Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer
Abstract
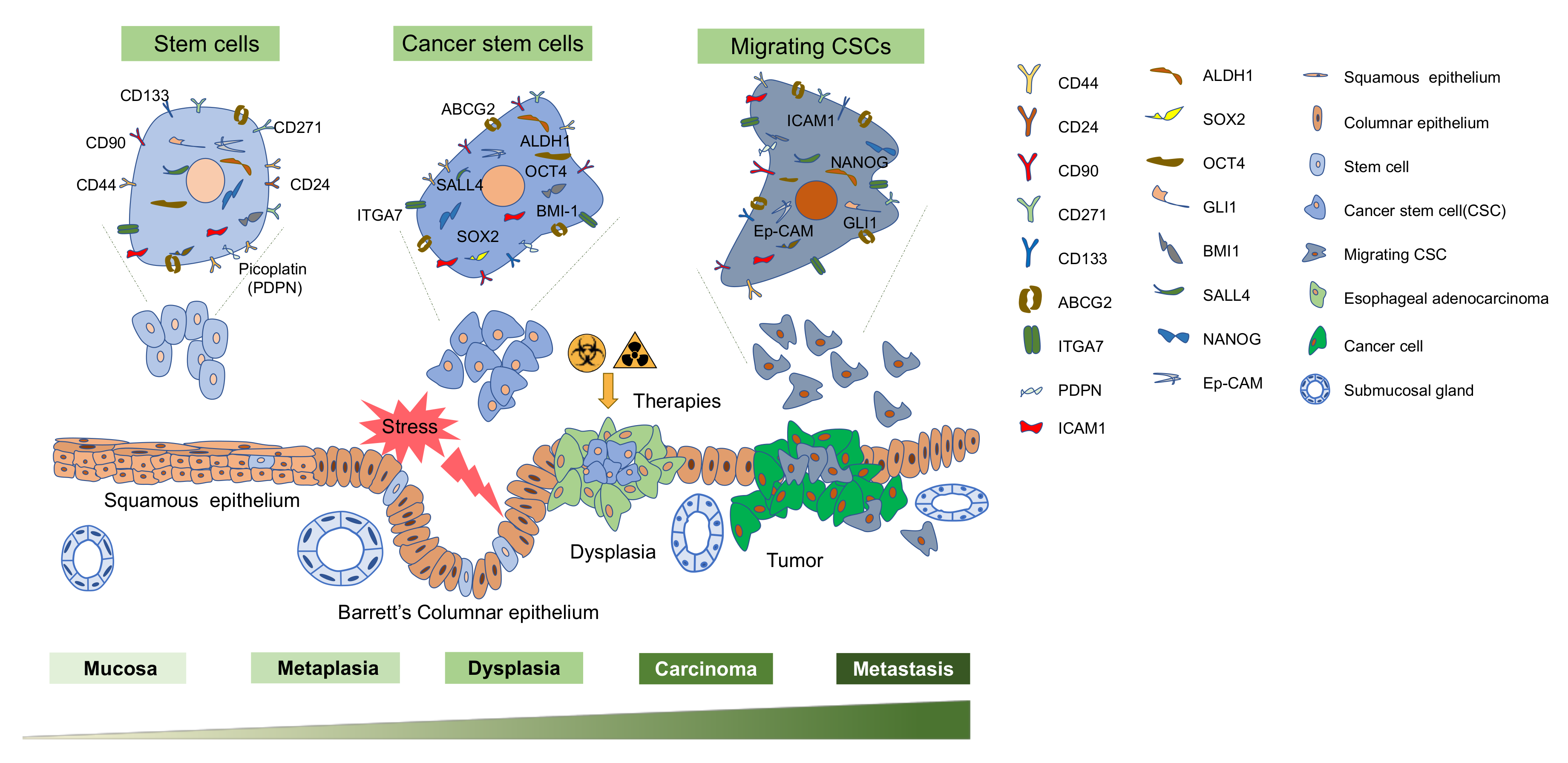
1. Introduction
2. Isolation of Esophageal Cancer Stem Cells
2.1. ECSC Isolation—Biomarker Based
2.2. ECSC Isolation—Biomarker-Free
2.3. Heterogeneity and Single-Cell Analysis of ECSCs
3. ECSC Signaling Pathways
4. Therapeutic Resistance and CSC in EC
5. Therapeutic Strategies Targeting CSC in EC
6. Conclusions and Future Perspectives
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ACAM | Attached-cell Aldefluor method |
| ALDHs | Aldehyde dehydrogenases |
| AML | Acute myeloid leukemia |
| BE | Barrett’s esophagus |
| CAFs | Cancer-associated fibroblasts |
| CAR | Chimeric antigen receptor |
| CSCs | Cancer stem cells |
| CRT | Chemoradiotherapy |
| DDR | DNA damage response |
| EAC | Esophageal adenocarcinoma |
| EC | Esophageal cancer |
| ECSCs | Esophageal cancer stem cells |
| EGFR | Epidermal growth factor receptor |
| EMT | Epithelial mesenchymal transition |
| ESCC | Esophageal squamous cell carcinoma |
| FACS | Fluorescence-activated cell sorting |
| GEJ | Gastroesophageal junction |
| GERD | Gastroesophageal reflux disease |
| Hh | Hedgehog |
| HIF-1α | Hypoxia-inducible-factor 1α |
| HPV | Human Papillomavirus |
| ICAM1 | Intercellular adhesion molecule1 |
| lncRNA | Long noncoding RNA |
| Gli-1 | Glioma-associated oncogene homolog 1 |
| mAb | Monoclonal antibodies |
| MRD | Minimal residual disease |
| nCRT | Neoadjuvant chemoradiation |
| MAML1 | Mastermind like1 |
| MDR | Multidrug resistance |
| MHC-I | MHC class I molecules |
| REGARD | Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma |
| PDPN | Podoplanin |
| PTCH1 | Patched 1 |
| ROS | Reactive oxygen species |
| RR | Reporter-responsive |
| scRNA-seq | Single-cell RNA sequencing |
| SCs | Stem cells |
| SHH | Sonic Hedgehog |
| SP | Side population |
| SRR2 | Sox2 regulatory region |
| TGF-β | Transforming growths factor-β |
| TME | Tumor microenvironment |
| T-ICs | Tumor-initiating cells |
| YAP | Yes-associated protein |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Lagergren, J.; Smyth, E.; Cunningham, D.; Lagergren, P. Oesophageal cancer. Lancet 2017, 390, 2383–2396. [Google Scholar] [CrossRef]
- Chung, C.S.; Lee, Y.C.; Wu, M.S. Prevention strategies for esophageal cancer: Perspectives of the East vs. West. Best Pract. Res. Clin. Gastroenterol. 2015, 29, 869–883. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Frankell, A.M.; Jammula, S.; Li, X.; Contino, G.; Killcoyne, S.; Abbas, S.; Perner, J.; Bower, L.; Devonshire, G.; Ococks, E.; et al. The landscape of selection in 551 esophageal adenocarcinomas defines genomic biomarkers for the clinic. Nat. Genet. 2019, 51, 506–516. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Chen, H.; Zhou, D.; Zhang, J.; Chen, Y.; Liu, Q.; Ai, D.; Zhu, H.; Chu, L.; Ren, W.; et al. Comparative genomic analysis of esophageal squamous cell carcinoma between Asian and Caucasian patient populations. Nat. Commun. 2017, 8, 1533. [Google Scholar] [CrossRef]
- Lin, Y.; Totsuka, Y.; He, Y.; Kikuchi, S.; Qiao, Y.; Ueda, J.; Wei, W.; Inoue, M.; Tanaka, H. Epidemiology of esophageal cancer in Japan and China. J. Epidemiol. 2013, 23, 233–242. [Google Scholar] [CrossRef]
- Abnet, C.C.; Arnold, M.; Wei, W.Q. Epidemiology of Esophageal Squamous Cell Carcinoma. Gastroenterology 2018, 154, 360–373. [Google Scholar] [CrossRef]
- Ajani, J.A.; D’Amico, T.A.; Bentrem, D.J.; Chao, J.; Corvera, C.; Das, P.; Denlinger, C.S.; Enzinger, P.C.; Fanta, P.; Farjah, F.; et al. Esophageal and Esophagogastric Junction Cancers, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 855–883. [Google Scholar] [CrossRef]
- Van Hagen, P.; Hulshof, M.C.; van Lanschot, J.J.; Steyerberg, E.W.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.; Richel, D.J.; Nieuwenhuijzen, G.A.; Hospers, G.A.; Bonenkamp, J.J.; et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. N. Engl. J. Med. 2012, 366, 2074–2084. [Google Scholar] [CrossRef]
- Al-Batran, S.-E.; Homann, N.; Pauligk, C.; Goetze, T.O.; Meiler, J.; Kasper, S.; Kopp, H.-G.; Mayer, F.; Haag, G.M.; Luley, K.; et al. Perioperative chemotherapy with fluorouracil plus leucovorin, oxaliplatin, and docetaxel versus fluorouracil or capecitabine plus cisplatin and epirubicin for locally advanced, resectable gastric or gastro-oesophageal junction adenocarcinoma (FLOT4): A randomised, phase 2/3 trial. Lancet 2019, 393, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Clarke, M.F. Clinical and Therapeutic Implications of Cancer Stem Cells. N. Engl. J. Med. 2019, 380, 2237–2245. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, D.; Dick, J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat. Med. 1997, 3, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Hawkins, C.; Clarke, I.D.; Squire, J.A.; Bayani, J.; Hide, T.; Henkelman, R.M.; Cusimano, M.D.; Dirks, P.B. Identification of human brain tumour initiating cells. Nature 2004, 432, 396–401. [Google Scholar] [CrossRef]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef]
- Da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Munoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef]
- Vermeulen, L.; de Sousa e Melo, F.; Richel, D.J.; Medema, J.P. The developing cancer stem-cell model: Clinical challenges and opportunities. Lancet Oncol. 2012, 13, e83–e89. [Google Scholar] [CrossRef]
- Batlle, E.; Clevers, H. Cancer stem cells revisited. Nat. Med. 2017, 23, 1124–1134. [Google Scholar] [CrossRef]
- Ando, N.; Iizuka, T.; Ide, H.; Ishida, K.; Shinoda, M.; Nishimaki, T.; Takiyama, W.; Watanabe, H.; Isono, K.; Aoyama, N.; et al. Surgery plus chemotherapy compared with surgery alone for localized squamous cell carcinoma of the thoracic esophagus: A Japan Clinical Oncology Group Study—JCOG9204. J. Clin. Oncol. 2003, 21, 4592–4596. [Google Scholar] [CrossRef]
- Lee, J.; Lee, K.E.; Im, Y.H.; Kang, W.K.; Park, K.; Kim, K.; Shim, Y.M. Adjuvant chemotherapy with 5-fluorouracil and cisplatin in lymph node-positive thoracic esophageal squamous cell carcinoma. Ann. Thorac. Surg. 2005, 80, 1170–1175. [Google Scholar] [CrossRef]
- Iizuka, T. A comparison of chemotherapy and radiotherapy as adjuvant treatment to surgery for esophageal carcinoma. Japanese Esophageal Oncology Group. Chest 1993, 104, 203–207. [Google Scholar] [CrossRef]
- Ando, N.; Kato, H.; Igaki, H.; Shinoda, M.; Ozawa, S.; Shimizu, H.; Nakamura, T.; Yabusaki, H.; Aoyama, N.; Kurita, A.; et al. A Randomized Trial Comparing Postoperative Adjuvant Chemotherapy with Cisplatin and 5-Fluorouracil Versus Preoperative Chemotherapy for Localized Advanced Squamous Cell Carcinoma of the Thoracic Esophagus (JCOG9907). Ann. Surg. Oncol. 2012, 19, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Klevebro, F.; Alexandersson von Dobeln, G.; Wang, N.; Johnsen, G.; Jacobsen, A.B.; Friesland, S.; Hatlevoll, I.; Glenjen, N.I.; Lind, P.; Tsai, J.A.; et al. A randomized clinical trial of neoadjuvant chemotherapy versus neoadjuvant chemoradiotherapy for cancer of the oesophagus or gastro-oesophageal junction. Ann. Oncol. 2016, 27, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, J.; van Lanschot, J.J.B.; Hulshof, M.C.C.M.; van Hagen, P.; van Berge Henegouwen, M.I.; Wijnhoven, B.P.L.; van Laarhoven, H.W.M.; Nieuwenhuijzen, G.A.P.; Hospers, G.A.P.; Bonenkamp, J.J.; et al. Neoadjuvant chemoradiotherapy plus surgery versus surgery alone for oesophageal or junctional cancer (CROSS): Long-term results of a randomised controlled trial. Lancet Oncol. 2015, 16, 1090–1098. [Google Scholar] [CrossRef]
- Stahl, M.; Walz, M.K.; Riera-Knorrenschild, J.; Stuschke, M.; Sandermann, A.; Bitzer, M.; Wilke, H.; Budach, W. Preoperative chemotherapy versus chemoradiotherapy in locally advanced adenocarcinomas of the oesophagogastric junction (POET): Long-term results of a controlled randomised trial. Eur. J. Cancer 2017, 81, 183–190. [Google Scholar] [CrossRef]
- Alderson, D.; Cunningham, D.; Nankivell, M.; Blazeby, J.M.; Griffin, S.M.; Crellin, A.; Grabsch, H.I.; Langer, R.; Pritchard, S.; Okines, A.; et al. Neoadjuvant cisplatin and fluorouracil versus epirubicin, cisplatin, and capecitabine followed by resection in patients with oesophageal adenocarcinoma (UK MRC OE05): An open-label, randomised phase 3 trial. Lancet Oncol. 2017, 18, 1249–1260. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Zöller, M. CD44: Can a cancer-initiating cell profit from an abundantly expressed molecule? Nat. Rev. Cancer 2011, 11, 254–267. [Google Scholar] [CrossRef]
- Zhao, J.-S.; Li, W.-J.; Ge, D.; Zhang, P.-J.; Li, J.-J.; Lu, C.-L.; Ji, X.-D.; Guan, D.-X.; Gao, H.; Xu, L.-Y.; et al. Tumor initiating cells in esophageal squamous cell carcinomas express high levels of CD44. PLoS ONE 2011, 6, e21419. [Google Scholar] [CrossRef]
- Schizas, D.; Moris, D.; Kanavidis, P.; Michalinos, A.; Sioulas, A.; Pavlakis, K.; Machairas, A.; Liakakos, T. The Prognostic Value of CD44 Expression in Epithelial-Mesenchymal Transition: Preliminary Data from Patients with Gastric and Esophageal Cancer. In Vivo 2016, 30, 939–944. [Google Scholar] [CrossRef][Green Version]
- Le Bras, G.F.; Allison, G.L.; Richards, N.F.; Ansari, S.S.; Washington, M.K.; Andl, C.D. CD44 upregulation in E-cadherin-negative esophageal cancers results in cell invasion. PLoS ONE 2011, 6, e27063. [Google Scholar] [CrossRef][Green Version]
- Sui, Y.-P.; Jian, X.-P.; Ma, L.I.; Xu, G.-Z.; Liao, H.-W.; Liu, Y.-P.; Wen, H.-C. Prognostic value of cancer stem cell marker CD133 expression in esophageal carcinoma: A meta-analysis. Mol. Clin. Oncol. 2016, 4, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Hang, D.; Dong, H.C.; Ning, T.; Dong, B.; Hou, D.L.; Xu, W.G. Prognostic value of the stem cell markers CD133 and ABCG2 expression in esophageal squamous cell carcinoma. Dis. Esophagus 2012, 25, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xu, W.W.; Han, L.; Chan, K.T.; Tsao, S.W.; Lee, N.P.Y.; Law, S.; Xu, L.Y.; Li, E.M.; Chan, K.W.; et al. MicroRNA-377 suppresses initiation and progression of esophageal cancer by inhibiting CD133 and VEGF. Oncogene 2017, 36, 3986–4000. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-D.; Yuan, Y.; Liu, X.-H.; Gong, D.-J.; Bai, C.-G.; Wang, F.; Luo, J.-H.; Xu, Z.-Y. Self-renewal and chemotherapy resistance of p75NTR positive cells in esophageal squamous cell carcinomas. BMC Cancer 2009, 9, 9. [Google Scholar] [CrossRef]
- Li, S.; Yue, D.; Chen, X.; Wang, L.; Li, J.; Ping, Y.; Gao, Q.; Wang, D.; Zhang, T.; Li, F.; et al. Epigenetic regulation of CD271, a potential cancer stem cell marker associated with chemoresistance and metastatic capacity. Oncol. Rep. 2015, 33, 425–432. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Okumura, T.; Hirano, K.; Watanabe, T.; Nagata, T.; Shimada, Y.; Tsukada, K. Detection of circulating tumor cells by p75NTR expression in patients with esophageal cancer. World J. Surg. Oncol. 2016, 14, 40. [Google Scholar] [CrossRef]
- Von Rahden, B.H.; Kircher, S.; Lazariotou, M.; Reiber, C.; Stuermer, L.; Otto, C.; Germer, C.T.; Grimm, M. LgR5 expression and cancer stem cell hypothesis: Clue to define the true origin of esophageal adenocarcinomas with and without Barrett’s esophagus? J. Exp. Clin. Cancer Res. 2011, 30, 23. [Google Scholar] [CrossRef]
- Becker, L.; Huang, Q.; Mashimo, H. Lgr5, an intestinal stem cell marker, is abnormally expressed in Barrett’s esophagus and esophageal adenocarcinoma. Dis. Esophagus 2010, 23, 168–174. [Google Scholar] [CrossRef]
- Lv, Z.; Yu, J.J.; Zhang, W.J.; Xiong, L.; Wang, F.; Li, L.F.; Zhou, X.L.; Gao, X.Y.; Ding, X.F.; Han, L.; et al. Expression and functional regulation of stemness gene Lgr5 in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 26492–26504. [Google Scholar] [CrossRef]
- Tang, K.H.; Dai, Y.D.; Tong, M.; Chan, Y.P.; Kwan, P.S.; Fu, L.; Qin, Y.R.; Tsao, S.W.; Lung, H.L.; Lung, M.L.; et al. A CD90(+) tumor-initiating cell population with an aggressive signature and metastatic capacity in esophageal cancer. Cancer Res. 2013, 73, 2322–2332. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Zhu, H.; Tang, J.; Zhang, S.; Luo, J.; Sun, X. CD90 positive cells exhibit aggressive radioresistance in esophageal squamous cell carcinoma. J. Thorac. Dis. 2017, 9, 610–620. [Google Scholar] [CrossRef] [PubMed]
- Ajani, J.A.; Wang, X.; Song, S.; Suzuki, A.; Taketa, T.; Sudo, K.; Wadhwa, R.; Hofstetter, W.L.; Komaki, R.; Maru, D.M.; et al. ALDH-1 expression levels predict response or resistance to preoperative chemoradiation in resectable esophageal cancer patients. Mol. Oncol. 2014, 8, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ren, Y.; Yu, X.; Qian, F.; Bian, B.S.; Xiao, H.L.; Wang, W.G.; Xu, S.L.; Yang, J.; Cui, W.; et al. ALDH1A1 defines invasive cancer stem-like cells and predicts poor prognosis in patients with esophageal squamous cell carcinoma. Mod. Pathol. 2014, 27, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.F.; Chen, P.T.; Lu, M.S.; Chen, W.C. Role of ALDH1 in the prognosis of esophageal cancer and its relationship with tumor microenvironment. Mol. Carcinog. 2018, 57, 78–88. [Google Scholar] [CrossRef]
- Huang, L.; Lu, Q.; Han, Y.; Li, Z.; Zhang, Z.; Li, X. ABCG2/V-ATPase was associated with the drug resistance and tumor metastasis of esophageal squamous cancer cells. Diagn Pathol. 2012, 7, 180. [Google Scholar] [CrossRef]
- Tsunoda, S.; Okumura, T.; Ito, T.; Kondo, K.; Ortiz, C.; Tanaka, E.; Watanabe, G.; Itami, A.; Sakai, Y.; Shimada, Y. ABCG2 expression is an independent unfavorable prognostic factor in esophageal squamous cell carcinoma. Oncology 2006, 71, 251–258. [Google Scholar] [CrossRef]
- Tsai, S.-T.; Wang, P.-J.; Liou, N.-J.; Lin, P.-S.; Chen, C.-H.; Chang, W.-C. ICAM1 Is a Potential Cancer Stem Cell Marker of Esophageal Squamous Cell Carcinoma. PLoS ONE 2015, 10, e0142834. [Google Scholar] [CrossRef]
- Ming, X.-Y.; Fu, L.; Zhang, L.-Y.; Qin, Y.-R.; Cao, T.-T.; Chan, K.W.; Ma, S.; Xie, D.; Guan, X.-Y. Integrin α7 is a functional cancer stem cell surface marker in oesophageal squamous cell carcinoma. Nat. Commun. 2016, 7, 13568. [Google Scholar] [CrossRef]
- Smit, J.K.; Faber, H.; Niemantsverdriet, M.; Baanstra, M.; Bussink, J.; Hollema, H.; van Os, R.P.; Plukker, J.T.M.; Coppes, R.P. Prediction of response to radiotherapy in the treatment of esophageal cancer using stem cell markers. Radiother. Oncol. 2013, 107, 434–441. [Google Scholar] [CrossRef]
- Okamoto, K.; Ninomiya, I.; Ohbatake, Y.; Hirose, A.; Tsukada, T.; Nakanuma, S.; Sakai, S.; Kinoshita, J.; Makino, I.; Nakamura, K.; et al. Expression status of CD44 and CD133 as a prognostic marker in esophageal squamous cell carcinoma treated with neoadjuvant chemotherapy followed by radical esophagectomy. Oncol. Rep. 2016, 36, 3333–3342. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lu, C.; Xu, F.; Gu, J.; Yuan, Y.; Zhao, G.; Yu, X.; Ge, D. Clinical and biological significance of stem-like CD133(+)CXCR4(+) cells in esophageal squamous cell carcinoma. J. Thorac. Cardiovasc. Surg. 2015, 150, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Bass, A.J.; Watanabe, H.; Mermel, C.H.; Yu, S.; Perner, S.; Verhaak, R.G.; Kim, S.Y.; Wardwell, L.; Tamayo, P.; Gat-Viks, I.; et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat. Genet. 2009, 41, 1238–1242. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; He, W.; Lu, C.; Wang, Z.; Wang, J.; Giercksky, K.E.; Nesland, J.M.; Suo, Z. Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res. 2009, 29, 1233–1241. [Google Scholar]
- Forghanifard, M.M.; Ardalan Khales, S.; Javdani-Mallak, A.; Rad, A.; Farshchian, M.; Abbaszadegan, M.R. Stemness state regulators SALL4 and SOX2 are involved in progression and invasiveness of esophageal squamous cell carcinoma. Med. Oncol. 2014, 31, 922. [Google Scholar] [CrossRef]
- Ten Kate, F.J.C.; van Olphen, S.H.; Bruno, M.J.; Wijnhoven, B.P.L.; van Lanschot, J.J.B.; Looijenga, L.H.J.; Fitzgerald, R.C.; Biermann, K. Loss of SRY-box2 (SOX2) expression and its impact on survival of patients with oesophageal adenocarcinoma. Br. J. Surg. 2017, 104, 1327–1337. [Google Scholar] [CrossRef]
- Chai, Y.; Li, Q.; Zhao, H.; Zhang, Z.; Yu, X.; Pang, L.; Liu, Z.; Zhao, J.; Wang, L.; Li, F. SOX2 antagonizes WWC1 to drive YAP1 activation in esophageal squamous cell carcinoma. Cancer Med. 2019, 8, 7055–7064. [Google Scholar] [CrossRef]
- Deng, L.; Xiang, X.; Yang, F.; Xiao, D.; Liu, K.; Chen, Z.; Zhang, R.; Feng, G. Functional evidence that the self-renewal gene NANOG regulates esophageal squamous cancer development. Biochem. Biophys. Res. Commun. 2017, 490, 161–168. [Google Scholar] [CrossRef]
- Du, Y.; Shi, L.; Wang, T.; Liu, Z.; Wang, Z. Nanog siRNA plus Cisplatin may enhance the sensitivity of chemotherapy in esophageal cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 1759–1767. [Google Scholar] [CrossRef]
- Shimada, Y.; Okumura, T.; Sekine, S.; Moriyama, M.; Sawada, S.; Matsui, K.; Yoshioka, I.; Hojo, S.; Yoshida, T.; Nagata, T.; et al. Expression analysis of iPS cell-inductive genes in esophageal squamous cell carcinoma by tissue microarray. Anticancer Res. 2012, 32, 5507–5514. [Google Scholar]
- He, X.-T.; Cao, X.-F.; Ji, L.; Zhu, B.; Lv, J.; Wang, D.-D.; Lu, P.-H.; Cui, H.-G. Association between Bmi1 and clinicopathological status of esophageal squamous cell carcinoma. World J. Gastroenterol. 2009, 15, 2389–2394. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Gao, Q.; Guo, L.; Zhang, C.; Jiang, W.; Li, H.; Wang, J.; Han, X.; Shi, Y.; Lu, S.H. Isolation and identification of cancer stem-like cells in esophageal carcinoma cell lines. Stem Cells Dev. 2009, 18, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, K.; Jiang, X.; Wang, X.; Chen, Y.; Cui, X.; Pang, L.; Li, S.; Liu, C.; Zou, H.; et al. Clinicopathological significance of Bmi-1 overexpression in esophageal cancer: A meta-analysis. Biomark Med. 2018, 12, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.X.; Sang, M.X.; Zhu, S.C.; Liu, Z.K.; Ma, M. Radiosensitization of esophageal carcinoma cells by the silencing of BMI-1. Oncol. Rep. 2016, 35, 3669–3678. [Google Scholar] [CrossRef][Green Version]
- Vaiphei, K.; Sinha, S.K.; Kochhar, R. Comparative analysis of Oct4 in different histological subtypes of esophageal squamous cell carcinomas in different clinical conditions. Asian Pac. J. Cancer Prev. 2014, 15, 3519–3524. [Google Scholar] [CrossRef]
- Li, Z.; Li, X.; Li, C.; Su, Y.; Fang, W.; Zhong, C.; Ji, W.; Zhang, Q.; Su, C. Transcription factor OCT4 promotes cell cycle progression by regulating CCND1 expression in esophageal carcinoma. Cancer Lett. 2014, 354, 77–86. [Google Scholar] [CrossRef]
- Stoecklein, N.H.; Siegmund, A.; Scheunemann, P.; Luebke, A.M.; Erbersdobler, A.; Verde, P.E.; Eisenberger, C.F.; Peiper, M.; Rehders, A.; Esch, J.S.A.; et al. Ep-CAM expression in squamous cell carcinoma of the esophagus: A potential therapeutic target and prognostic marker. BMC Cancer 2006, 6, 165. [Google Scholar] [CrossRef]
- Liu, Z.; Lu, Y.; Pu, Y.; Liu, J.; Liu, B.; Yu, B.; Chen, K.; Fu, T.; Yang, C.J.; Liu, H.; et al. Using aptamers to elucidate esophageal cancer clinical samples. Sci. Rep. 2015, 5, 18516. [Google Scholar] [CrossRef]
- Matsuda, T.; Takeuchi, H.; Matsuda, S.; Hiraiwa, K.; Miyasho, T.; Okamoto, M.; Kawasako, K.; Nakamura, R.; Takahashi, T.; Wada, N.; et al. EpCAM, a potential therapeutic target for esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2014, 21 (Suppl. 3), S356–S364. [Google Scholar] [CrossRef]
- Wadhwa, R.; Wang, X.; Baladandayuthapani, V.; Liu, B.; Shiozaki, H.; Shimodaira, Y.; Lin, Q.; Elimova, E.; Hofstetter, W.L.; Swisher, S.G.; et al. Nuclear expression of Gli-1 is predictive of pathologic complete response to chemoradiation in trimodality treated oesophageal cancer patients. Br. J. Cancer 2017, 117, 648–655. [Google Scholar] [CrossRef]
- Sims-Mourtada, J.; Izzo, J.G.; Apisarnthanarax, S.; Wu, T.T.; Malhotra, U.; Luthra, R.; Liao, Z.; Komaki, R.; van der Kogel, A.; Ajani, J.; et al. Hedgehog: An attribute to tumor regrowth after chemoradiotherapy and a target to improve radiation response. Clin. Cancer Res. 2006, 12, 6565–6572. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Cui, Y.; Ni, W.; Kim, S.; Xuan, Y. Gli1, a potential regulator of esophageal cancer stem cell, is identified as an independent adverse prognostic factor in esophageal squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2017, 143, 243–254. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhou, M.; Chen, X.; Yue, D.; Yang, L.; Qin, G.; Zhang, Z.; Gao, Q.; Wang, D.; Zhang, C.; et al. Inhibition of SALL4 reduces tumorigenicity involving epithelial-mesenchymal transition via Wnt/beta-catenin pathway in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 98. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.-K.; Chuang, W.-Y.; Yeh, C.-J.; Wu, Y.-C.; Liu, Y.-H.; Hsieh, M.-J.; Cheng, A.-J.; Hsueh, C.; Liu, H.-P. Prognostic significance of high podoplanin expression after chemoradiotherapy in esophageal squamous cell carcinoma patients. J. Surg. Oncol. 2012, 105, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Wang, K.; Yang, S.; Wang, J.; Tan, B.; Bai, B.; Wang, N.; Jia, Y.; Jia, M.; Cheng, Y. Clinicopathology significance of podoplanin immunoreactivity in esophageal squamous cell carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 2361–2371. [Google Scholar] [PubMed]
- Li, J.C.; Li, Y.; Ai, J.Y.; Chen, K.; Zhu, Y.H.; Fu, L.; Qin, Y.R.; Wang, L.J.; Guan, X.Y. Podoplaninpositive cancer cells at the edge of esophageal squamous cell carcinomas are involved in invasion. Mol. Med. Rep. 2014, 10, 1513–1518. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, Q.; Yan, X.; Kato, Y.; Tanaka, M.; Inokuchi, S.; Yoshizawa, T.; Morohashi, S.; Kijima, H. Podoplanin-mediated TGF-beta-induced epithelial-mesenchymal transition and its correlation with bHLH transcription factor DEC in TE-11 cells. Int. J. Oncol. 2016, 48, 2310–2320. [Google Scholar] [CrossRef]
- Hadnagy, A.; Gaboury, L.; Beaulieu, R.; Balicki, D. SP analysis may be used to identify cancer stem cell populations. Exp. Cell Res. 2006, 312, 3701–3710. [Google Scholar] [CrossRef]
- Gross, E.; L’Faqihi-Olive, F.E.; Ysebaert, L.; Brassac, M.; Struski, S.; Kheirallah, S.; Fournié, J.J.; Laurent, G.; Quillet-Mary, A. B-chronic lymphocytic leukemia chemoresistance involves innate and acquired leukemic side population cells. Leukemia 2010, 24, 1885–1892. [Google Scholar] [CrossRef]
- Du, J.; Liu, S.; He, J.; Liu, X.; Qu, Y.; Yan, W.; Fan, J.; Li, R.; Xi, H.; Fu, W.; et al. MicroRNA-451 regulates stemness of side population cells via PI3K/Akt/mTOR signaling pathway in multiple myeloma. Oncotarget 2015, 6, 14993–15007. [Google Scholar] [CrossRef]
- Britton, K.M.; Kirby, J.A.; Lennard, T.W.; Meeson, A.P. Cancer stem cells and side population cells in breast cancer and metastasis. Cancers 2011, 3, 2106–2130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Komaki, R.; Wang, L.; Fang, B.; Chang, J.Y. Treatment of radioresistant stem-like esophageal cancer cells by an apoptotic gene-armed, telomerase-specific oncolytic adenovirus. Clin. Cancer Res. 2008, 14, 2813–2823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Ma, L.; Xie, Y.K.; Miao, X.B.; Jin, C. Esophageal cancer tumorspheres involve cancer stem-like populations with elevated aldehyde dehydrogenase enzymatic activity. Mol. Med. Rep. 2012, 6, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Bao, Q.; Schwarz, B.; Zhao, L.; Mysliwietz, J.; Ellwart, J.; Renner, A.; Hirner, H.; Niess, H.; Camaj, P.; et al. Stem cell-like side populations in esophageal cancer: A source of chemotherapy resistance and metastases. Stem Cells Dev. 2014, 23, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Möbest, D.; Goan, S.R.; Junghahn, I.; Winkler, J.; Fichtner, I.; Hermann, M.; Becker, M.; de Lima-Hahn, E.; Henschler, R. Differential kinetics of primitive hematopoietic cells assayed in vitro and in vivo during serum-free suspension culture of CD34+ blood progenitor cells. Stem Cells 1999, 17, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Trisciuoglio, D.; Tupone, M.G.; Desideri, M.; Di Martile, M.; Gabellini, C.; Buglioni, S.; Pallocca, M.; Alessandrini, G.; D’Aguanno, S.; Del Bufalo, D. BCL-X(L) overexpression promotes tumor progression-associated properties. Cell Death Dis. 2017, 8, 3216. [Google Scholar] [CrossRef]
- Xiao, G.; Li, X.; Li, G.; Zhang, B.; Xu, C.; Qin, S.; Du, N.; Wang, J.; Tang, S.C.; Zhang, J.; et al. MiR-129 blocks estrogen induction of NOTCH signaling activity in breast cancer stem-like cells. Oncotarget 2017, 8, 103261–103273. [Google Scholar] [CrossRef]
- Li, J.C.; Liu, D.; Yang, Y.; Wang, X.Y.; Pan, D.L.; Qiu, Z.D.; Su, Y.; Pan, J.J. Growth, clonability, and radiation resistance of esophageal carcinoma-derived stem-like cells. Asian Pac. J. Cancer Prev. 2013, 14, 4891–4896. [Google Scholar] [CrossRef][Green Version]
- Wang, J.L.; Yu, J.P.; Sun, Z.Q.; Sun, S.P. Radiobiological characteristics of cancer stem cells from esophageal cancer cell lines. World J. Gastroenterol. 2014, 20, 18296–18305. [Google Scholar] [CrossRef]
- Xu, D.D.; Zhou, P.J.; Wang, Y.; Zhang, L.; Fu, W.Y.; Ruan, B.B.; Xu, H.P.; Hu, C.Z.; Tian, L.; Qin, J.H.; et al. Reciprocal activation between STAT3 and miR-181b regulates the proliferation of esophageal cancer stem-like cells via the CYLD pathway. Mol. Cancer 2016, 15, 40. [Google Scholar] [CrossRef]
- Pastrana, E.; Silva-Vargas, V.; Doetsch, F. Eyes wide open: A critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 2011, 8, 486–498. [Google Scholar] [CrossRef] [PubMed]
- Lanzardo, S.; Conti, L.; Rooke, R.; Ruiu, R.; Accart, N.; Bolli, E.; Arigoni, M.; Macagno, M.; Barrera, G.; Pizzimenti, S.; et al. Immunotargeting of Antigen xCT Attenuates Stem-like Cell Behavior and Metastatic Progression in Breast Cancer. Cancer Res. 2016, 76, 62–72. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.C.; Tsai, J.T.; Chao, T.Y.; Ma, H.I.; Liu, W.H. The STAT3/Slug Axis Enhances Radiation-Induced Tumor Invasion and Cancer Stem-like Properties in Radioresistant Glioblastoma. Cancers 2018, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.L.; Li, Y.J.; Liao, K.; Shi, L.; Zhang, N.; Liu, S.; Hu, Y.Y.; Li, S.L.; Wang, Y. 2-Methoxyestradiol inhibits the proliferation and migration and reduces the radioresistance of nasopharyngeal carcinoma CNE-2 stem cells via NF-κB/HIF-1 signaling pathway inactivation and EMT reversal. Oncol. Rep. 2017, 37, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Lynam-Lennon, N.; Heavey, S.; Sommerville, G.; Bibby, B.A.; Ffrench, B.; Quinn, J.; Gasch, C.; O’Leary, J.J.; Gallagher, M.F.; Reynolds, J.V.; et al. MicroRNA-17 is downregulated in esophageal adenocarcinoma cancer stem-like cells and promotes a radioresistant phenotype. Oncotarget 2017, 8, 11400–11413. [Google Scholar] [CrossRef]
- Che, S.M.; Zhang, X.Z.; Liu, X.L.; Chen, X.; Hou, L. The radiosensitization effect of NS398 on esophageal cancer stem cell-like radioresistant cells. Dis. Esophagus 2011, 24, 265–273. [Google Scholar] [CrossRef]
- Almanaa, T.N.; Geusz, M.E.; Jamasbi, R.J. A new method for identifying stem-like cells in esophageal cancer cell lines. J. Cancer 2013, 4, 536–548. [Google Scholar] [CrossRef]
- Nguyen, L.V.; Vanner, R.; Dirks, P.; Eaves, C.J. Cancer stem cells: An evolving concept. Nat. Rev. Cancer 2012, 12, 133–143. [Google Scholar] [CrossRef]
- Wu, S.S.; Lee, J.H.; Koo, B.K. Lineage Tracing: Computational Reconstruction Goes Beyond the Limit of Imaging. Mol. Cells 2019, 42, 104–112. [Google Scholar] [CrossRef]
- Shimokawa, M.; Ohta, Y.; Nishikori, S.; Matano, M.; Takano, A.; Fujii, M.; Date, S.; Sugimoto, S.; Kanai, T.; Sato, T. Visualization and targeting of LGR5(+) human colon cancer stem cells. Nature 2017, 545, 187–192. [Google Scholar] [CrossRef]
- Jiang, M.; Li, H.; Zhang, Y.; Yang, Y.; Lu, R.; Liu, K.; Lin, S.; Lan, X.; Wang, H.; Wu, H.; et al. Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature 2017, 550, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Giroux, V.; Lento, A.A.; Islam, M.; Pitarresi, J.R.; Kharbanda, A.; Hamilton, K.E.; Whelan, K.A.; Long, A.; Rhoades, B.; Tang, Q.; et al. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J. Clin. Investig. 2017, 127, 2378–2391. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Math, M.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-cell genome sequencing: Current state of the science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yu, J.; Li, Y.; Hou, Q.; Zhou, R.; Zhang, N.; Jing, Z.; Jiang, M.; Li, Z.; Hua, Y.; et al. Single-cell RNA sequencing reveals diverse intratumoral heterogeneities and gene signatures of two types of esophageal cancers. Cancer Lett. 2018, 438, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, S.; Yu, J.; Li, Y.; Zhang, X.Y.; Yang, L.; Zhang, H.; Hou, Q.; Jiang, M.; Brunicardi, F.C.; et al. Single-cell Transcriptome Analyses Reveal Molecular Signals to Intrinsic and Acquired Paclitaxel Resistance in Esophageal Squamous Cancer Cells. Cancer Lett. 2018, 420, 156–167. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Hou, Q.; Huang, M.; Zhang, H.; Jiang, Z.; Yue, J.; Wu, S. Single-cell RNA-seq of esophageal squamous cell carcinoma cell line with fractionated irradiation reveals radioresistant gene expression patterns. BMC Genom. 2019, 20, 611. [Google Scholar] [CrossRef]
- Wu, H.; Yu, J.; Kong, D.; Xu, Y.; Zhang, Z.; Shui, J.; Li, Z.; Luo, H.; Wang, K. Population and singlecell transcriptome analyses reveal diverse transcriptional changes associated with radioresistance in esophageal squamous cell carcinoma. Int. J. Oncol. 2019, 55, 1237–1248. [Google Scholar] [CrossRef]
- Wu, H.; Li, Y.; Hou, Q.; Zhou, R.; Li, Z.; Wu, S.; Yu, J.; Jiang, M. Single-cell intratumoral stemness analysis reveals the involvement of cell cycle and DNA damage repair in two different types of esophageal cancer. Oncol. Rep. 2019, 41, 3201–3208. [Google Scholar] [CrossRef]
- Zheng, H.; Pomyen, Y.; Hernandez, M.O.; Li, C.; Livak, F.; Tang, W.; Dang, H.; Greten, T.F.; Davis, J.L.; Zhao, Y.; et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology 2018, 68, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Shema, E.; Bernstein, B.E.; Buenrostro, J.D. Single-cell and single-molecule epigenomics to uncover genome regulation at unprecedented resolution. Nat. Genet. 2019, 51, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Marx, V. A dream of single-cell proteomics. Nat. Methods 2019, 16, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Girardi, D.; Barrichello, A.; Fernandes, G.; Pereira, A. Targeting the Hedgehog Pathway in Cancer: Current Evidence and Future Perspectives. Cells 2019, 8, 153. [Google Scholar] [CrossRef]
- Hoffmeyer, K.; Raggioli, A.; Rudloff, S.; Anton, R.; Hierholzer, A.; Del Valle, I.; Hein, K.; Vogt, R.; Kemler, R. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 2012, 336, 1549–1554. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Mo, J.S.; Park, H.W.; Guan, K.L. The Hippo signaling pathway in stem cell biology and cancer. EMBO Rep. 2014, 15, 642–656. [Google Scholar] [CrossRef]
- Kim, S.Y.; Baek, K.H. TGF-β signaling pathway mediated by deubiquitinating enzymes. Cell Mol. Life Sci. 2019, 76, 653–665. [Google Scholar] [CrossRef]
- Zhang, H.F.; Wu, C.; Alshareef, A.; Gupta, N.; Zhao, Q.; Xu, X.E.; Jiao, J.W.; Li, E.M.; Xu, L.Y.; Lai, R. The PI3K/AKT/c-MYC Axis Promotes the Acquisition of Cancer Stem-Like Features in Esophageal Squamous Cell Carcinoma. Stem Cells 2016, 34, 2040–2051. [Google Scholar] [CrossRef]
- Long, A.; Giroux, V.; Whelan, K.A.; Hamilton, K.E.; Tétreault, M.P.; Tanaka, K.; Lee, J.S.; Klein-Szanto, A.J.; Nakagawa, H.; Rustgi, A.K. WNT10A promotes an invasive and self-renewing phenotype in esophageal squamous cell carcinoma. Carcinogenesis 2015, 36, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Liu, R.D.; Chen, X.Q.; Wang, B.; Li, L.F.; Guo, Y.S.; Chen, X.J.; Ren, X.Q. HIF-1α promotes the stemness of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin pathway. Oncol. Rep. 2019, 42, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Zhu, J.; Wu, G.; Cao, L.; Tan, Z.; Zhang, S.; Jiang, L.; Wu, J.; Li, M.; Song, L.; et al. Antagonizing miR-455-3p inhibits chemoresistance and aggressiveness in esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 106. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Zhang, Z.; Li, J.; Chen, X.; Ping, Y.; Liu, S.; Shi, X.; Li, L.; Wang, L.; Huang, L.; et al. Transforming growth factor-beta1 promotes the migration and invasion of sphere-forming stem-like cell subpopulations in esophageal cancer. Exp. Cell Res. 2015, 336, 141–149. [Google Scholar] [CrossRef]
- Moghbeli, M.; Abbaszadegan, M.R.; Golmakani, E.; Forghanifard, M.M. Correlation of Wnt and NOTCH pathways in esophageal squamous cell carcinoma. J. Cell Commun. Signal. 2016, 10, 129–135. [Google Scholar] [CrossRef]
- Forghanifard, M.M.; Azaraz, S.; Ardalan Khales, S.; Morshedi Rad, D.; Abbaszadegan, M.R. MAML1 promotes ESCC aggressiveness through upregulation of EMT marker TWIST1. Mol. Biol. Rep. 2020. [Google Scholar] [CrossRef]
- Moghbeli, M.; Mosannen Mozaffari, H.; Memar, B.; Forghanifard, M.M.; Gholamin, M.; Abbaszadegan, M.R. Role of MAML1 in targeted therapy against the esophageal cancer stem cells. J. Transl. Med. 2019, 17, 126. [Google Scholar] [CrossRef]
- Wang, Z.; Da Silva, T.G.; Jin, K.; Han, X.; Ranganathan, P.; Zhu, X.; Sanchez-Mejias, A.; Bai, F.; Li, B.; Fei, D.L.; et al. Notch signaling drives stemness and tumorigenicity of esophageal adenocarcinoma. Cancer Res. 2014, 74, 6364–6374. [Google Scholar] [CrossRef]
- Yoshikawa, R.; Nakano, Y.; Tao, L.; Koishi, K.; Matsumoto, T.; Sasako, M.; Tsujimura, T.; Hashimoto-Tamaoki, T.; Fujiwara, Y. Hedgehog signal activation in oesophageal cancer patients undergoing neoadjuvant chemoradiotherapy. Br. J. Cancer 2008, 98, 1670–1674. [Google Scholar] [CrossRef]
- Isohata, N.; Aoyagi, K.; Mabuchi, T.; Daiko, H.; Fukaya, M.; Ohta, H.; Ogawa, K.; Yoshida, T.; Sasaki, H. Hedgehog and epithelial-mesenchymal transition signaling in normal and malignant epithelial cells of the esophagus. Int. J. Cancer 2009, 125, 1212–1221. [Google Scholar] [CrossRef]
- Wang, D.; Nagle, P.W.; Wang, H.H.; Smit, J.K.; Faber, H.; Baanstra, M.; Karrenbeld, A.; Chiu, R.K.; Plukker, J.T.M.; Coppes, R.P. Hedgehog Pathway as a Potential Intervention Target in Esophageal Cancer. Cancers 2019, 11, 821. [Google Scholar] [CrossRef] [PubMed]
- Giraud, J.; Molina-Castro, S.; Seeneevassen, L.; Sifré, E.; Izotte, J.; Tiffon, C.; Staedel, C.; Boeuf, H.; Fernandez, S.; Barthelemy, P.; et al. Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int. J. Cancer 2020, 146, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.J.; Lin, C.Y.; Liao, W.Y.; Hour, T.C.; Wang, H.D.; Chuu, C.P. CD44 Promotes Migration and Invasion of Docetaxel-Resistant Prostate Cancer Cells Likely via Induction of Hippo-Yap Signaling. Cells 2019, 8, 295. [Google Scholar] [CrossRef] [PubMed]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Z.; Yu, X.; Huang, X.; Liu, Z.; Chai, Y.; Yang, L.; Wang, Q.; Li, M.; Zhao, J.; et al. Unbalanced YAP-SOX9 circuit drives stemness and malignant progression in esophageal squamous cell carcinoma. Oncogene 2019, 38, 2042–2055. [Google Scholar] [CrossRef]
- Song, S.; Ajani, J.A.; Honjo, S.; Maru, D.M.; Chen, Q.; Scott, A.W.; Heallen, T.R.; Xiao, L.; Hofstetter, W.L.; Weston, B.; et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014, 74, 4170–4182. [Google Scholar] [CrossRef]
- Song, S.; Honjo, S.; Jin, J.; Chang, S.S.; Scott, A.W.; Chen, Q.; Kalhor, N.; Correa, A.M.; Hofstetter, W.L.; Albarracin, C.T.; et al. The Hippo Coactivator YAP1 Mediates EGFR Overexpression and Confers Chemoresistance in Esophageal Cancer. Clin. Cancer Res. 2015, 21, 2580–2590. [Google Scholar] [CrossRef]
- Gao, J.; Xia, R.; Chen, J.; Gao, J.; Luo, X.; Ke, C.; Ren, C.; Li, J.; Mi, Y. Inhibition of esophageal-carcinoma cell proliferation by genistein via suppression of JAK1/2-STAT3 and AKT/MDM2/p53 signaling pathways. Aging 2020, 12. [Google Scholar] [CrossRef]
- Lu, Z.; Lu, C.; Li, C.; Jiao, Y.; Li, Y.; Zhang, G. Dracorhodin perchlorate induces apoptosis and G2/M cell cycle arrest in human esophageal squamous cell carcinoma through inhibition of the JAK2/STAT3 and AKT/FOXO3a pathways. Mol. Med. Rep. 2019, 20, 2091–2100. [Google Scholar] [CrossRef]
- Ha, Y.N.E.; Dai, Y.; Wufuer, D.; Pidayi, M.; Anasihan, G.; Chen, L. Second-generation Src/Abl inhibitor bosutinib effectively induces apoptosis in human esophageal squamous cell carcinoma (ESCC) cells via inhibiting Src/Abl signaling. Neoplasma 2020, 67, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Bossler, F.; Kuhn, B.J.; Gunther, T.; Kraemer, S.J.; Khalkar, P.; Adrian, S.; Lohrey, C.; Holzer, A.; Shimobayashi, M.; Durst, M.; et al. Repression of Human Papillomavirus Oncogene Expression under Hypoxia Is Mediated by PI3K/mTORC2/AKT Signaling. MBio 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Hufbauer, M.; Maltseva, M.; Meinrath, J.; Lechner, A.; Beutner, D.; Huebbers, C.U.; Akgul, B. HPV16 increases the number of migratory cancer stem cells and modulates their miRNA expression profile in oropharyngeal cancer. Int. J. Cancer 2018, 143, 1426–1439. [Google Scholar] [CrossRef] [PubMed]
- Reid, P.; Marcu, L.G.; Olver, I.; Moghaddasi, L.; Staudacher, A.H.; Bezak, E. Diversity of cancer stem cells in head and neck carcinomas: The role of HPV in cancer stem cell heterogeneity, plasticity and treatment response. Radiother. Oncol. 2019, 135, 1–12. [Google Scholar] [CrossRef]
- Prieto-Vila, M.; Takahashi, R.U.; Usuba, W.; Kohama, I.; Ochiya, T. Drug Resistance Driven by Cancer Stem Cells and Their Niche. Int. J. Mol. Sci. 2017, 18, 2574. [Google Scholar] [CrossRef]
- Lytle, N.K.; Barber, A.G.; Reya, T. Stem cell fate in cancer growth, progression and therapy resistance. Nat. Rev. Cancer 2018, 18, 669–680. [Google Scholar] [CrossRef]
- Carnero, A.; Garcia-Mayea, Y.; Mir, C.; Lorente, J.; Rubio, I.T.; ME, L.L. The cancer stem-cell signaling network and resistance to therapy. Cancer Treat. Rev. 2016, 49, 25–36. [Google Scholar] [CrossRef]
- Gasch, C.; Ffrench, B.; O’Leary, J.J.; Gallagher, M.F. Catching moving targets: Cancer stem cell hierarchies, therapy-resistance & considerations for clinical intervention. Mol. Cancer 2017, 16, 43. [Google Scholar] [CrossRef]
- Cheung, T.H.; Rando, T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013, 14, 329–340. [Google Scholar] [CrossRef]
- Li, L.; Bhatia, R. Stem cell quiescence. Clin. Cancer Res. 2011, 17, 4936–4941. [Google Scholar] [CrossRef]
- Cho, I.J.; Lui, P.P.; Obajdin, J.; Riccio, F.; Stroukov, W.; Willis, T.L.; Spagnoli, F.; Watt, F.M. Mechanisms, Hallmarks, and Implications of Stem Cell Quiescence. Stem Cell Rep. 2019, 12, 1190–1200. [Google Scholar] [CrossRef] [PubMed]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Blanpain, C.; Rossi, D.J. DNA damage response in adult stem cells: Pathways and consequences. Nat. Rev. Mol. Cell Biol. 2011, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.; Lyle, S. Quiescent, slow-cycling stem cell populations in cancer: A review of the evidence and discussion of significance. J. Oncol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Danes, A.; Larsimont, J.C.; Liagre, M.; Munoz-Couselo, E.; Lapouge, G.; Brisebarre, A.; Dubois, C.; Suppa, M.; Sukumaran, V.; Del Marmol, V.; et al. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature 2018, 562, 434–438. [Google Scholar] [CrossRef]
- Chen, Y.; Li, D.; Wang, D.; Liu, X.; Yin, N.; Song, Y.; Lu, S.H.; Ju, Z.; Zhan, Q. Quiescence and attenuated DNA damage response promote survival of esophageal cancer stem cells. J. Cell Biochem. 2012, 113, 3643–3652. [Google Scholar] [CrossRef]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Bugde, P.; Biswas, R.; Merien, F.; Lu, J.; Liu, D.-X.; Chen, M.; Zhou, S.; Li, Y. The therapeutic potential of targeting ABC transporters to combat multi-drug resistance. Expert Opin. Ther. Targets 2017, 21, 511–530. [Google Scholar] [CrossRef]
- Kathawala, R.J.; Gupta, P.; Ashby, C.R., Jr.; Chen, Z.S. The modulation of ABC transporter-mediated multidrug resistance in cancer: A review of the past decade. Drug Resist. Updat. 2015, 18, 1–17. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Westover, D.; Ling, X.; Lam, H.; Welch, J.; Jin, C.; Gongora, C.; Del Rio, M.; Wani, M.; Li, F. FL118, a novel camptothecin derivative, is insensitive to ABCG2 expression and shows improved efficacy in comparison with irinotecan in colon and lung cancer models with ABCG2-induced resistance. Mol. Cancer 2015, 14, 92. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, K.; Katayama, K.; Mitsuhashi, J.; Sugimoto, Y. Functions of the breast cancer resistance protein (BCRP/ABCG2) in chemotherapy. Adv. Drug Deliv. Rev. 2009, 61, 26–33. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Yu, L.; Liu, S.; Fu, J.; Cho, C.H. Constitutive AhR activation leads to concomitant ABCG2-mediated multidrug resistance in cisplatin-resistant esophageal carcinoma cells. Mol. Carcinog. 2012, 51, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Mathur, A.; Zhang, Y.; Xi, S.; Atay, S.; Hong, J.A.; Datrice, N.; Upham, T.; Kemp, C.D.; Ripley, R.T.; et al. Mithramycin represses basal and cigarette smoke-induced expression of ABCG2 and inhibits stem cell signaling in lung and esophageal cancer cells. Cancer Res. 2012, 72, 4178–4192. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, J.Q.; Assaraf, Y.G.; Ren, L.; Gupta, P.; Wei, L.; Ashby, C.R., Jr.; Yang, D.H.; Chen, Z.S. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018, 41, 1–25. [Google Scholar] [CrossRef]
- Wang, H.; Gao, Z.; Liu, X.; Agarwal, P.; Zhao, S.; Conroy, D.W.; Ji, G.; Yu, J.; Jaroniec, C.P.; Liu, Z.; et al. Targeted production of reactive oxygen species in mitochondria to overcome cancer drug resistance. Nat. Commun. 2018, 9, 562. [Google Scholar] [CrossRef]
- Diehn, M.; Cho, R.W.; Lobo, N.A.; Kalisky, T.; Dorie, M.J.; Kulp, A.N.; Qian, D.; Lam, J.S.; Ailles, L.E.; Wong, M.; et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 2009, 458, 780–783. [Google Scholar] [CrossRef]
- Dinavahi, S.S.; Bazewicz, C.G.; Gowda, R.; Robertson, G.P. Aldehyde Dehydrogenase Inhibitors for Cancer Therapeutics. Trends Pharmacol. Sci. 2019, 40, 774–789. [Google Scholar] [CrossRef]
- Mizuno, T.; Suzuki, N.; Makino, H.; Furui, T.; Morii, E.; Aoki, H.; Kunisada, T.; Yano, M.; Kuji, S.; Hirashima, Y.; et al. Cancer stem-like cells of ovarian clear cell carcinoma are enriched in the ALDH-high population associated with an accelerated scavenging system in reactive oxygen species. Gynecol. Oncol. 2015, 137, 299–305. [Google Scholar] [CrossRef]
- Xu, X.; Chai, S.; Wang, P.; Zhang, C.; Yang, Y.; Yang, Y.; Wang, K. Aldehyde dehydrogenases and cancer stem cells. Cancer Lett. 2015, 369, 50–57. [Google Scholar] [CrossRef]
- Voog, J.; Jones, D.L. Stem cells and the niche: A dynamic duo. Cell Stem Cell 2010, 6, 103–115. [Google Scholar] [CrossRef] [PubMed]
- Kise, K.; Kinugasa-Katayama, Y.; Takakura, N. Tumor microenvironment for cancer stem cells. Adv. Drug Deliv. Rev. 2016, 99, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef] [PubMed]
- Karakasheva, T.A.; Lin, E.W.; Tang, Q.; Qiao, E.; Waldron, T.J.; Soni, M.; Klein-Szanto, A.J.; Sahu, V.; Basu, D.; Ohashi, S.; et al. IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment. Cancer Res. 2018, 78, 4957–4970. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Hua, Y.; Jiang, Z.; Yue, J.; Shi, M.; Zhen, X.; Zhang, X.; Yang, L.; Zhou, R.; Wu, S. Cancer-associated Fibroblast-promoted LncRNA DNM3OS Confers Radioresistance by Regulating DNA Damage Response in Esophageal Squamous Cell Carcinoma. Clin. Cancer Res. 2019, 25, 1989–2000. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Lin, E.W.; Karakasheva, T.A.; Hicks, P.D.; Bass, A.J.; Rustgi, A.K. The tumor microenvironment in esophageal cancer. Oncogene 2016, 35, 5337–5349. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, J.; Tao, L.; Huang, G.; Chu, X.; Song, H.; Chen, L. Wnt signaling induces radioresistance through upregulating HMGB1 in esophageal squamous cell carcinoma. Cell Death Dis. 2018, 9, 433. [Google Scholar] [CrossRef]
- Su, H.; Jin, X.; Zhang, X.; Zhao, L.; Lin, B.; Li, L.; Fei, Z.; Shen, L.; Fang, Y.; Pan, H.; et al. FH535 increases the radiosensitivity and reverses epithelial-to-mesenchymal transition of radioresistant esophageal cancer cell line KYSE-150R. J. Transl. Med. 2015, 13, 104. [Google Scholar] [CrossRef]
- Das, B.; Tsuchida, R.; Malkin, D.; Koren, G.; Baruchel, S.; Yeger, H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells 2008, 26, 1818–1830. [Google Scholar] [CrossRef]
- Melzer, C.; von der Ohe, J.; Lehnert, H.; Ungefroren, H.; Hass, R. Cancer stem cell niche models and contribution by mesenchymal stroma/stem cells. Mol. Cancer 2017, 16, 28. [Google Scholar] [CrossRef] [PubMed]
- Colwell, N.; Larion, M.; Giles, A.J.; Seldomridge, A.N.; Sizdahkhani, S.; Gilbert, M.R.; Park, D.M. Hypoxia in the glioblastoma microenvironment: Shaping the phenotype of cancer stem-like cells. Neuro Oncol. 2017, 19, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Vey, N.; Delaunay, J.; Martinelli, G.; Fiedler, W.; Raffoux, E.; Prebet, T.; Gomez-Roca, C.; Papayannidis, C.; Kebenko, M.; Paschka, P.; et al. Phase I clinical study of RG7356, an anti-CD44 humanized antibody, in patients with acute myeloid leukemia. Oncotarget 2016, 7, 32532–32542. [Google Scholar] [CrossRef]
- Justilien, V.; Fields, A.P. Molecular pathways: Novel approaches for improved therapeutic targeting of Hedgehog signaling in cancer stem cells. Clin. Cancer Res. 2015, 21, 505–513. [Google Scholar] [CrossRef]
- Pan, Q.; Li, Q.; Liu, S.; Ning, N.; Zhang, X.; Xu, Y.; Chang, A.E.; Wicha, M.S. Concise Review: Targeting Cancer Stem Cells Using Immunologic Approaches. Stem Cells 2015, 33, 2085–2092. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]
- Satoh, T.; Lee, K.H.; Rha, S.Y.; Sasaki, Y.; Park, S.H.; Komatsu, Y.; Yasui, H.; Kim, T.-Y.; Yamaguchi, K.; Fuse, N.; et al. Randomized phase II trial of nimotuzumab plus irinotecan versus irinotecan alone as second-line therapy for patients with advanced gastric cancer. Gastric. Cancer 2015, 18, 824–832. [Google Scholar] [CrossRef]
- Hanahan, D.; Coussens, L.M. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell 2012, 21, 309–322. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef]
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer Stem Cells and Targeting Strategies. Cells 2019, 8, 926. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Daniel, S.K.; Seo, Y.D.; Pillarisetty, V.G. The CXCL12-CXCR4/CXCR7 axis as a mechanism of immune resistance in gastrointestinal malignancies. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Sleightholm, R.L.; Neilsen, B.K.; Li, J.; Steele, M.M.; Singh, R.K.; Hollingsworth, M.A.; Oupicky, D. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol. Ther. 2017, 179, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Łukaszewicz-Zając, M.; Mroczko, B.; Szmitkowski, M. Chemokines and their receptors in esophageal cancer—The systematic review and future perspectives. Tumour. Biol. 2015, 36, 5707–5714. [Google Scholar] [CrossRef]
- Singh, S.; Srivastava, S.K.; Bhardwaj, A.; Owen, L.B.; Singh, A.P. CXCL12-CXCR4 signalling axis confers gemcitabine resistance to pancreatic cancer cells: A novel target for therapy. Br. J. Cancer 2010, 103, 1671–1679. [Google Scholar] [CrossRef]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef]
- Gockel, I.; Schimanski, C.C.; Heinrich, C.; Wehler, T.; Frerichs, K.; Drescher, D.; von Langsdorff, C.; Domeyer, M.; Biesterfeld, S.; Galle, P.R.; et al. Expression of chemokine receptor CXCR4 in esophageal squamous cell and adenocarcinoma. BMC Cancer 2006, 6, 290. [Google Scholar] [CrossRef][Green Version]
- Gros, S.J.; Kurschat, N.; Drenckhan, A.; Dohrmann, T.; Forberich, E.; Effenberger, K.; Reichelt, U.; Hoffman, R.M.; Pantel, K.; Kaifi, J.T.; et al. Involvement of CXCR4 chemokine receptor in metastastic HER2-positive esophageal cancer. PLoS ONE 2012, 7, e47287. [Google Scholar] [CrossRef]
- Fuchs, C.S.; Tomasek, J.; Yong, C.J.; Dumitru, F.; Passalacqua, R.; Goswami, C.; Safran, H.; dos Santos, L.V.; Aprile, G.; Ferry, D.R.; et al. Ramucirumab monotherapy for previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (REGARD): An international, randomised, multicentre, placebo-controlled, phase 3 trial. Lancet 2014, 383, 31–39. [Google Scholar] [CrossRef]
- Sayin, S.I.; Baumeister, T.; Wang, T.C.; Quante, M. Origins of Metaplasia in the Esophagus: Is This a GE Junction Stem Cell Disease? Dig. Dis. Sci. 2018, 63, 2013–2021. [Google Scholar] [CrossRef] [PubMed]
- Raufi, A.G.; Klempner, S.J. Immunotherapy for advanced gastric and esophageal cancer: Preclinical rationale and ongoing clinical investigations. J. Gastrointest Oncol. 2015, 6, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell 2015, 27, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.K.; Boku, N.; Satoh, T.; Ryu, M.H.; Chao, Y.; Kato, K.; Chung, H.C.; Chen, J.S.; Muro, K.; Kang, W.K.; et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 2461–2471. [Google Scholar] [CrossRef]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef] [PubMed]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef]
- Tallerico, R.; Garofalo, C.; Carbone, E. A New Biological Feature of Natural Killer Cells: The Recognition of Solid Tumor-Derived Cancer Stem Cells. Front. Immunol. 2016, 7, 179. [Google Scholar] [CrossRef]
- Raulet, D.H.; Vance, R.E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006, 6, 520–531. [Google Scholar] [CrossRef]
- Tallerico, R.; Todaro, M.; Di Franco, S.; Maccalli, C.; Garofalo, C.; Sottile, R.; Palmieri, C.; Tirinato, L.; Pangigadde, P.N.; La Rocca, R.; et al. Human NK cells selective targeting of colon cancer-initiating cells: A role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 2013, 190, 2381–2390. [Google Scholar] [CrossRef]
- Castriconi, R.; Daga, A.; Dondero, A.; Zona, G.; Poliani, P.L.; Melotti, A.; Griffero, F.; Marubbi, D.; Spaziante, R.; Bellora, F.; et al. NK cells recognize and kill human glioblastoma cells with stem cell-like properties. J. Immunol. 2009, 182, 3530–3539. [Google Scholar] [CrossRef]
- Yin, T.; Wang, G.; He, S.; Liu, Q.; Sun, J.; Wang, Y. Human cancer cells with stem cell-like phenotype exhibit enhanced sensitivity to the cytotoxicity of IL-2 and IL-15 activated natural killer cells. Cell. Immunol. 2016, 300, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; O’Brien, C.A.; van Galen, P.; Gan, O.I.; Notta, F.; Brown, A.M.; Ng, K.; Ma, J.; Wienholds, E.; Dunant, C.; et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013, 339, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, Y.; Yu, T.S.; McKay, R.M.; Burns, D.K.; Kernie, S.G.; Parada, L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature 2012, 488, 522–526. [Google Scholar] [CrossRef]
- Ranjan, T.; Howard, C.M.; Yu, A.; Xu, L.; Aziz, K.; Jho, D.; Leonardo, J.; Hameed, M.A.; Karlovits, S.M.; Wegner, R.E.; et al. Cancer Stem Cell Chemotherapeutics Assay for Prospective Treatment of Recurrent Glioblastoma and Progressive Anaplastic Glioma: A Single-Institution Case Series. Transl. Oncol. 2020, 13, 100755. [Google Scholar] [CrossRef]
| Markers | Cancer Type EAC/ESCC | Results | Marker for Diagnosis or Prognosis | Reference |
|---|---|---|---|---|
| Single marker | ||||
| CD44 | EAC ESCC | Cell surface protein: contributes to tumor invasion and regulates EMT | High CD44 expression correlates to positive lymph node ratio and lymph vascular invasion | [29,30,31] |
| ABCG2 | ESCC | ATP-binding cassette transporter (membrane transporter) is associated with the drug resistance and metastasis | The presence of ABCG2-positive cells was associated with poor survival independent of primary tumor size and positive lymph node metastasis | [33,46,47] |
| ALDH1 | EAC ESCC | Intracellular enzyme oxidizing aldehydes: ALDH1+ cancer cells possess highly invasive and metastatic capabilities with EMT phenotype and are associated with therapy resistances | Positive ALDH1 staining was relevant to higher clinical stage and shorter survival time | [43,44,45] |
| CD133 | ESCC | Cell surface protein: promotes tumor initiation and self-renewal capacity as well as chemoresistance. | The presence of CD133+ cancer cells was associated with tumor cell differentiation | [32,33,34] |
| CD271 | ESCC | Cell surface protein: CD271+ cancer cells possess higher self-renewal activity and are associated with therapy-resistance and lymphnode metastasis | Ep-CAM+ CD271(p75NTR)+ tumor cells in peripheral blood correlate with clinically diagnosed metastasis and venous invasion | [35,36,37] |
| LgR5 | EAC | Cell surface protein: promotes proliferation, migration and invasion ability | High LgR5 was associated with worse survival | [38,39,40] |
| CD90 | ESCC | Cell surface protein: CD90+ cells possess higher self-renewal activity and metastatic potential, and are more resistant to chemotherapy | Higher CD90 expression exhibit more local invasion and distant metastasis, indicating a poor prognosis | [41,42] |
| ITGA7 | ESCC | Cell surface receptor: ITGA7 contributes to tumor innitiation and drug resistance, it promotes metastasis via inducing EMT together with an anti-apoptosis function. | More ITGA7+ cells in ESCC tissues predict a worse prognosis | [49] |
| ICAM1 | ESCC | Intercellular adhesion molecule1: promotes cancer cell migration, invasion, EMT, sphere formation, tumorigenesis and drug resistance | [48] | |
| SOX2 | ESCC | Transcription factor: promotes cancer cells migration and invasion as well as chemoresistance to cisplatin | Controversial results exist regarding the prognostic value of SOX2 because of opposite conclusion among studies | [53,54,55,56,57] |
| NANOG | ESCC | Transcriptional regulator: regulates cancer cells proliferation and drug resistance | [58,59,60] | |
| BMI-1 | ESCC | Transcriptional regulator: regulates radiosensitivity of tumor cells and inbitits cell growth and invasion | Overexpression of BMI-1 is associated with progression and invasion of EC | [61,62,63,64] |
| OCT-4 | ESCC | Transcriptional regulator: promotes cell cycle progression and accelerates proliferation and invasion of esophageal cancer cells | Overexpression of OCT-4 is significantly associated with higher histological grade and poorer survival | [54,62,65,66] |
| Ep-CAM | ESCC | Transmembrane glycoprotein: Ep-CAM contributes to cell proliferation and tumorigenesis | Expression level of Ep-CAM inversely correlates with degree of differentiation | [67,68,69] |
| Gli-1 | EAC ESCC | Transcription factor: promotes cell proliferation and is associated with chemoradiation resistance | Gli-1 is positively associated with distant metastasis, indicates poor outcome | [70,71,72] |
| SALL4 | ESCC | Transcription factor: promotes cell proliferation, migration and invasion as well as chemoresistance to cisplatin, contributes to tumorigenesis in vivo | Overexpression of SALL4 was found in a majority of ESCC tissues and correlates with poor survival | [55,73] |
| Podoplanin (PDPN) | ESCC | Transmembrane protein: accelerates the proliferation and regulates tumor EMT | PDPN expression at the edge of cancer cell nest associates with tumor invasion and poor prognosis | [74,75,76,77] |
| Combined markers | ||||
| CD44+/CD24− | EAC and ESCC | CD44+/CD24− EC cells exert a higher proliferation rate and mediate therapy resistance | [50] | |
| CD44+/CD133+ | ESCC | Strong expression of CD44 and CD133 indicates a poor prognosis regardless of chemotherapy in ESCC | [51] | |
| CD133+/CXCR4+ | ESCC | CD133+CXCR4+cells regulate tumor invasion and show high proliferative capacity | Concomitant CD133-CXCR4 expression heralds impaired disease-free survival and overall survival | [52] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, C.; Fan, N.; Liu, F.; Fang, N.; Plum, P.S.; Thieme, R.; Gockel, I.; Gromnitza, S.; Hillmer, A.M.; Chon, S.-H.; et al. Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer. Cells 2020, 9, 1481. https://doi.org/10.3390/cells9061481
Zhou C, Fan N, Liu F, Fang N, Plum PS, Thieme R, Gockel I, Gromnitza S, Hillmer AM, Chon S-H, et al. Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer. Cells. 2020; 9(6):1481. https://doi.org/10.3390/cells9061481
Chicago/Turabian StyleZhou, Chenghui, Ningbo Fan, Fanyu Liu, Nan Fang, Patrick S. Plum, René Thieme, Ines Gockel, Sascha Gromnitza, Axel M. Hillmer, Seung-Hun Chon, and et al. 2020. "Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer" Cells 9, no. 6: 1481. https://doi.org/10.3390/cells9061481
APA StyleZhou, C., Fan, N., Liu, F., Fang, N., Plum, P. S., Thieme, R., Gockel, I., Gromnitza, S., Hillmer, A. M., Chon, S.-H., Schlösser, H. A., Bruns, C. J., & Zhao, Y. (2020). Linking Cancer Stem Cell Plasticity to Therapeutic Resistance-Mechanism and Novel Therapeutic Strategies in Esophageal Cancer. Cells, 9(6), 1481. https://doi.org/10.3390/cells9061481









