therapeutic in medullary thyroid cancer. Cancer Med. 2014, 4, 174–182. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, H.; Zheng, H.; Zhou, X.; Shen, G.; Teng, X.; Liu, X.; Zhang, J.; Wei, X.; Hu, Z.; et al. Autophagy-Based Unconventional Secretion of Hmgb1 by Keratinocytes Plays a Pivotal Role in Psoriatic Skin inflammation. Autophagy 2020, 1–24. [Google Scholar] [CrossRef]
- Shao, Y.; Chong, L.; Lin, P.; Li, H.; Zhu, L.; Wu, Q.; Li, C.-C. MicroRNA-133a alleviates airway remodeling in asthtama through PI3K/AKT/mTOR signaling pathway by targeting IGF1R. J. Cell. Physiol. 2018, 234, 4068–4080. [Google Scholar] [CrossRef]
- Krane, J.F.; Gottlieb, A.B.; Carter, D.M.; Krueger, J.G. The insulin-like growth factor I receptor is overexpressed in psoriatic epidermis, but is differentially regulated from the epidermal growth factor receptor. J. Exp. Med. 1992, 175, 1081–1090. [Google Scholar] [CrossRef][Green Version]
- Wraight, C.J.; White, P.J.; Mckean, S.C.; Fogarty, R.D.; Venables, D.J.; Liepe, I.J.; Edmondson, S.R.; Werther, G.A. Reversal of epidermal hyperproliferation in psoriasis by insulin-like growth factor I receptor antisense oligonucleotides. Nat. Biotechnol. 2000, 18, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.-M.; Zhu, H.; Bai, W.-D.; Wang, T.; Wang, L.; Chen, Y.; Yang, A.; Jia, L. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer 2014, 14, 134. [Google Scholar] [CrossRef]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef]
- Ding, L.; Xu, Y.; Zhang, W.; Deng, Y.; Si, M.; Du, Y.; Yao, H.; Liu, X.; Ke, Y.; Si, J.; et al. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010, 20, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Besler, C.; Urban, D.; Watzka, S.; Lang, D.; Rommel, K.-P.; Kandolf, R.; Klingel, K.; Thiele, H.; Linke, A.; Schuler, G.; et al. Endomyocardial miR-133a levels correlate with myocardial inflammation, improved left ventricular function, and clinical outcome in patients with inflammatory cardiomyopathy. Eur. J. Heart Fail. 2016, 18, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Li, J.; Yang, H.; Zhang, H.; Zhou, Z.; Deng, T.; Zhu, K.; Ning, T.; Fan, Q.; Ying, G.; et al. miR-135b Delivered by Gastric Tumor Exosomes Inhibits FOXO1 Expression in Endothelial Cells and Promotes Angiogenesis. Mol. Ther. 2019, 27, 1772–1783. [Google Scholar] [CrossRef]
- Umezu, T.; Tadokoro, H.; Azuma, K.; Yoshizawa, S.; Ohyashiki, K.; Ohyashiki, J.H. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 2014, 124, 3748–3757. [Google Scholar] [CrossRef]
- Hedrick, S.M.; Michelini, R.H.; Doedens, A.; Goldrath, A.W.; Stone, E.L. FOXO transcription factors throughout T cell biology. Nat. Rev. Immunol. 2012, 12, 649–661. [Google Scholar] [CrossRef]
- Ouyang, W.; Liao, W.; Luo, C.T.; Yin, N.; Huse, M.; Kim, M.V.; Peng, M.; Chan, P.; Ma, Q.; Mo, Y.; et al. Novel Foxo1-dependent transcriptional programs control T(reg) cell function. Nature 2012, 491, 554–559. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Lei, J.; Yang, L.; Gao, C.; Dang, E.; Cao, T.; Xue, K.; Zhuang, Y.; Shao, S.; Zhi, D.; et al. Dysregulation of Akt-FOXO1 Pathway Leads to Dysfunction of Regulatory T Cells in Patients with Psoriasis. J. Investig. Dermatol. 2019, 139, 2098–2107. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, H.; Gyulai, R.; Toichi, E.; Garaczi, E.; Shimada, S.; Stevens, S.R.; McCormick, T.S.; Cooper, K.D. Dysfunctional blood and target tissue CD4+CD25high regulatory T cells in psoriasis: Mechanism underlying unrestrained pathogenic effector T cell proliferation. J. Immunol. 2005, 174, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Keijsers, R.; Van Der Velden, H.; Van Erp, P.; Joosten, I.; Koenen, H.; Van De Kerkhof, P.; Huizen, R.D.B.-V. Balance of Treg vs. T-helper cells in the transition from symptomless to lesional psoriatic skin. Br. J. Dermatol. 2013, 168, 1294–1302. [Google Scholar] [CrossRef] [PubMed]
- Kotb, I.S.; Lewis, B.J.; Barker, R.; Ormerod, A.D. Differential effects of phototherapy, adalimumab and betamethasone-calcipotriol on effector and regulatory T cells in psoriasis. Br. J. Dermatol. 2018, 179, 127–135. [Google Scholar] [CrossRef]
- Hartwig, T.; Zwicky, P.; Schreiner, B.; Yawalkar, N.; Cheng, P.F.; Navarini, A.; Dummer, R.; Flatz, L.; Conrad, C.; Schlapbach, C.; et al. Regulatory T Cells Restrain Pathogenic T Helper Cells during Skin Inflammation. Cell Rep. 2018, 25, 3564–3572. [Google Scholar] [CrossRef]
- Chen, L.; Shen, Z.; Wang, G.; Fan, P.; Liu, Y. Dynamic frequency of CD4+CD25+Foxp3+ Treg cells in Psoriasis vulgaris. J. Dermatol. Sci. 2008, 51, 200–203. [Google Scholar] [CrossRef]
- Richetta, A.G.; Mattozzi, C.; Salvi, M.; Giancristoforo, S.; D’Epiro, S.; Milana, B.; Carboni, V.; Zampetti, M.; Calvieri, S.; Morrone, S. CD4+ CD25+ T-regulatory cells in psoriasis. Correlation between their numbers and biologics-induced clinical improvement. Eur. J. Dermatol. 2011, 21, 344–348. [Google Scholar] [CrossRef]
- Shimizu, T.; Kamata, M.; Fukaya, S.; Hayashi, K.; Fukuyasu, A.; Tanaka, T.; Ishikawa, T.; Ohnishi, T.; Tada, Y. Anti-IL-17A and IL-23p19 antibodies but not anti-TNFα antibody induce expansion of regulatory T cells and restoration of their suppressive function in imiquimod-induced psoriasiform dermatitis. J. Dermatol. Sci. 2019, 95, 90–98. [Google Scholar] [CrossRef]
- Matsuyama, H.; Suzuki, H.I.; Nishimori, H.; Noguchi, M.; Yao, T.; Komatsu, N.; Mano, H.; Sugimoto, K.; Miyazono, K. miR-135b mediates NPM-ALK–driven oncogenicity and renders IL-17–producing immunophenotype to anaplastic large cell lymphoma. Blood 2011, 118, 6881–6892. [Google Scholar] [CrossRef]

| β Coefficient (95% CI) | p-Value | |
|---|---|---|
| DM | −10.36 (−20.13–−0.59) | 0.039 |
| * creatinine levels | 21.49 (8.21–34.7) | 0.003 |
| ** miRNA-9-5p | 7.35 (2.44–12.25) | 0.005 |
| OR (95% CI) | p-Value | |
|---|---|---|
| Treatment * anti-IL12/IL23 anti-TNFa | ||
| 0.31 (0.05–1.88) | 0.200 | |
| 0.55 (0.08–3.83) | 0.550 | |
| Age | 0.95 (0.90–1.0) | 0.078 |
| NL miRNA-146a | 2.33 (1.25–4.58) | 0.015 |
| L miRNA-135b | 6.06 (1.57–23.33) | 0.009 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chicharro, P.; Rodríguez-Jiménez, P.; Llamas-Velasco, M.; Montes, N.; Sanz-García, A.; Cibrian, D.; Vara, A.; Gómez, M.J.; Jiménez-Fernández, M.; Martínez-Fleta, P.; et al. Expression of miR-135b in Psoriatic Skin and Its Association with Disease Improvement. Cells 2020, 9, 1603. https://doi.org/10.3390/cells9071603
Chicharro P, Rodríguez-Jiménez P, Llamas-Velasco M, Montes N, Sanz-García A, Cibrian D, Vara A, Gómez MJ, Jiménez-Fernández M, Martínez-Fleta P, et al. Expression of miR-135b in Psoriatic Skin and Its Association with Disease Improvement. Cells. 2020; 9(7):1603. https://doi.org/10.3390/cells9071603
Chicago/Turabian StyleChicharro, Pablo, Pedro Rodríguez-Jiménez, Mar Llamas-Velasco, Nuria Montes, Ancor Sanz-García, Danay Cibrian, Alicia Vara, Manuel J Gómez, María Jiménez-Fernández, Pedro Martínez-Fleta, and et al. 2020. "Expression of miR-135b in Psoriatic Skin and Its Association with Disease Improvement" Cells 9, no. 7: 1603. https://doi.org/10.3390/cells9071603
APA StyleChicharro, P., Rodríguez-Jiménez, P., Llamas-Velasco, M., Montes, N., Sanz-García, A., Cibrian, D., Vara, A., Gómez, M. J., Jiménez-Fernández, M., Martínez-Fleta, P., Sánchez-García, I., Lozano-Prieto, M., Triviño, J. C., Miñambres, R., Sánchez-Madrid, F., de la Fuente, H., & Dauden, E. (2020). Expression of miR-135b in Psoriatic Skin and Its Association with Disease Improvement. Cells, 9(7), 1603. https://doi.org/10.3390/cells9071603






