Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Antibodies
2.3. Recombinant Protein Purification
2.4. Pull-Down, Immunoprecipitation and Western Blots
2.5. Proteomic Analysis
2.5.1. Protein Digestion
2.5.2. Nano-Scale Chromatographic Tandem Mass Spectrometry (nLC-MS 2) Analysis
2.5.3. Data Analysis
2.6. Indirect Immunofluorescence Microscopy
2.7. Electron Microscopy
3. Results
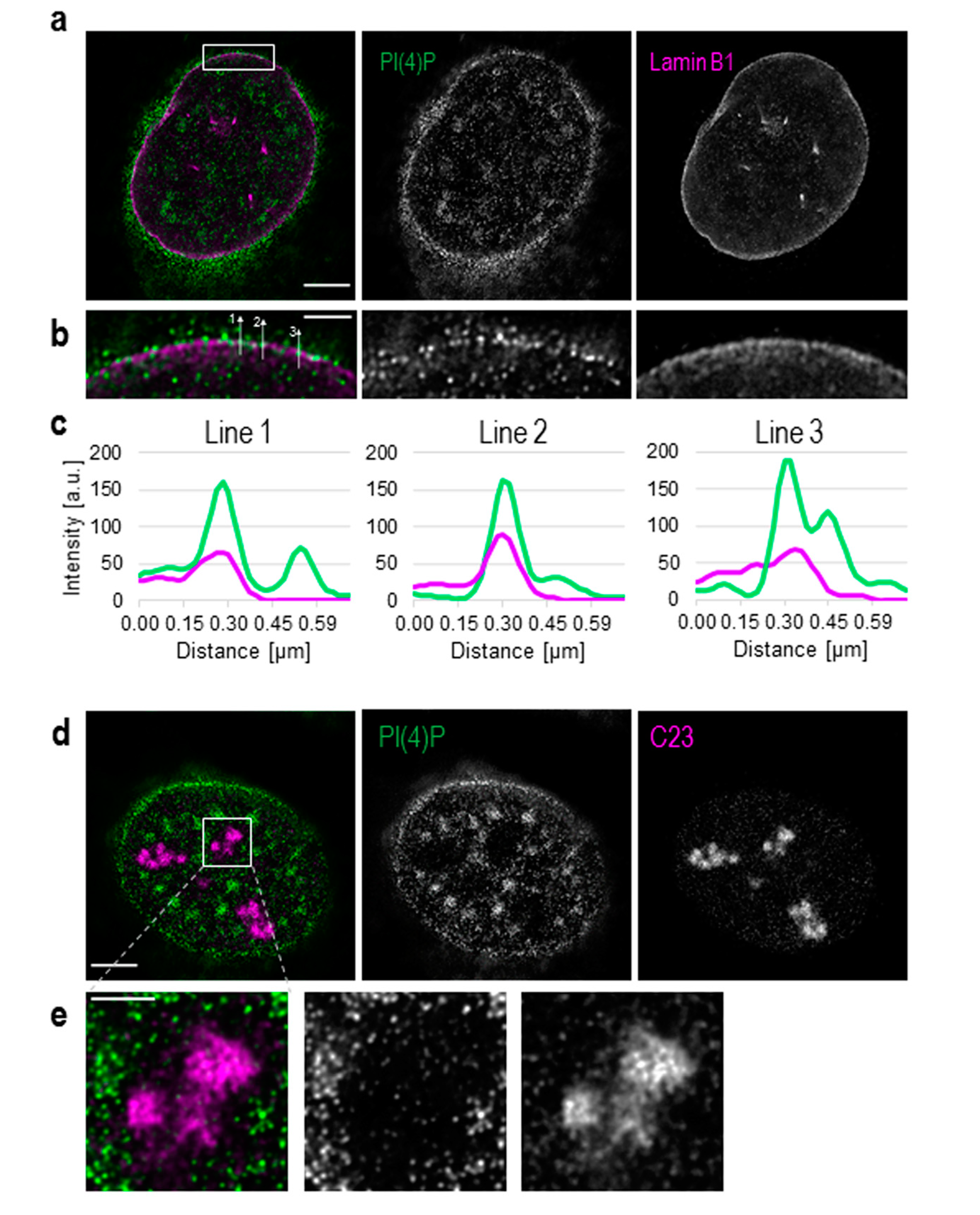
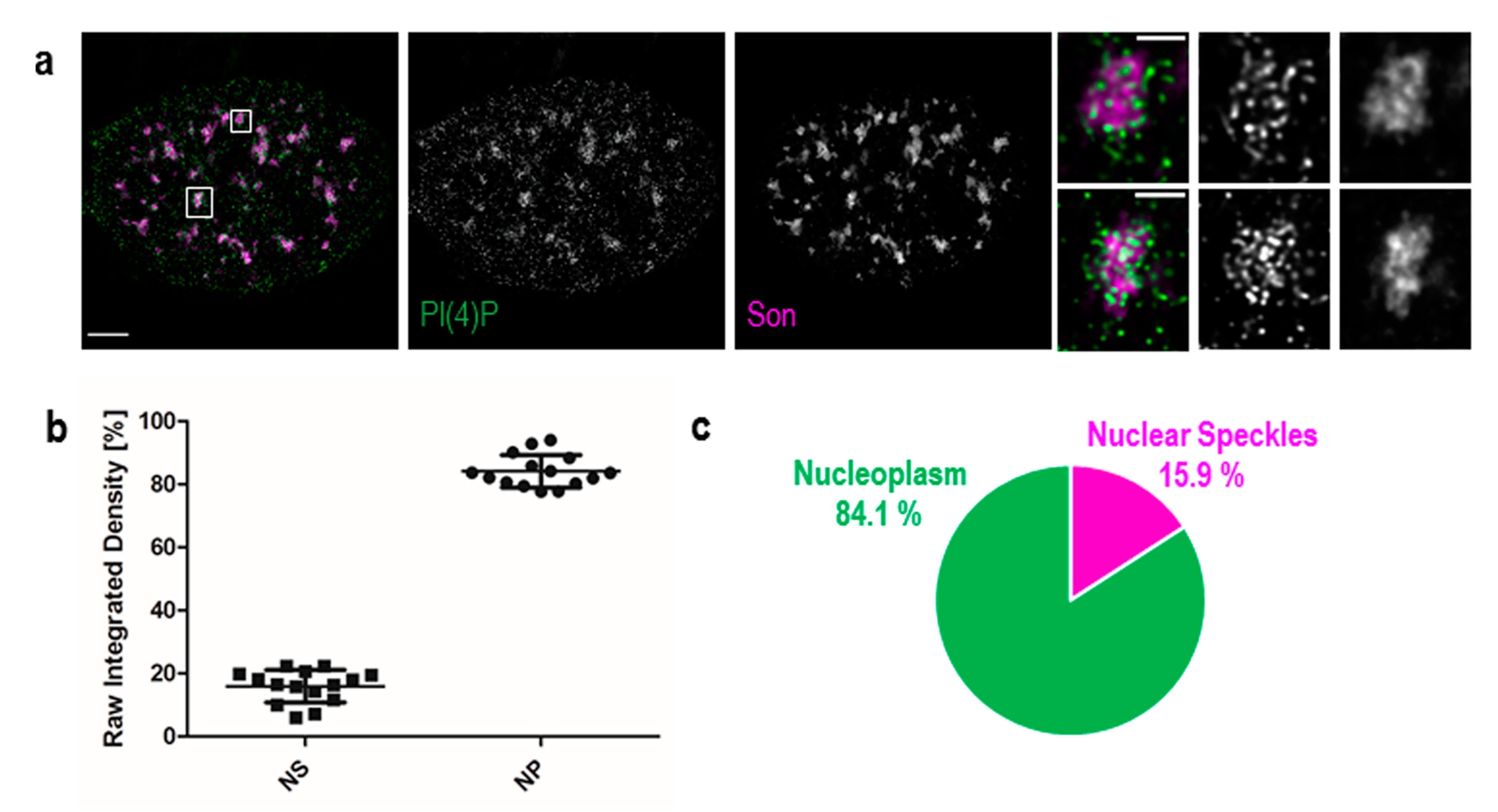
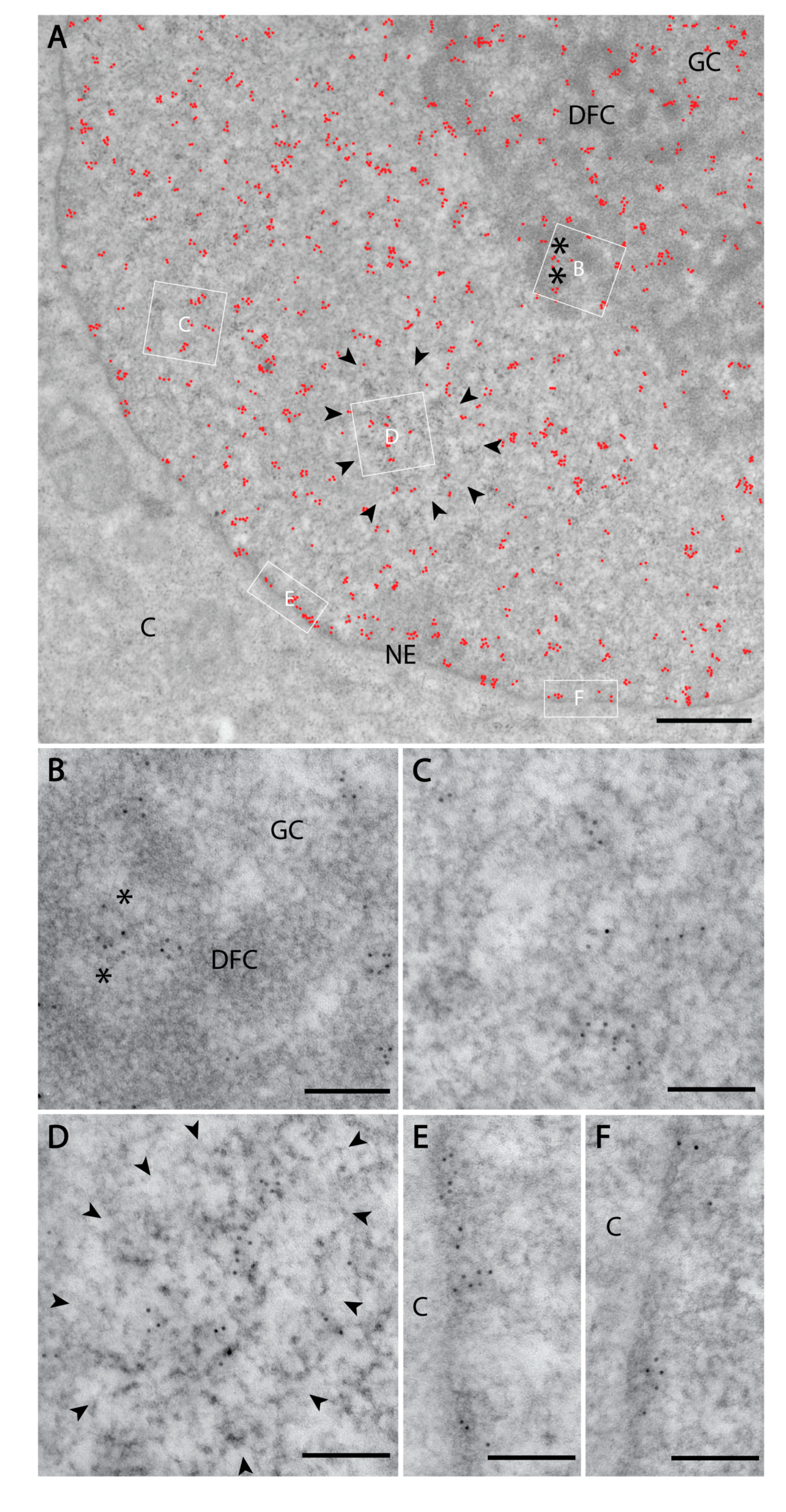
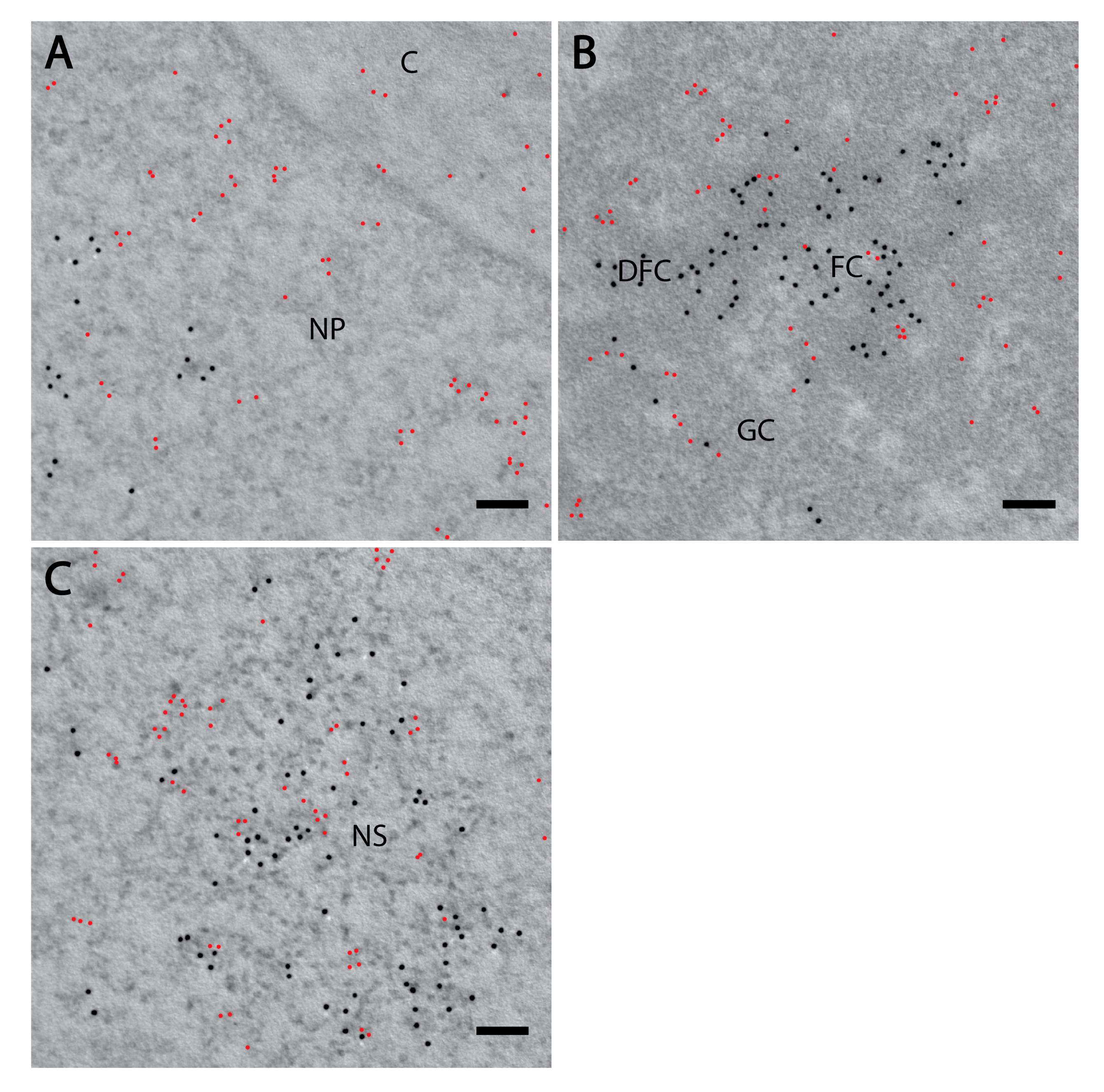
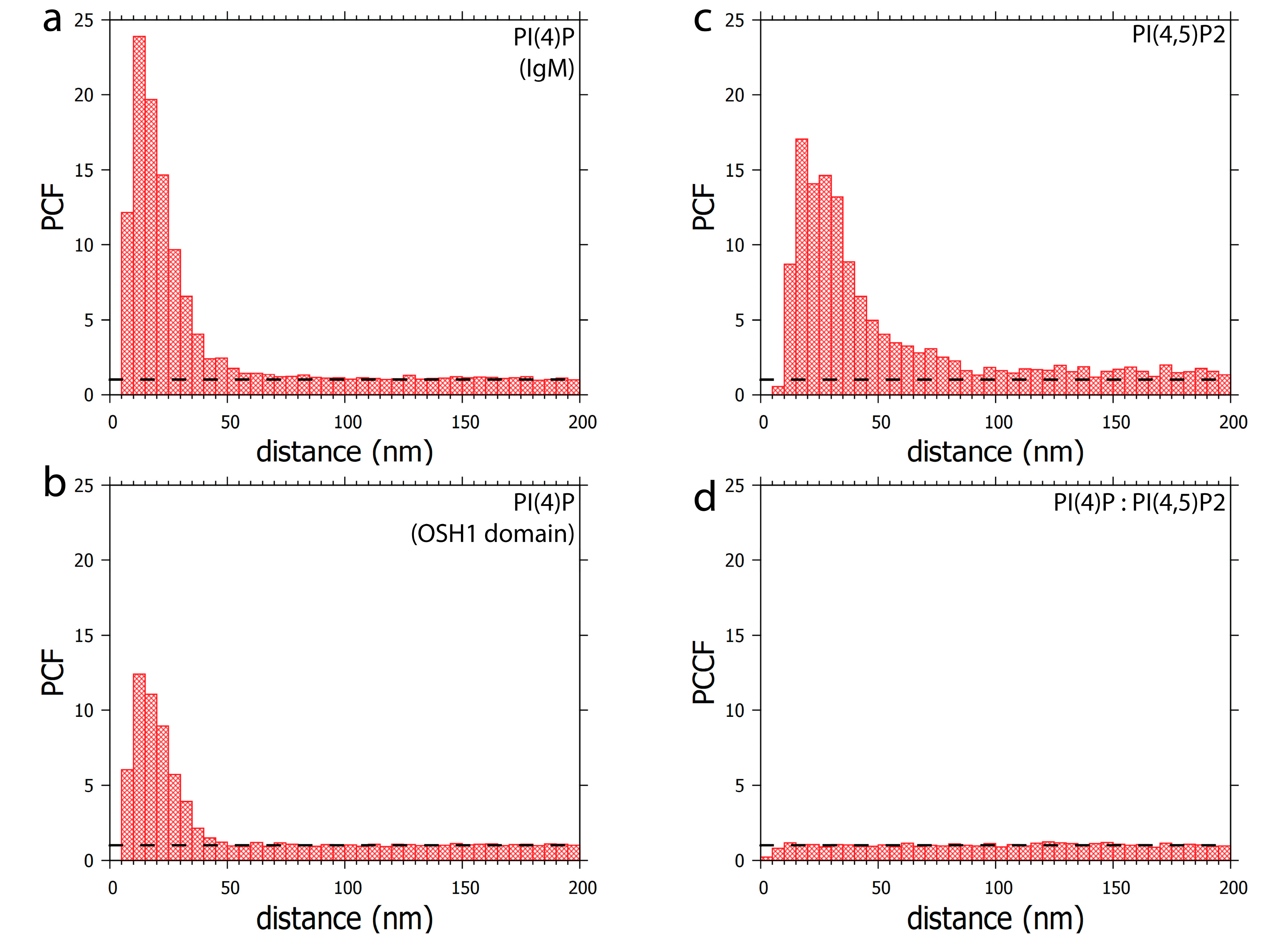
3.1. PI(4)P Localises to the Nuclear Envelope, Nucleoli, Nuclear Speckles and Nucleoplasmic Foci
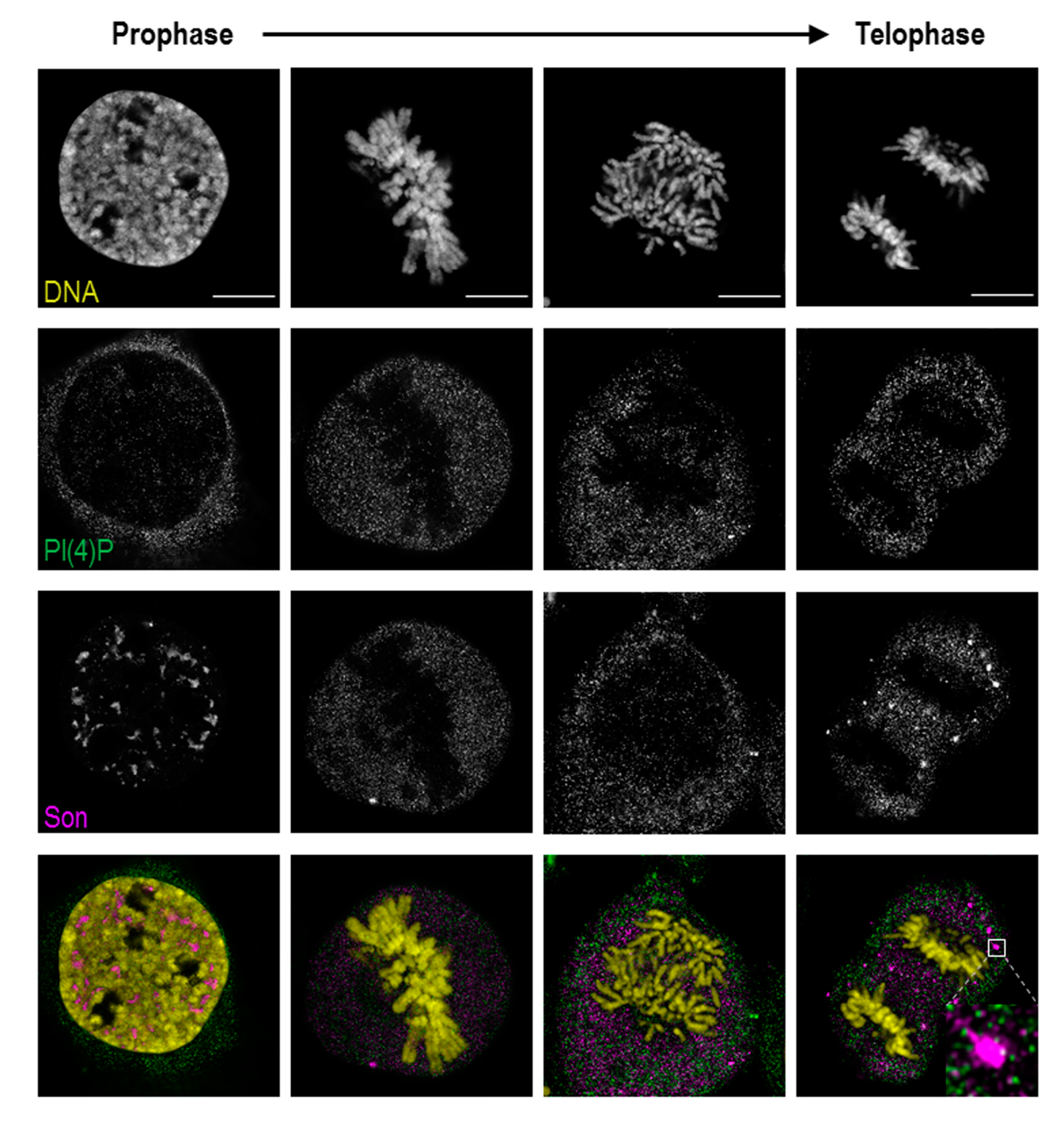
3.2. Localisation of PI(4)P through the Cell Cycle
3.3. PI(4)P is in Complex with a Number of Nuclear Proteins
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Viaud, J.; Mansour, R.; Antkowiak, A.; Mujalli, A.; Valet, C.; Chicanne, G.; Xuereb, J.M.; Terrisse, A.D.; Séverin, S.; Gratacap, M.P.; et al. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 2016, 125, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Cauvin, C.; Echard, A. Phosphoinositides: Lipids with informative heads and mastermind functions in cell division. Biochim. Biophys. Acta 2015, 1851, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.R.; Elong Edimo, W.; Erneux, C. Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved. Adv. Biol. Regul. 2018, 67, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Vann, L.R.; Wooding, F.B.; Irvine, R.F.; Divecha, N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 1997, 327, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Mazzotti, G.; Zini, N.; Rizzi, E.; Rizzoli, R.; Galanzi, A.; Ognibene, A.; Santi, S.; Matteucci, A.; Martelli, A.M.; Maraldi, N.M. Immunocytochemical detection of phosphatidylinositol 4,5-bisphosphate localization sites within the nucleus. J. Histochem. Cytochem. 1995, 43, 181–191. [Google Scholar] [CrossRef]
- Boronenkov, I.V.; Loijens, J.C.; Umeda, M.; Anderson, R.A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 1998, 9, 3547–3560. [Google Scholar]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef]
- Osborne, S.L.; Thomas, C.L.; Gschmeissner, S.; Schiavo, G. Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001, 114, 2501–2511. [Google Scholar]
- Watt, S.A.; Kular, G.; Fleming, I.N.; Downes, C.P.; Lucocq, J.M. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 2002, 363, 657–666. [Google Scholar] [CrossRef]
- Lindsay, Y.; McCoull, D.; Davidson, L.; Leslie, N.R.; Fairservice, A.; Gray, A.; Lucocq, J.; Downes, C.P. Localization of agonist-sensitive PtdIns(3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 2006, 119, 5160–5168. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.R.; Sim, Y.; Lagnado, L.; Irvine, R.F. Reversible binding and rapid diffusion of proteins in complex with inositol lipids serves to coordinate free movement with spatial information. J. Cell Biol. 2009, 184, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, S.; Castano, E.; Sobol, M.; Philimonenko, V.V.; Dzijak, R.; Venit, T.; Hozák, P. Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J. Cell Sci. 2013, 126, 2730–2739. [Google Scholar] [CrossRef] [PubMed]
- Castano, E.; Yildirim, S.; Faberova, V.; Krausova, A.; Ulicna, L.; Paprckova, D.; Sztacho, M.; Hozák, P. Nuclear phosphoinositides-versatile regulators of genome functions. Cells 2019, 8, 649. [Google Scholar] [CrossRef]
- Jacobsen, R.G.; Mazloumi Gavgani, F.; Edson, A.J.; Goris, M.; Altankhuyag, A.; Lewis, A.E. Polyphosphoinositides in the nucleus: Roadmap of their effectors and mechanisms of interaction. Adv. Biol. Regul. 2019, 72, 7–21. [Google Scholar] [CrossRef]
- Blind, R.D.; Suzawa, M.; Ingraham, H.A. Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Sci. Signal. 2012, 5, ra44. [Google Scholar] [CrossRef]
- Toska, E.; Campbell, H.A.; Shandilya, J.; Goodfellow, S.J.; Shore, P.; Medler, K.F.; Roberts, S.G. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2012, 2, 462–469. [Google Scholar] [CrossRef]
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marasek, P.; Castano, E.; Hozak, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486. [Google Scholar] [CrossRef]
- Sobol, M.; Krausova, A.; Yildirim, S.; Kalasova, I.; Faberova, V.; Vrkoslav, V.; Philimonenko, V.; Marášek, P.; Pastorek, L.; Čapek, M.; et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Stijf-Bultsma, Y.; Sommer, L.; Tauber, M.; Baalbaki, M.; Giardoglou, P.; Jones, D.R.; Gelato, K.A.; van Pelt, J.; Shah, Z.; Rahnamoun, H.; et al. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell 2015, 58, 453–467. [Google Scholar] [CrossRef]
- Rando, O.J.; Zhao, K.; Janmey, P.; Crabtree, G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2002, 99, 2824–2829. [Google Scholar] [CrossRef] [PubMed]
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 2014, 54, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Ulicna, L.; Kalendova, A.; Kalasova, I.; Vacik, T.; Hozak, P. PIP2 epigenetically represses rRNA genes transcription interacting with PHF8. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Ulicna, L.; Paprckova, D.; Faberova, V.; Hozak, P. Phospholipids and inositol phosphates linked to the epigenome. Histochem. Cell Biol. 2018, 150, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Graham, T.R.; Burd, C.G. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011, 21, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Hammond, G.R.; Schiavo, G.; Irvine, R.F. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem. J. 2009, 422, 23–35. [Google Scholar] [CrossRef]
- Kalasova, I.; Faberova, V.; Kalendova, A.; Yildirim, S.; Ulicna, L.; Venit, T.; Hozák, P. Tools for visualization of phosphoinositides in the cell nucleus. Histochem. Cell Biol. 2016, 145, 485–496. [Google Scholar] [CrossRef]
- Andersen, J.S.; Lyon, C.E.; Fox, A.H.; Leung, A.K.; Lam, Y.W.; Steen, H.; Mann, M.; Lamond, A.I. Directed proteomic analysis of the human nucleolus. Curr. Biol. 2002, 12, 1–11. [Google Scholar] [CrossRef]
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Hebert, A.S.; Richards, A.L.; Bailey, D.J.; Ulbrich, A.; Coughlin, E.E.; Westphall, M.S.; Coon, J.J. The one hour yeast proteome. Mol. Cell Proteom. 2014, 13, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Stradalova, V.; Gaplovska-Kysela, K.; Hozak, P. Ultrastructural and nuclear antigen preservation after high-pressure freezing/freeze-substitution and low-temperature LR White embedding of HeLa cells. Histochem. Cell Biol. 2008, 130, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Philimonenko, A.A.; Janacek, J.; Hozak, P. Statistical evaluation of colocalization patterns in immunogold labeling experiments. J. Struct. Biol. 2000, 132, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Takata, H.; Shibahara, K.; Bubulya, A.; Bubulya, P.A. Son is essential for nuclear speckle organization and cell cycle progression. Mol. Biol. Cell 2010, 21, 650–663. [Google Scholar] [CrossRef] [PubMed]
- Huen, M.S.; Sy, S.M.; Leung, K.M.; Ching, Y.P.; Tipoe, G.L.; Man, C.; Dong, S.; Chen, J. SON is a spliceosome-associated factor required for mitotic progression. Cell Cycle 2010, 9, 2679–2685. [Google Scholar] [CrossRef]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011, 3. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 2018, 174, 744–757. [Google Scholar] [CrossRef]
- Kim, J.; Han, K.Y.; Khanna, N.; Ha, T.; Belmont, A.S. Nuclear speckle fusion via long-range directional motion regulates speckle morphology after transcriptional inhibition. J. Cell Sci. 2019, 132. [Google Scholar] [CrossRef]
- Yao, R.W.; Xu, G.; Wang, Y.; Shan, L.; Luan, P.F.; Wang, Y.; Wu, M.; Yang, L.Z.; Xing, Y.H.; Yang, L.; et al. Nascent Pre-rRNA Sorting via phase separation drives the assembly of dense fibrillar components in the human nucleolus. Mol. Cell 2019, 76, 767–783. [Google Scholar]
- Prasanth, K.V.; Sacco-Bubulya, P.A.; Prasanth, S.G.; Spector, D.L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 2003, 14, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Gomes, X.V.; Wold, M.S. Functional domains of the 70-kilodalton subunit of human replication protein A. Biochemistry 1996, 35, 10558–10568. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Wells, W.W. Phosphorylation of rat liver nuclear envelopes. II. Characterization of in vitro lipid phosphorylation. J. Biol. Chem. 1983, 258, 9368–9373. [Google Scholar]
- Cocco, L.; Gilmour, R.S.; Ognibene, A.; Letcher, A.J.; Manzoli, F.A.; Irvine, R.F. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 1987, 248, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lee, M.; Fairn, G.D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 2018, 293, 6230–6240. [Google Scholar] [CrossRef] [PubMed]
- Vance, J.E. Phospholipid synthesis and transport in mammalian cells. Traffic 2015, 16, 1–18. [Google Scholar] [CrossRef]
- de Graaf, P.; Klapisz, E.E.; Schulz, T.K.; Cremers, A.F.; Verkleij, A.J.; van Bergen en Henegouwen, P.M. Nuclear localization of phosphatidylinositol 4-kinase beta. J. Cell Sci. 2002, 115, 1769–1775. [Google Scholar]
- Strahl, T.; Hama, H.; DeWald, D.B.; Thorner, J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J. Cell Biol. 2005, 171, 967–979. [Google Scholar] [CrossRef]
- Szivak, I.; Lamb, N.; Heilmeyer, L.M. Subcellular localization and structural function of endogenous phosphorylated phosphatidylinositol 4-kinase (PI4K92). J. Biol. Chem. 2006, 281, 16740–16749. [Google Scholar] [CrossRef]
- Kakuk, A.; Friedlander, E.; Vereb, G., Jr.; Kasa, A.; Balla, A.; Balla, T.; Heilmeyer, L.M., Jr.; Gergely, P.; Vereb, G. Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytometry A 2006, 69, 1174–1183. [Google Scholar] [CrossRef]
- Lachyankar, M.B.; Sultana, N.; Schonhoff, C.M.; Mitra, P.; Poluha, W.; Lambert, S.; Quesenberry, P.J.; Litofsky, N.S.; Recht, L.D.; Nabi, R.; et al. A role for nuclear PTEN in neuronal differentiation. J. Neurosci. 2000, 20, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Gimm, O.; Perren, A.; Weng, L.P.; Marsh, D.J.; Yeh, J.J.; Ziebold, U.; Gil, E.; Hinze, R.; Delbridge, L.; Lees, J.A.; et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 2000, 156, 1693–1700. [Google Scholar] [CrossRef]
- Deleris, P.; Bacqueville, D.; Gayral, S.; Carrez, L.; Salles, J.P.; Perret, B.; Breton-Douillon, M. SHIP-2 and PTEN are expressed and active in vascular smooth muscle cell nuclei, but only SHIP-2 is associated with nuclear speckles. J. Biol. Chem. 2003, 278, 38884–38891. [Google Scholar] [CrossRef] [PubMed]
- Elong Edimo, W.; Derua, R.; Janssens, V.; Nakamura, T.; Vanderwinden, J.M.; Waelkens, E.; Erneux, C. Evidence of SHIP2 Ser132 phosphorylation, its nuclear localization and stability. Biochem. J. 2011, 439, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Nalaskowski, M.M.; Metzner, A.; Brehm, M.A.; Labiadh, S.; Brauer, H.; Grabinski, N.; Mayr, G.W.; Jücker, M. The inositol 5-phosphatase SHIP1 is a nucleo-cytoplasmic shuttling protein and enzymatically active in cell nuclei. Cell Signal. 2012, 24, 621–628. [Google Scholar] [CrossRef]
- Ehm, P.; Nalaskowski, M.M.; Wundenberg, T.; Jucker, M. The tumor suppressor SHIP1 colocalizes in nucleolar cavities with p53 and components of PML nuclear bodies. Nucleus 2015, 6, 154–164. [Google Scholar] [CrossRef]
- Mellman, D.L.; Gonzales, M.L.; Song, C.; Barlow, C.A.; Wang, P.; Kendziorski, C.; Anderson, R.A. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 2008, 451, 1013–1017. [Google Scholar] [CrossRef]
- Kakuk, A.; Friedlander, E.; Vereb, G., Jr.; Lisboa, D.; Bagossi, P.; Toth, G.; Gergely, P.; Vereb, G. Nuclear and nucleolar localization signals and their targeting function in phosphatidylinositol 4-kinase PI4K230. Exp. Cell Res. 2008, 314, 2376–8238. [Google Scholar] [CrossRef]
- Balla, A.; Kim, Y.J.; Varnai, P.; Szentpetery, Z.; Knight, Z.; Shokat, K.M.; Balla, T. Maintenance of hormone-sensitive phosphoinositide pools in the plasma membrane requires phosphatidylinositol 4-kinase IIIalpha. Mol. Biol. Cell 2008, 19, 711–721. [Google Scholar] [CrossRef]
- Harak, C.; Radujkovic, D.; Taveneau, C.; Reiss, S.; Klein, R.; Bressanelli, S.; Lohmann, V. Mapping of functional domains of the lipid kinase phosphatidylinositol 4-kinase type III alpha involved in enzymatic activity and hepatitis C virus replication. J. Virol. 2014, 88, 9909–9926. [Google Scholar] [CrossRef]
- Izquierdo, J.M.; Majos, N.; Bonnal, S.; Martinez, C.; Castelo, R.; Guigo, R.; Bilbao, D.; Valcárcel, J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol. Cell 2005, 19, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Busso, C.; Gonczy, P. NuMA interacts with phosphoinositides and links the mitotic spindle with the plasma membrane. EMBO J. 2014, 33, 1815–1830. [Google Scholar] [CrossRef] [PubMed]
- Kukalev, A.; Nord, Y.; Palmberg, C.; Bergman, T.; Percipalle, P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat. Struct. Mol. Biol. 2005, 12, 238–244. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M.; Hung, M.L.; Golovanov, A.P.; Lian, L.Y.; Wilson, S.A. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA 2008, 105, 5154–5159. [Google Scholar] [CrossRef] [PubMed]
- Sztacho, M.; Sobol, M.; Balaban, C.; Escudeiro Lopes, S.E.; Hozak, P. NucSlear phosphoinositides and phase separation: Important players in nuclear compartmentalization. Adv. Biol. Regul. 2019, 71, 111–117. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fáberová, V.; Kalasová, I.; Krausová, A.; Hozák, P. Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome. Cells 2020, 9, 1191. https://doi.org/10.3390/cells9051191
Fáberová V, Kalasová I, Krausová A, Hozák P. Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome. Cells. 2020; 9(5):1191. https://doi.org/10.3390/cells9051191
Chicago/Turabian StyleFáberová, Veronika, Ilona Kalasová, Alžběta Krausová, and Pavel Hozák. 2020. "Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome" Cells 9, no. 5: 1191. https://doi.org/10.3390/cells9051191
APA StyleFáberová, V., Kalasová, I., Krausová, A., & Hozák, P. (2020). Super-Resolution Localisation of Nuclear PI(4)P and Identification of Its Interacting Proteome. Cells, 9(5), 1191. https://doi.org/10.3390/cells9051191





