Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Optical Microscopy
2.3. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
2.4. Western Blotting
2.5. Confocal Immuno-Staining Sample Preparation
2.6. Terminal Deoxynucleotidyl Transferase-Mediated Deoxyuridine Tri-Phosphate (dUTP) Nick and Labeling Assay
2.7. Library Preparation and Sequencing
2.8. Gene Expression Data Analysis
2.9. Electron Microscopy
2.10. Flow Cytometry
2.11. Statistical Analysis
3. Results
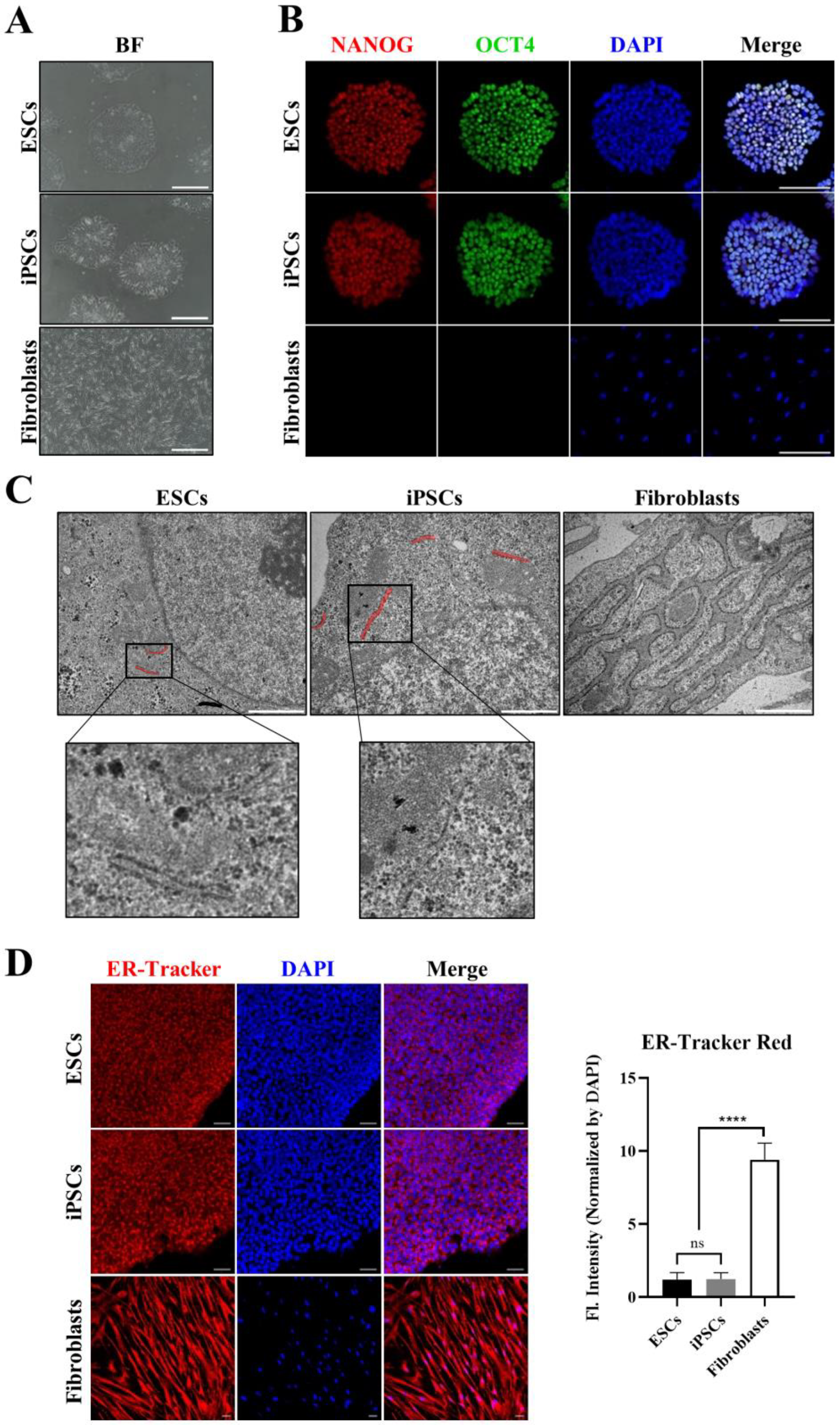
3.1. Morphological Differences in ER between hPSCs and Somatic Cells
3.2. Expression Profiles of ER-Related Genes in hPSCs Differ from Those in Fibroblasts
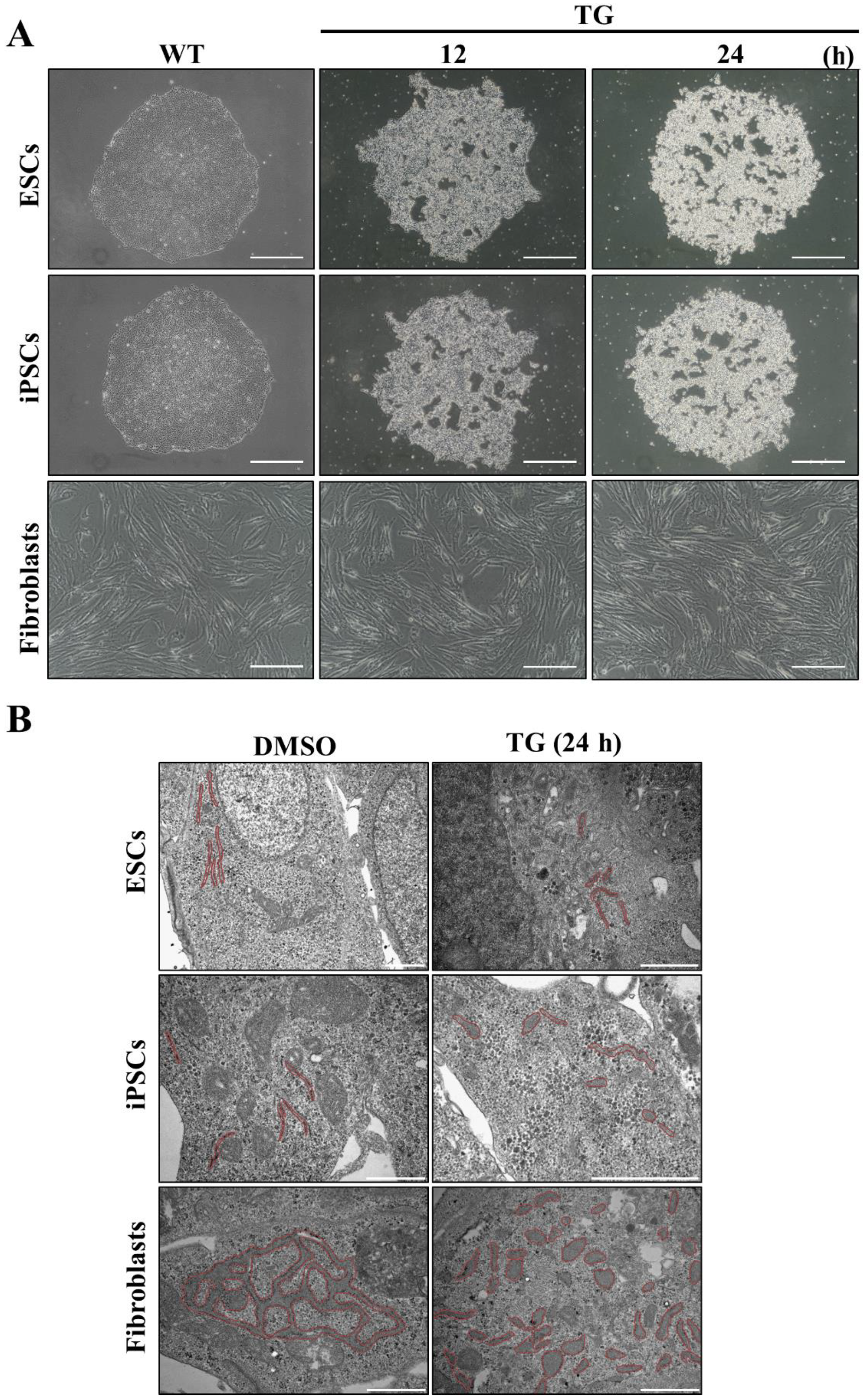
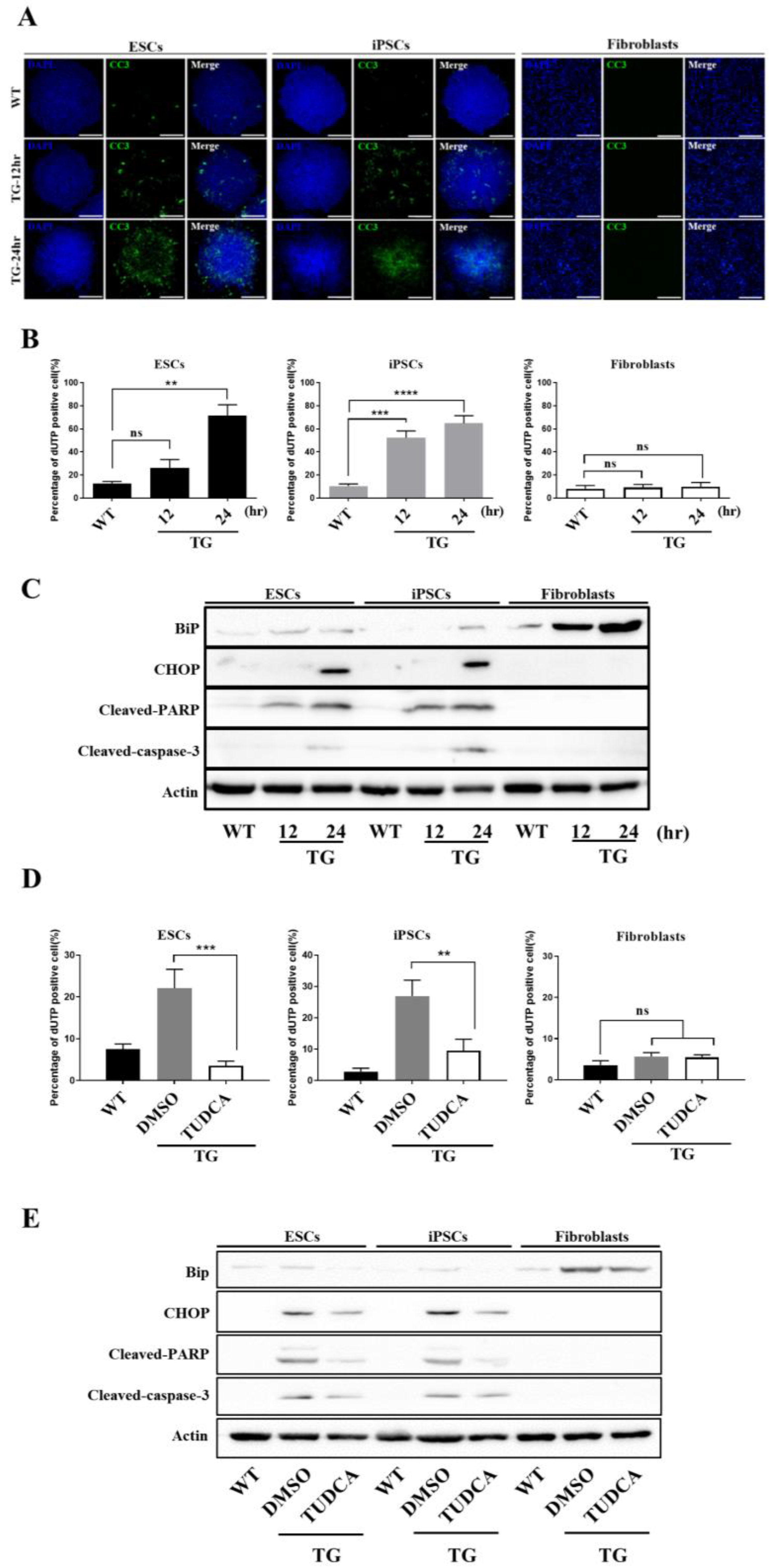
3.3. Embryonic Stem Cells Are Relatively Vulnerable to ER Stress, Resulting in More Severe Damage Compared to in Fibroblasts
3.4. Apoptotic Response Predominated in ESCs upon ER Stress
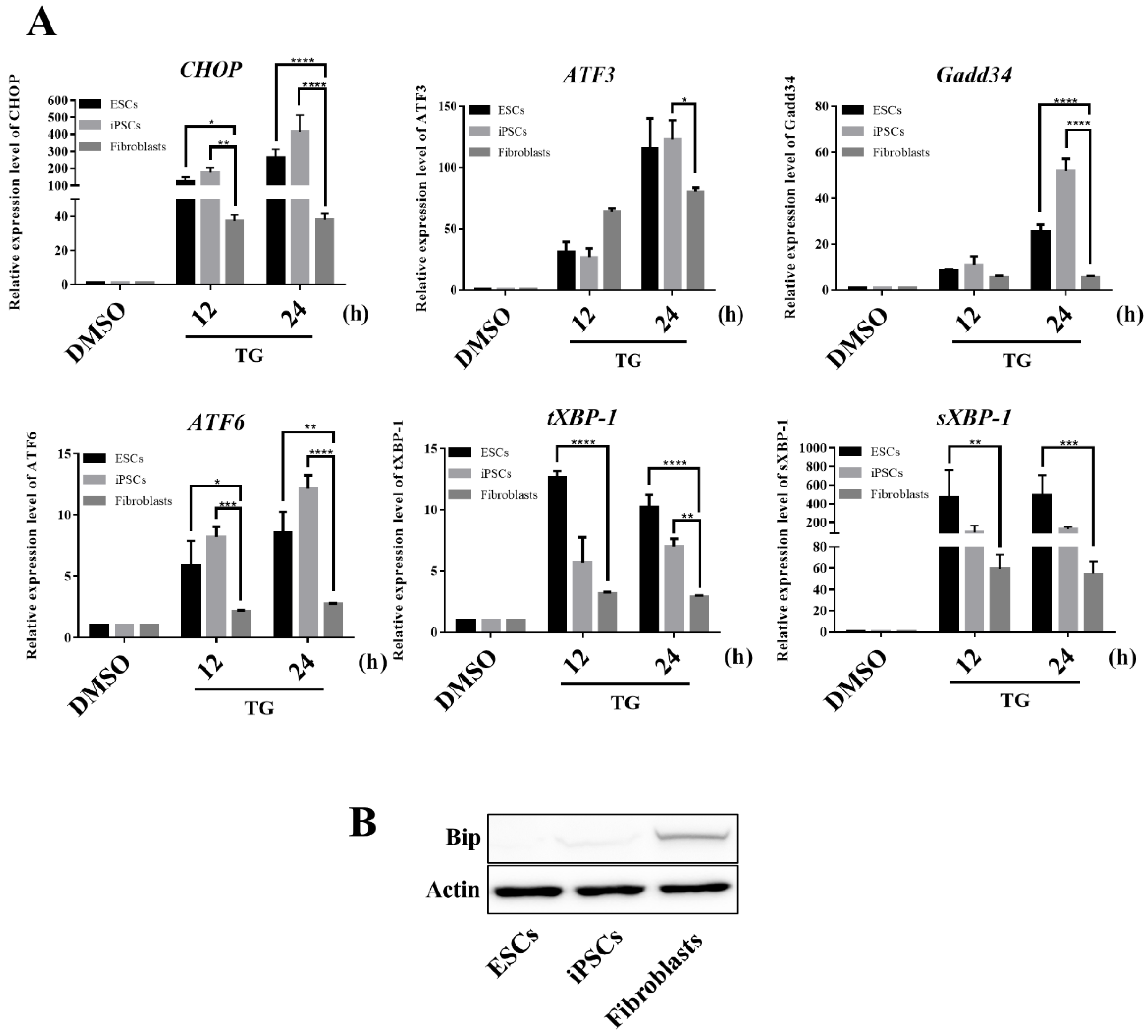
3.5. hESCs and iPSCs Displayed Abundant Induction of CHOP and Lacked BiP
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science (N. Y. NY) 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Vazin, T.; Freed, W.J. Human embryonic stem cells: Derivation, culture, and differentiation: A review. Restor. Neurol. Neurosci. 2010, 28, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131. [Google Scholar] [CrossRef] [PubMed]
- Araki, R.; Uda, M.; Hoki, Y.; Sunayama, M.; Nakamura, M.; Ando, S.; Sugiura, M.; Ideno, H.; Shimada, A.; Nifuji, A.; et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 2013, 494, 100–104. [Google Scholar] [CrossRef]
- Martin, U. Therapeutic Application of Pluripotent Stem Cells: Challenges and Risks. Front. Med. 2017, 4, 229. [Google Scholar] [CrossRef]
- Prigione, A.; Ruiz-Perez, M.V.; Bukowiecki, R.; Adjaye, J. Metabolic restructuring and cell fate conversion. Cell. Mol. Life Sci. CMLS 2015, 72, 1759–1777. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Duan, S.; Yi, F.; Ocampo, A.; Liu, G.H.; Izpisua Belmonte, J.C. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013, 18, 325–332. [Google Scholar] [CrossRef]
- Shibata, Y.; Voeltz, G.K.; Rapoport, T.A. Rough sheets and smooth tubules. Cell 2006, 126, 435–439. [Google Scholar] [CrossRef]
- Larner, S.F.; Hayes, R.L.; Wang, K.K. Unfolded protein response after neurotrauma. J. Neurotrauma 2006, 23, 807–829. [Google Scholar] [CrossRef]
- Xu, C.; Bailly-Maitre, B.; Reed, J.C. Endoplasmic reticulum stress: Cell life and death decisions. J. Clin. Investig. 2005, 115, 2656–2664. [Google Scholar] [CrossRef]
- Ozcan, U.; Cao, Q.; Yilmaz, E.; Lee, A.H.; Iwakoshi, N.N.; Ozdelen, E.; Tuncman, G.; Gorgun, C.; Glimcher, L.H.; Hotamisligil, G.S. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science (N. Y. NY) 2004, 306, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science (N. Y. NY) 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S.; Gorman, A.M.; Hori, O.; Samali, A. Cellular stress responses: Cell survival and cell death. Int. J. Cell Biol. 2010, 2010, 214074. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Rozpedek, W.; Pytel, D.; Mucha, B.; Leszczynska, H.; Diehl, J.A.; Majsterek, I. The Role of the PERK/eIF2alpha/ATF4/CHOP Signaling Pathway in Tumor Progression during Endoplasmic Reticulum Stress. Curr. Mol. Med. 2016, 16, 533–544. [Google Scholar] [CrossRef]
- Sano, R.; Reed, J.C. ER stress-induced cell death mechanisms. Biochim. Biophys. Acta 2013, 1833, 3460–3470. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Chiang, W.C.; Kurt, T.D.; Sigurdson, C.J.; Lin, J.H. Multiple Mechanisms of Unfolded Protein Response-Induced Cell Death. Am. J. Pathol. 2015, 185, 1800–1808. [Google Scholar] [CrossRef]
- Mimura, N.; Fulciniti, M.; Gorgun, G.; Tai, Y.T.; Cirstea, D.; Santo, L.; Hu, Y.; Fabre, C.; Minami, J.; Ohguchi, H.; et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood 2012, 119, 5772–5781. [Google Scholar] [CrossRef]
- Takayanagi, S.; Fukuda, R.; Takeuchi, Y.; Tsukada, S.; Yoshida, K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones 2013, 18, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Ye, J.; Rawson, R.B.; Komuro, R.; Chen, X.; Dave, U.P.; Prywes, R.; Brown, M.S.; Goldstein, J.L. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 2000, 6, 1355–1364. [Google Scholar] [CrossRef]
- Shen, J.; Prywes, R. Dependence of site-2 protease cleavage of ATF6 on prior site-1 protease digestion is determined by the size of the luminal domain of ATF6. J. Biol. Chem. 2004, 279, 43046–43051. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Lee, S.A.; Prasain, N.; Bae, D.; Kang, H.; Ha, T.; Kim, J.S.; Hong, K.S.; Mantel, C.; Moon, S.H.; et al. Metabolome Profiling of Partial and Fully Reprogrammed Induced Pluripotent Stem Cells. Stem Cells Dev. 2017, 26, 734–742. [Google Scholar] [CrossRef]
- Cao, S.S.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress in cell fate decision and human disease. Antioxid. Redox Signal. 2014, 21, 396–413. [Google Scholar] [CrossRef]
- Marei, W.F.A.; Van Raemdonck, G.; Baggerman, G.; Bols, P.E.J.; Leroy, J. Proteomic changes in oocytes after in vitro maturation in lipotoxic conditions are different from those in cumulus cells. Sci. Rep. 2019, 9, 3673. [Google Scholar] [CrossRef] [PubMed]
- Rogers, T.B.; Inesi, G.; Wade, R.; Lederer, W.J. Use of thapsigargin to study Ca2+ homeostasis in cardiac cells. Biosci. Rep. 1995, 15, 341–349. [Google Scholar] [CrossRef]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics (Oxf. Engl.) 2009, 25, 1105–1111. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics (Oxf. Engl.) 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Murtagh, F.; Legendre, P. Ward’s Hierarchical Agglomerative Clustering Method: Which Algorithms Implement Ward’s Criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Cui, L.H.; Joo, H.J.; Kim, D.H.; Seo, H.R.; Kim, J.S.; Choi, S.C.; Huang, L.H.; Na, J.E.; Lim, I.R.; Kim, J.H.; et al. Manipulation of the response of human endothelial colony-forming cells by focal adhesion assembly using gradient nanopattern plates. Acta Biomater. 2018, 65, 272–282. [Google Scholar] [CrossRef]
- Kim, J.J.; Khalid, O.; Namazi, A.; Tu, T.G.; Elie, O.; Lee, C.; Kim, Y. Discovery of consensus gene signature and intermodular connectivity defining self-renewal of human embryonic stem cells. Stem Cells (Dayt. Ohio) 2014, 32, 1468–1479. [Google Scholar] [CrossRef]
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Barkho, B.Z.; Ruiz, S.; Diep, D.; Qu, J.; Yang, S.L.; Panopoulos, A.D.; Suzuki, K.; Kurian, L.; Walsh, C.; et al. Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 2011, 472, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Booth, H.D.E.; Wessely, F.; Connor-Robson, N.; Rinaldi, F.; Vowles, J.; Browne, C.; Evetts, S.G.; Hu, M.T.; Cowley, S.A.; Webber, C.; et al. RNA sequencing reveals MMP2 and TGFB1 downregulation in LRRK2 G2019S Parkinson’s iPSC-derived astrocytes. Neurobiol. Dis. 2019, 129, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.; Yeganeh, B.; Stelmack, G.L.; Kashani, H.H.; Sharma, P.; Cunnington, R.; Rattan, S.; Bathe, K.; Klonisch, T.; Dixon, I.M.; et al. Apoptosis, autophagy and ER stress in mevalonate cascade inhibition-induced cell death of human atrial fibroblasts. Cell Death Dis. 2012, 3, e330. [Google Scholar] [CrossRef] [PubMed]
- Nishitoh, H. CHOP is a multifunctional transcription factor in the ER stress response. J. Biochem. 2012, 151, 217–219. [Google Scholar] [CrossRef]
- Liu, H.; Bowes, R.C., 3rd; van de Water, B.; Sillence, C.; Nagelkerke, J.F.; Stevens, J.L. Endoplasmic reticulum chaperones GRP78 and calreticulin prevent oxidative stress, Ca2+ disturbances, and cell death in renal epithelial cells. J. Biol. Chem. 1997, 272, 21751–21759. [Google Scholar] [CrossRef] [PubMed]
- Little, E.; Lee, A.S. Generation of a mammalian cell line deficient in glucose-regulated protein stress induction through targeted ribozyme driven by a stress-inducible promoter. J. Biol. Chem. 1995, 270, 9526–9534. [Google Scholar] [CrossRef]
- Morris, J.A.; Dorner, A.J.; Edwards, C.A.; Hendershot, L.M.; Kaufman, R.J. Immunoglobulin binding protein (BiP) function is required to protect cells from endoplasmic reticulum stress but is not required for the secretion of selective proteins. J. Biol. Chem. 1997, 272, 4327–4334. [Google Scholar] [CrossRef]
- Skottman, H.; Stromberg, A.M.; Matilainen, E.; Inzunza, J.; Hovatta, O.; Lahesmaa, R. Unique gene expression signature by human embryonic stem cells cultured under serum-free conditions correlates with their enhanced and prolonged growth in an undifferentiated stage. Stem Cells (Dayt. Ohio) 2006, 24, 151–167. [Google Scholar] [CrossRef]
- Kashyap, V.; Rezende, N.C.; Scotland, K.B.; Shaffer, S.M.; Persson, J.L.; Gudas, L.J.; Mongan, N.P. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009, 18, 1093–1108. [Google Scholar] [CrossRef] [PubMed]
- Filion, T.M.; Qiao, M.; Ghule, P.N.; Mandeville, M.; van Wijnen, A.J.; Stein, J.L.; Lian, J.B.; Altieri, D.C.; Stein, G.S. Survival responses of human embryonic stem cells to DNA damage. J. Cell. Physiol. 2009, 220, 586–592. [Google Scholar] [CrossRef]
- Maynard, S.; Swistowska, A.M.; Lee, J.W.; Liu, Y.; Liu, S.T.; Da Cruz, A.B.; Rao, M.; de Souza-Pinto, N.C.; Zeng, X.; Bohr, V.A. Human embryonic stem cells have enhanced repair of multiple forms of DNA damage. Stem Cells (Dayt. Ohio) 2008, 26, 2266–2274. [Google Scholar] [CrossRef]
- Bottone, M.G.; Santin, G.; Aredia, F.; Bernocchi, G.; Pellicciari, C.; Scovassi, A.I. Morphological Features of Organelles during Apoptosis: An Overview. Cells 2013, 2, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Denechaud, P.D.; Lopez-Mejia, I.C.; Giralt, A.; Lai, Q.; Blanchet, E.; Delacuisine, B.; Nicolay, B.N.; Dyson, N.J.; Bonner, C.; Pattou, F.; et al. E2F1 mediates sustained lipogenesis and contributes to hepatic steatosis. J. Clin. Investig. 2016, 126, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Teodoro-Morrison, T.; Schuiki, I.; Zhang, L.; Belsham, D.D.; Volchuk, A. GRP78 overproduction in pancreatic beta cells protects against high-fat-diet-induced diabetes in mice. Diabetologia 2013, 56, 1057–1067. [Google Scholar] [CrossRef]
- Wang, X.; Bi, X.; Zhang, G.; Deng, Y.; Luo, X.; Xu, L.; Scherer, P.E.; Ferdous, A.; Fu, G.; Gillette, T.G.; et al. Glucose-regulated protein 78 is essential for cardiac myocyte survival. Cell Death Differ. 2018, 25, 2181–2194. [Google Scholar] [CrossRef]
- Suzuki, T.; Lu, J.; Zahed, M.; Kita, K.; Suzuki, N. Reduction of GRP78 expression with siRNA activates unfolded protein response leading to apoptosis in HeLa cells. Arch. Biochem. Biophys. 2007, 468, 1–14. [Google Scholar] [CrossRef]
- So, J.S. Roles of Endoplasmic Reticulum Stress in Immune Responses. Mol. Cells 2018, 41, 705–716. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, T.W.; Jeong, J.H.; Shin, H.; Kim, H.K.; Im, J.S.; Song, B.H.; Hanna, J.; Oh, J.S.; Woo, D.-H.; Han, J.; et al. Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress. Cells 2020, 9, 1078. https://doi.org/10.3390/cells9051078
Ha TW, Jeong JH, Shin H, Kim HK, Im JS, Song BH, Hanna J, Oh JS, Woo D-H, Han J, et al. Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress. Cells. 2020; 9(5):1078. https://doi.org/10.3390/cells9051078
Chicago/Turabian StyleHa, Tae Won, Ji Hun Jeong, HyeonSeok Shin, Hyun Kyu Kim, Jeong Suk Im, Byung Hoo Song, Jacob Hanna, Jae Sang Oh, Dong-Hun Woo, Jaeseok Han, and et al. 2020. "Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress" Cells 9, no. 5: 1078. https://doi.org/10.3390/cells9051078
APA StyleHa, T. W., Jeong, J. H., Shin, H., Kim, H. K., Im, J. S., Song, B. H., Hanna, J., Oh, J. S., Woo, D.-H., Han, J., & Lee, M. R. (2020). Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress. Cells, 9(5), 1078. https://doi.org/10.3390/cells9051078





