HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Primary Material
2.2. TCR Sequences and Cloning
2.3. Flow Cytometry and Antibodies
2.4. Retroviral Transduction and RNA Electroporation of T Cells
2.5. In Vitro Assays of TCR Modified T Cells
2.6. AML Xenograft Mouse Model
2.7. Statistical Analysis
3. Results
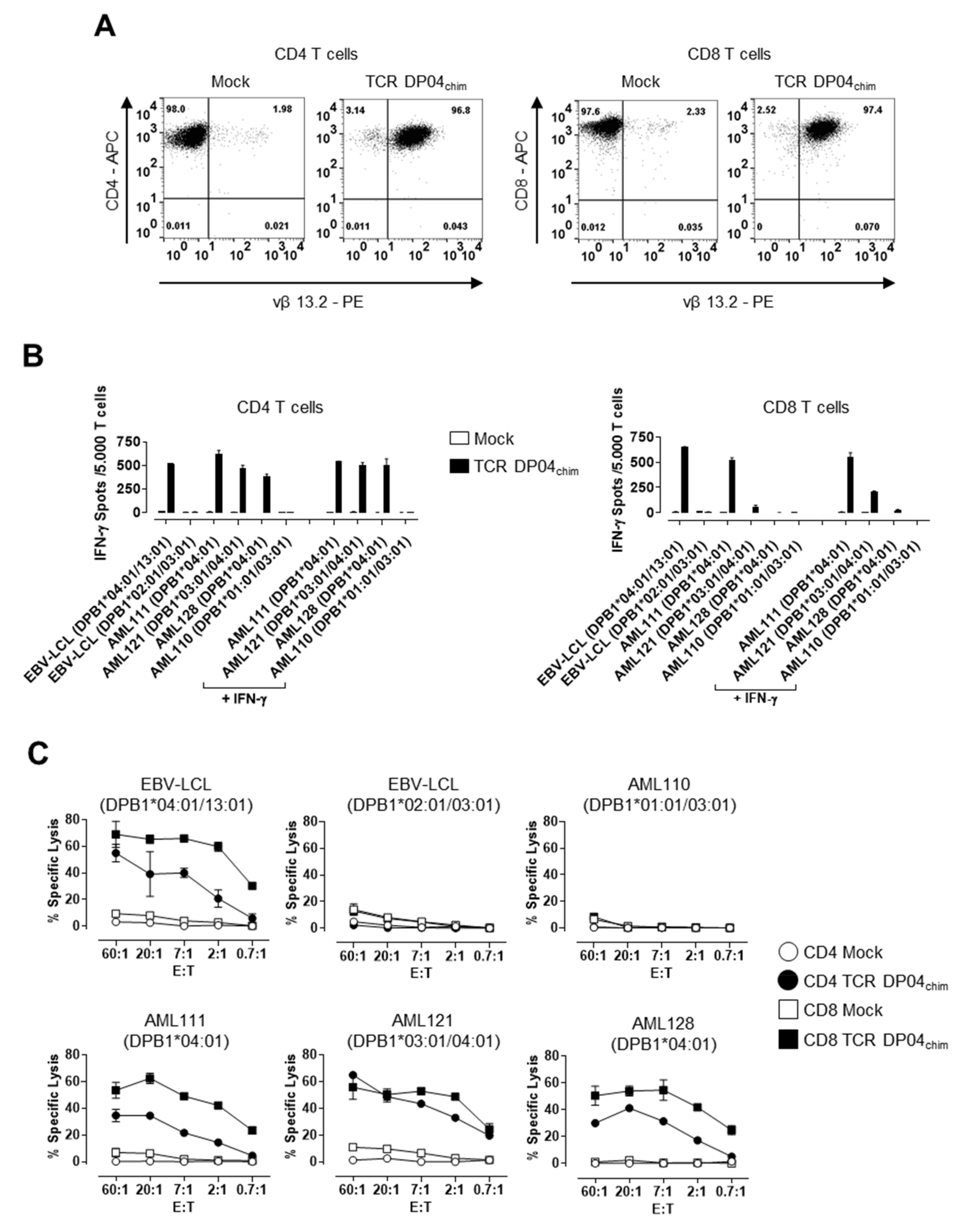
3.1. TCR DP04 Triggers Specific Recognition and Lysis of AML Blasts by CD4 and CD8 T Cells
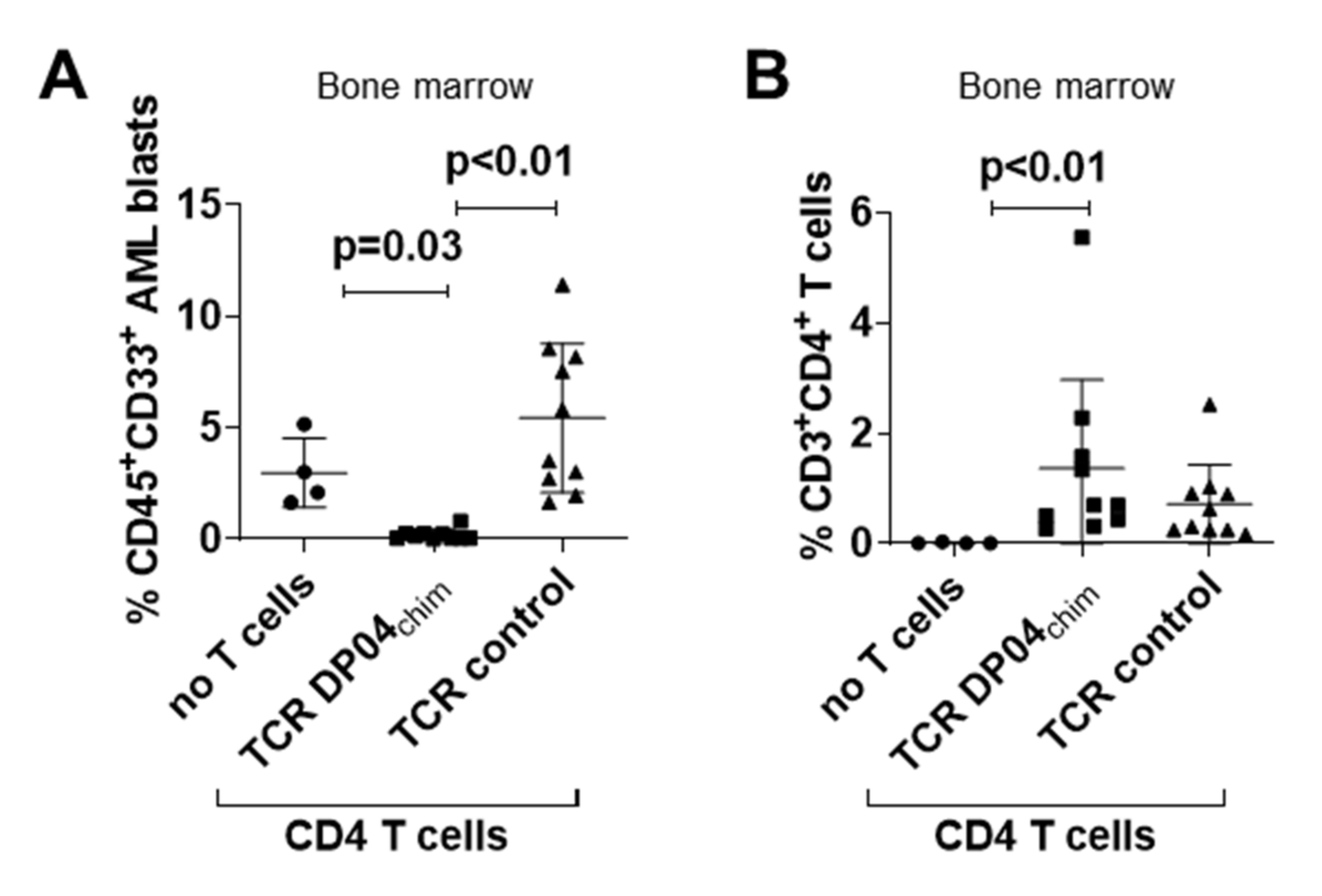
3.2. TCR DP04chim Modified CD4 T Cells Effectively Eliminate Human AML Blasts in NSG Mice
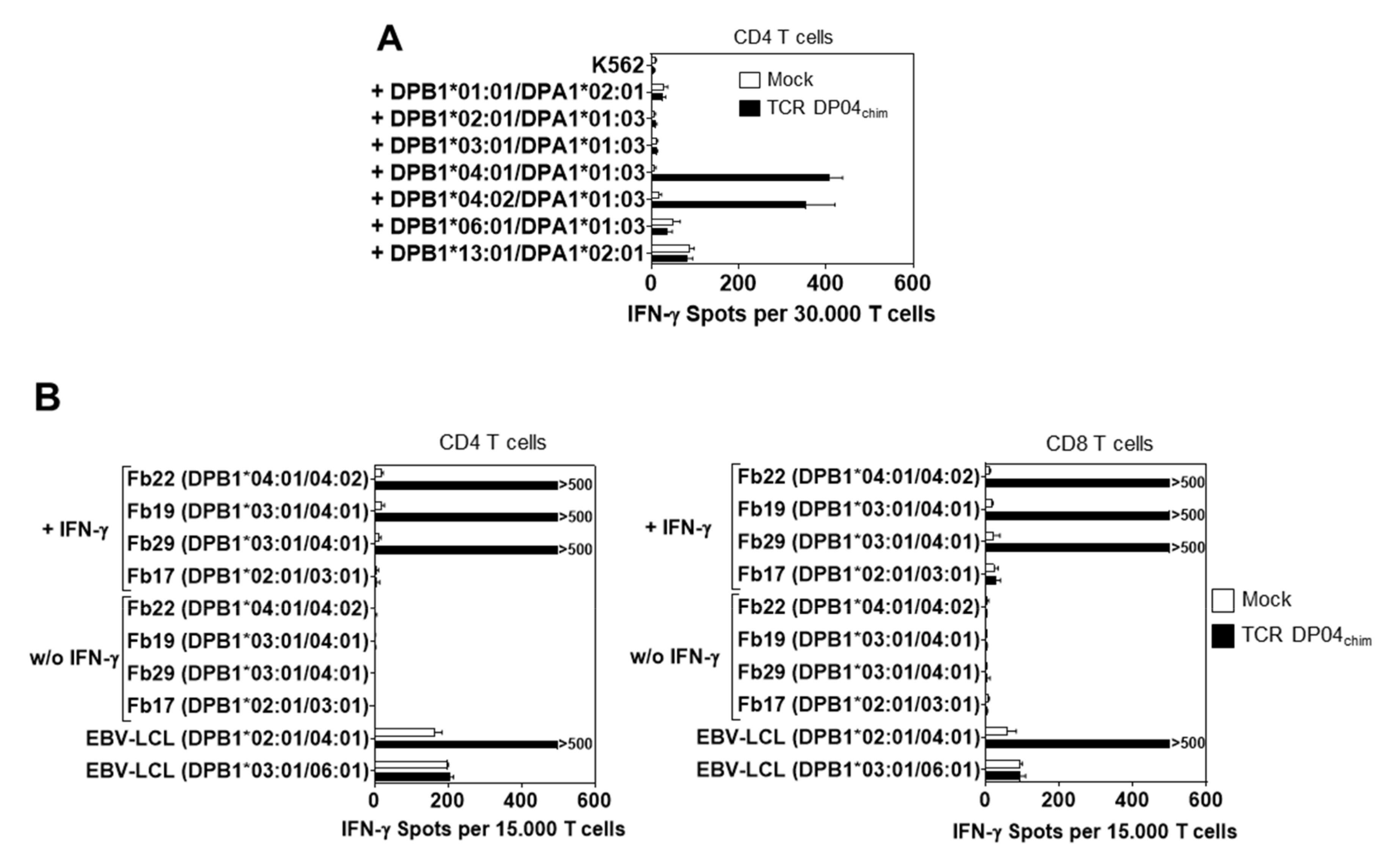
3.3. Reactivity of TCR DP04chim is Restricted to HLA-DPB1*04:01 Positive Cells, but not to Hematopoietic Cells
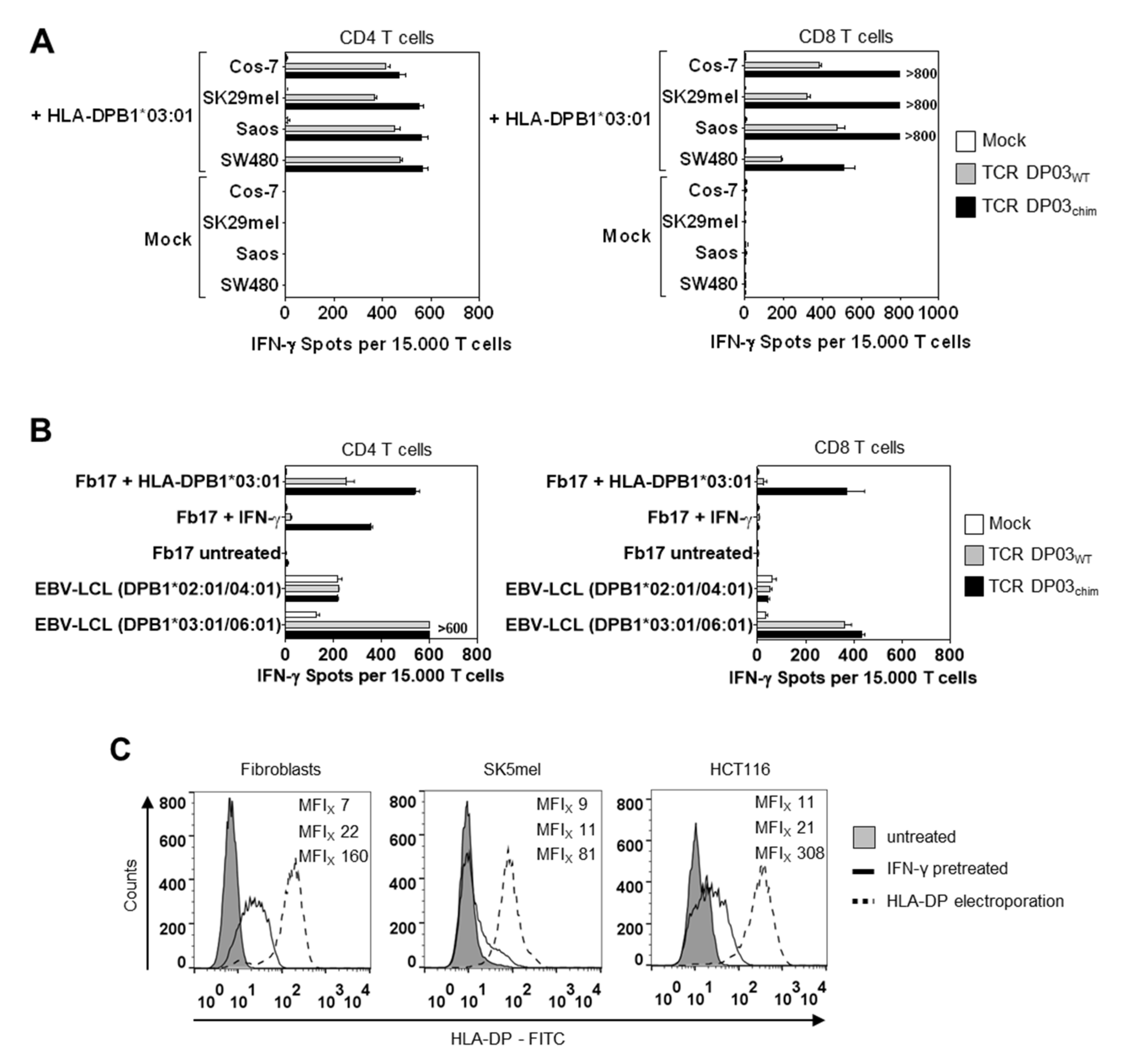
3.4. HLA-DPB1*03:01 Specific TCR Recognizes AML Blasts, but not Fibroblasts under Physiological Conditions
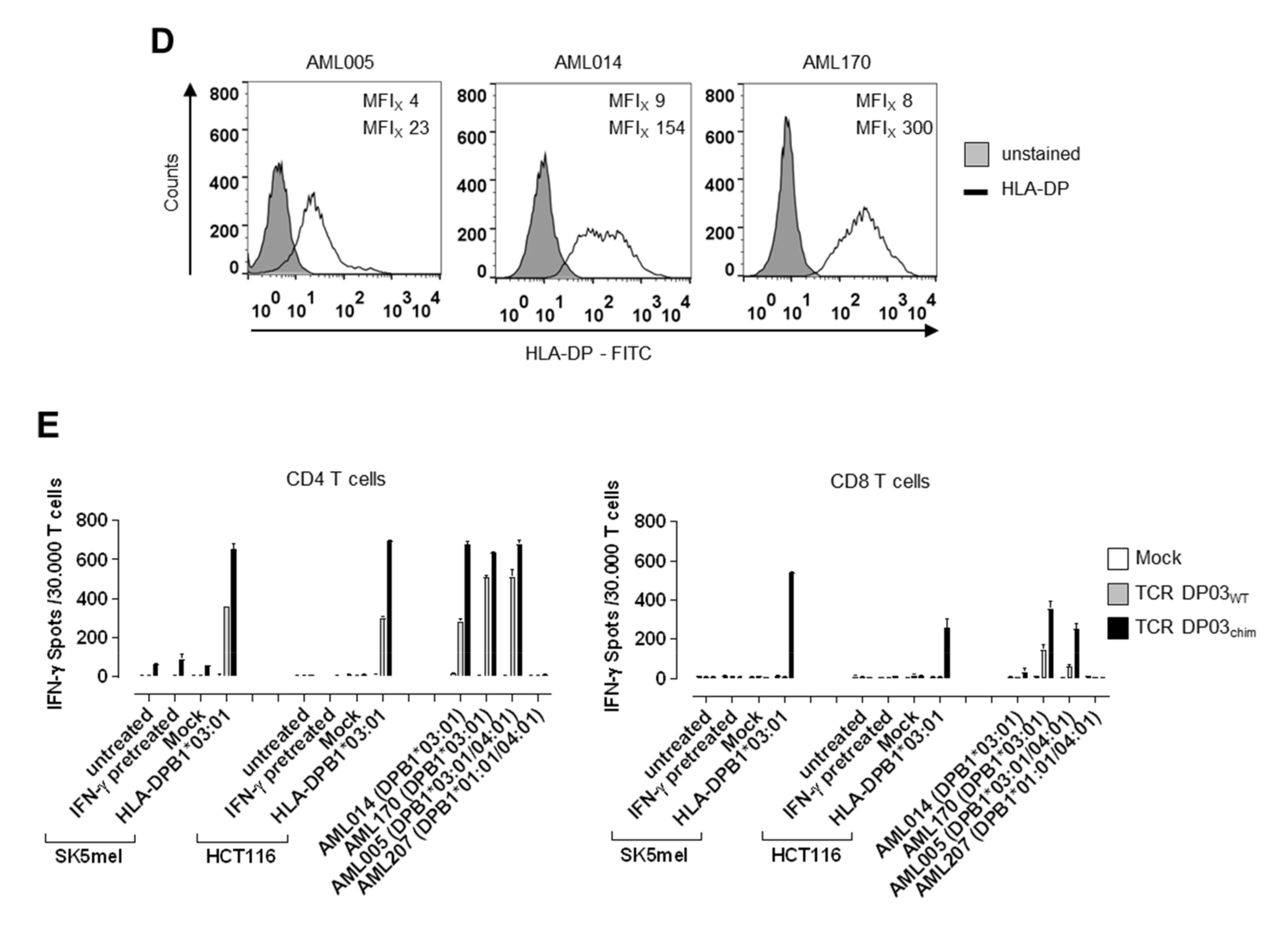
3.5. Expression Level of HLA-DPB1 Determines Recognition of Target Cells by TCR DP03
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Van Bergen, C.A.; van Luxemburg-Heijs, S.A.; de Wreede, L.C.; Eefting, M.; von dem Borne, P.A.; van Balen, P.; Heemskerk, M.H.; Mulder, A.; Claas, F.H.; Navarrete, M.A.; et al. Selective graft-versus-leukemia depends on magnitude and diversity of the alloreactive T cell response. J. Clin. Invest. 2017, 127, 517–529. [Google Scholar] [CrossRef]
- Loke, J.; Malladi, R.; Moss, P.; Craddock, C. The role of allogeneic stem cell transplantation in the management of acute myeloid leukaemia: A triumph of hope and experience. Br. J. Haematol 2019. [Google Scholar] [CrossRef] [Green Version]
- Chapuis, A.G.; Egan, D.N.; Bar, M.; Schmitt, T.M.; McAfee, M.S.; Paulson, K.G.; Voillet, V.; Gottardo, R.; Ragnarsson, G.B.; Bleakley, M.; et al. T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 2019, 25, 1064–1072. [Google Scholar] [CrossRef]
- Majzner, R.G.; Mackall, C.L. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med. 2019, 25, 1341–1355. [Google Scholar] [CrossRef]
- Petersdorf, E.W.; Gooley, T.; Malkki, M.; Anasetti, C.; Martin, P.; Woolfrey, A.; Smith, A.; Mickelson, E.; Hansen, J.A. The biological significance of HLA-DP gene variation in haematopoietic cell transplantation. Br. J. Haematol. 2001, 112, 988–994. [Google Scholar] [CrossRef]
- Petersdorf, E.W.; Malkki, M.; O’HUigin, C.; Carrington, M.; Gooley, T.; Haagenson, M.D.; Horowitz, M.M.; Spellman, S.R.; Wang, T.; Stevenson, P. High HLA-DP Expression and Graft-versus-Host Disease. N. Engl. J. Med. 2015, 373, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Fleischhauer, K.; Locatelli, F.; Zecca, M.; Orofino, M.G.; Giardini, C.; De Stefano, P.; Pession, A.; Iannone, A.M.; Carcassi, C.; Zino, E.; et al. Graft rejection after unrelated donor hematopoietic stem cell transplantation for thalassemia is associated with nonpermissive HLA-DPB1 disparity in host-versus-graft direction. Blood 2006, 107, 2984–2992. [Google Scholar] [CrossRef] [Green Version]
- Fleischhauer, K.; Shaw, B.E.; Gooley, T.; Malkki, M.; Bardy, P.; Bignon, J.D.; Dubois, V.; Horowitz, M.M.; Madrigal, J.A.; Morishima, Y.; et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: A retrospective study. Lancet. Oncol. 2012, 13, 366–374. [Google Scholar] [CrossRef] [Green Version]
- Fleischhauer, K.; Shaw, B.E. HLA-DP in unrelated hematopoietic cell transplantation revisited: Challenges and opportunities. Blood 2017, 130, 1089–1096. [Google Scholar] [CrossRef] [Green Version]
- Zino, E.; Frumento, G.; Marktel, S.; Sormani, M.P.; Ficara, F.; Di Terlizzi, S.; Parodi, A.M.; Sergeant, R.; Martinetti, M.; Bontadini, A.; et al. A T-cell epitope encoded by a subset of HLA-DPB1 alleles determines nonpermissive mismatches for hematologic stem cell transplantation. Blood 2004, 103, 1417–1424. [Google Scholar] [CrossRef] [Green Version]
- Christopher, M.J.; Petti, A.A.; Rettig, M.P.; Miller, C.A.; Chendamarai, E.; Duncavage, E.J.; Klco, J.M.; Helton, N.M.; O’Laughlin, M.; Fronick, C.C.; et al. Immune Escape of Relapsed AML Cells after Allogeneic Transplantation. N. Engl. J. Med. 2018, 379, 2330–2341. [Google Scholar] [CrossRef]
- Toffalori, C.; Zito, L.; Gambacorta, V.; Riba, M.; Oliveira, G.; Bucci, G.; Barcella, M.; Spinelli, O.; Greco, R.; Crucitti, L.; et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat. Med. 2019, 25, 603–611. [Google Scholar] [CrossRef]
- Herr, W.; Eichinger, Y.; Beshay, J.; Bloetz, A.; Vatter, S.; Mirbeth, C.; Distler, E.; Hartwig, U.F.; Thomas, S. HLA-DPB1 mismatch alleles represent powerful leukemia rejection antigens in CD4 T-cell immunotherapy after allogeneic stem-cell transplantation. Leukemia 2017, 31, 434–445. [Google Scholar] [CrossRef]
- Vatter, S.; Schmid, M.; Gebhard, C.; Mirbeth, C.; Klobuch, S.; Rehli, M.; Herr, W.; Thomas, S. In-vitro blockade of the CD4 receptor co-signal in antigen-specific T-cell stimulation cultures induces the outgrowth of potent CD4 independent T-cell effectors. J. Immunol. Methods 2018, 454, 80–85. [Google Scholar] [CrossRef]
- Nonn, M.; Herr, W.; Khan, S.; Todorova, M.; Link, I.; Thies, J.; Distler, E.; Kaltwasser, M.; Hoffmann, J.; Huber, C.; et al. Selective depletion of alloreactive T lymphocytes using patient-derived nonhematopoietic stimulator cells in allograft engineering. Transplantation 2008, 86, 1427–1435. [Google Scholar] [CrossRef]
- Cohen, C.J.; Zhao, Y.; Zheng, Z.; Rosenberg, S.A.; Morgan, R.A. Enhanced antitumor activity of murine-human hybrid T-cell receptor (TCR) in human lymphocytes is associated with improved pairing and TCR/CD3 stability. Cancer Res. 2006, 66, 8878–8886. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Klobuch, S.; Podlech, J.; Plachter, B.; Hoffmann, P.; Renzaho, A.; Theobald, M.; Reddehase, M.J.; Herr, W.; Lemmermann, N.A. Evaluating Human T-Cell Therapy of Cytomegalovirus Organ Disease in HLA-Transgenic Mice. PLOS Pathog. 2015, 11, e1005049. [Google Scholar] [CrossRef]
- Thomas, S.; Klobuch, S.; Besold, K.; Plachter, B.; Dorrie, J.; Schaft, N.; Theobald, M.; Herr, W. Strong and sustained effector function of memory- versus naive-derived T cells upon T-cell receptor RNA transfer: Implications for cellular therapy. Eur. J. Immunol. 2012, 42, 3442–3453. [Google Scholar] [CrossRef]
- Dorrschuck, A.; Schmidt, A.; Schnurer, E.; Gluckmann, M.; Albrecht, C.; Wolfel, C.; Lennerz, V.; Lifke, A.; Di Natale, C.; Ranieri, E.; et al. CD8+ cytotoxic T lymphocytes isolated from allogeneic healthy donors recognize HLA class Ia/Ib-associated renal carcinoma antigens with ubiquitous or restricted tissue expression. Blood 2004, 104, 2591–2599. [Google Scholar] [CrossRef] [Green Version]
- Giudicelli, V.; Chaume, D.; Lefranc, M.P. IMGT/V-QUEST, an integrated software program for immunoglobulin and T cell receptor V-J and V-D-J rearrangement analysis. Nucleic. Acids. Res. 2004, 32, W435–W440. [Google Scholar] [CrossRef] [Green Version]
- Matsushita, M.; Yamazaki, R.; Ikeda, H.; Mori, T.; Sumimoto, H.; Fujita, T.; Okamoto, S.; Ikeda, Y.; Kawakami, Y. Possible involvement of allogeneic antigens recognised by donor-derived CD4 cytotoxic T cells in selective GVL effects after stem cell transplantation of patients with haematological malignancy. Br. J. Haematol. 2006, 132, 56–65. [Google Scholar] [CrossRef]
- Stevanovic, S.; Griffioen, M.; Nijmeijer, B.A.; van Schie, M.L.; Stumpf, A.N.; Rutten, C.E.; Willemze, R.; Falkenburg, J.H. Human allo-reactive CD4+ T cells as strong mediators of anti-tumor immunity in NOD/scid mice engrafted with human acute lymphoblastic leukemia. Leukemia 2012, 26, 312–322. [Google Scholar] [CrossRef] [Green Version]
- Markley, J.C.; Sadelain, M. IL-7 and IL-21 are superior to IL-2 and IL-15 in promoting human T cell-mediated rejection of systemic lymphoma in immunodeficient mice. Blood 2010, 115, 3508–3519. [Google Scholar] [CrossRef] [Green Version]
- Britten, C.M.; Meyer, R.G.; Kreer, T.; Drexler, I.; Wolfel, T.; Herr, W. The use of HLA-A*0201-transfected K562 as standard antigen-presenting cells for CD8(+) T lymphocytes in IFN-gamma ELISPOT assays. J. Immunol. Methods 2002, 259, 95–110. [Google Scholar] [CrossRef]
- Hollenbach, J.A.; Madbouly, A.; Gragert, L.; Vierra-Green, C.; Flesch, S.; Spellman, S.; Begovich, A.; Noreen, H.; Trachtenberg, E.; Williams, T.; et al. A combined DPA1~DPB1 amino acid epitope is the primary unit of selection on the HLA-DP heterodimer. Immunogenetics 2012, 64, 559–569. [Google Scholar] [CrossRef] [Green Version]
- Stevanovic, S.; van Bergen, C.A.; van Luxemburg-Heijs, S.A.; van der Zouwen, B.; Jordanova, E.S.; Kruisselbrink, A.B.; van de Meent, M.; Harskamp, J.C.; Claas, F.H.; Marijt, E.W.; et al. HLA class II upregulation during viral infection leads to HLA-DP-directed graft-versus-host disease after CD4+ donor lymphocyte infusion. Blood 2013, 122, 1963–1973. [Google Scholar] [CrossRef] [Green Version]
- Reith, W.; LeibundGut-Landmann, S.; Waldburger, J.M. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005, 5, 793–806. [Google Scholar] [CrossRef]
- Rutten, C.E.; van Luxemburg-Heijs, S.A.; van der Meijden, E.D.; Griffioen, M.; Oudshoorn, M.; Willemze, R.; Falkenburg, J.H. HLA-DPB1 mismatching results in the generation of a full repertoire of HLA-DPB1-specific CD4+ T cell responses showing immunogenicity of all HLA-DPB1 alleles. Biol. Blood Marrow. Transplant. 2010, 16, 1282–1292. [Google Scholar] [CrossRef] [Green Version]
- Rutten, C.E.; van Luxemburg-Heijs, S.A.; van der Meijden, E.D.; Griffioen, M.; Oudshoorn, M.; Willemze, R.; Falkenburg, J.H. Both permissive and nonpermissive HLA-DPB1 mismatches can induce polyclonal HLA-DPB1 specific immune responses in vivo and in vitro. Blood 2010, 115, 151–153. [Google Scholar] [CrossRef] [Green Version]
- Rutten, C.E.; van Luxemburg-Heijs, S.A.; Halkes, C.J.; van Bergen, C.A.; Marijt, E.W.; Oudshoorn, M.; Griffioen, M.; Falkenburg, J.H. Patient HLA-DP-specific CD4+ T cells from HLA-DPB1-mismatched donor lymphocyte infusion can induce graft-versus-leukemia reactivity in the presence or absence of graft-versus-host disease. Biol. Blood Marrow Transplant. 2013, 19, 40–48. [Google Scholar] [CrossRef] [Green Version]
- Van Balen, P.; van Bergen, C.A.M.; van Luxemburg-Heijs, S.A.P.; de Klerk, W.; van Egmond, E.H.M.; Veld, S.A.J.; Halkes, C.J.M.; Zwaginga, J.J.; Griffioen, M.; Jedema, I.; et al. CD4 Donor Lymphocyte Infusion Can Cause Conversion of Chimerism Without GVHD by Inducing Immune Responses Targeting Minor Histocompatibility Antigens in HLA Class II. Front. Immunol. 2018, 9, 3016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkenburg, W.J.; Melenhorst, J.J.; van de Meent, M.; Kester, M.G.; Hombrink, P.; Heemskerk, M.H.; Hagedoorn, R.S.; Gostick, E.; Price, D.A.; Falkenburg, J.H.; et al. Allogeneic HLA-A*02-restricted WT1-specific T cells from mismatched donors are highly reactive but show off-target promiscuity. J. Immunol. 2011, 187, 2824–2833. [Google Scholar] [CrossRef] [Green Version]
- Bassani-Sternberg, M.; Coukos, G. Mass spectrometry-based antigen discovery for cancer immunotherapy. Curr. Opin. Immunol. 2016, 41, 9–17. [Google Scholar] [CrossRef]
- Caron, E.; Kowalewski, D.J.; Chiek Koh, C.; Sturm, T.; Schuster, H.; Aebersold, R. Analysis of Major Histocompatibility Complex (MHC) Immunopeptidomes Using Mass Spectrometry. Mol. Cell Proteomics. 2015, 14, 3105–3117. [Google Scholar] [CrossRef]
- Rudolph, M.G.; Stanfield, R.L.; Wilson, I.A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 2006, 24, 419–466. [Google Scholar] [CrossRef]
- Laghmouchi, A.; Hoogstraten, C.; van Balen, P.; Falkenburg, J.H.F.; Jedema, I. The allogeneic HLA-DP-restricted T-cell repertoire provoked by allogeneic dendritic cells contains T cells that show restricted recognition of hematopoietic cells including primary malignant cells. Haematologica 2019, 104, 197–206. [Google Scholar] [CrossRef]
- Wang, X.; Chang, W.C.; Wong, C.W.; Colcher, D.; Sherman, M.; Ostberg, J.R.; Forman, S.J.; Riddell, S.R.; Jensen, M.C. A transgene-encoded cell surface polypeptide for selection, in vivo tracking, and ablation of engineered cells. Blood 2011, 118, 1255–1263. [Google Scholar] [CrossRef]
- Wu, C.Y.; Roybal, K.T.; Puchner, E.M.; Onuffer, J.; Lim, W.A. Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 2015, 350, aab4077. [Google Scholar] [CrossRef] [Green Version]
- Roybal, K.T.; Williams, J.Z.; Morsut, L.; Rupp, L.J.; Kolinko, I.; Choe, J.H.; Walker, W.J.; McNally, K.A.; Lim, W.A. Engineering T Cells with Customized Therapeutic Response Programs Using Synthetic Notch Receptors. Cell 2016, 167, 419–432. [Google Scholar] [CrossRef] [Green Version]
- Di Stasi, A.; Tey, S.K.; Dotti, G.; Fujita, Y.; Kennedy-Nasser, A.; Martinez, C.; Straathof, K.; Liu, E.; Durett, A.G.; Grilley, B.; et al. Inducible apoptosis as a safety switch for adoptive cell therapy. N. Engl. J. Med. 2011, 365, 1673–1683. [Google Scholar] [CrossRef] [Green Version]
- Spear, T.T.; Evavold, B.D.; Baker, B.M.; Nishimura, M.I. Understanding TCR affinity, antigen specificity, and cross-reactivity to improve TCR gene-modified T cells for cancer immunotherapy. Cancer Immunol. Immunother. 2019, 68, 1881–1889. [Google Scholar] [CrossRef] [PubMed]
- Kuball, J.; Dossett, M.L.; Wolfl, M.; Ho, W.Y.; Voss, R.H.; Fowler, C.; Greenberg, P.D. Facilitating matched pairing and expression of TCR chains introduced into human T cells. Blood 2007, 109, 2331–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linette, G.P.; Stadtmauer, E.A.; Maus, M.V.; Rapoport, A.P.; Levine, B.L.; Emery, L.; Litzky, L.; Bagg, A.; Carreno, B.M.; Cimino, P.J.; et al. Cardiovascular toxicity and titin cross-reactivity of affinity-enhanced T cells in myeloma and melanoma. Blood 2013, 122, 863–871. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klobuch, S.; Hammon, K.; Vatter-Leising, S.; Neidlinger, E.; Zwerger, M.; Wandel, A.; Neuber, L.M.; Heilmeier, B.; Fichtner, R.; Mirbeth, C.; et al. HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation. Cells 2020, 9, 1264. https://doi.org/10.3390/cells9051264
Klobuch S, Hammon K, Vatter-Leising S, Neidlinger E, Zwerger M, Wandel A, Neuber LM, Heilmeier B, Fichtner R, Mirbeth C, et al. HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation. Cells. 2020; 9(5):1264. https://doi.org/10.3390/cells9051264
Chicago/Turabian StyleKlobuch, Sebastian, Kathrin Hammon, Sarah Vatter-Leising, Elisabeth Neidlinger, Michael Zwerger, Annika Wandel, Laura Maria Neuber, Bernhard Heilmeier, Regina Fichtner, Carina Mirbeth, and et al. 2020. "HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation" Cells 9, no. 5: 1264. https://doi.org/10.3390/cells9051264
APA StyleKlobuch, S., Hammon, K., Vatter-Leising, S., Neidlinger, E., Zwerger, M., Wandel, A., Neuber, L. M., Heilmeier, B., Fichtner, R., Mirbeth, C., Herr, W., & Thomas, S. (2020). HLA-DPB1 Reactive T Cell Receptors for Adoptive Immunotherapy in Allogeneic Stem Cell Transplantation. Cells, 9(5), 1264. https://doi.org/10.3390/cells9051264




