Dysregulated FAM215A Stimulates LAMP2 Expression to Confer Drug-Resistant and Malignant in Human Liver Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Human HCC Specimens and Cancer Cell Lines
2.2. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
2.3. Rapid Amplification of cDNA Ends (RACE)
2.4. Fractionation of Nuclear and Cytoplasmic RNA
2.5. Establishment of FAM215A-Overexpressing Cells
2.6. Establishment of FAM215A-Knockdown Cells
2.7. Establishment of LAMP2-Overexpressing and -Knockdown Cells
2.8. Wound-Healing Assay
2.9. Invasion and Migration Assay
2.10. Cell Proliferation Assay
2.11. Western Blot Analysis
2.12. Ubiquitination assay
2.13. Drug Resistance Assay
2.14. Lysosomal Content Assay
2.15. RNA Immunoprecipitation (RIP)
2.16. Mouse Xenograft Model
2.17. Statistical Analysis
3. Results
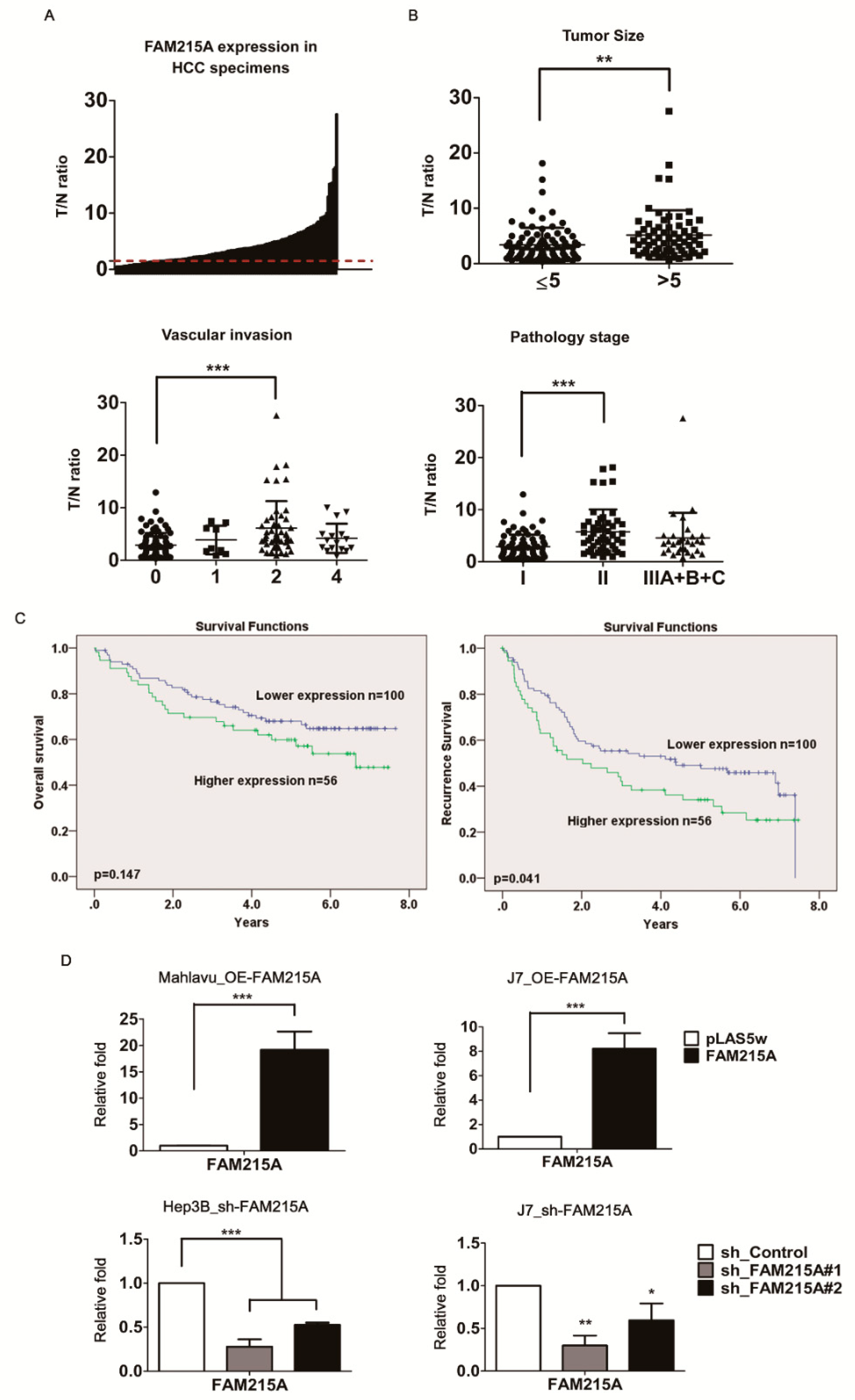
3.1. lncRNA FAM215A Is Up-Regulated in Hepatocellular Carcinoma and Is Associated with Tumor Progression and Metastasis
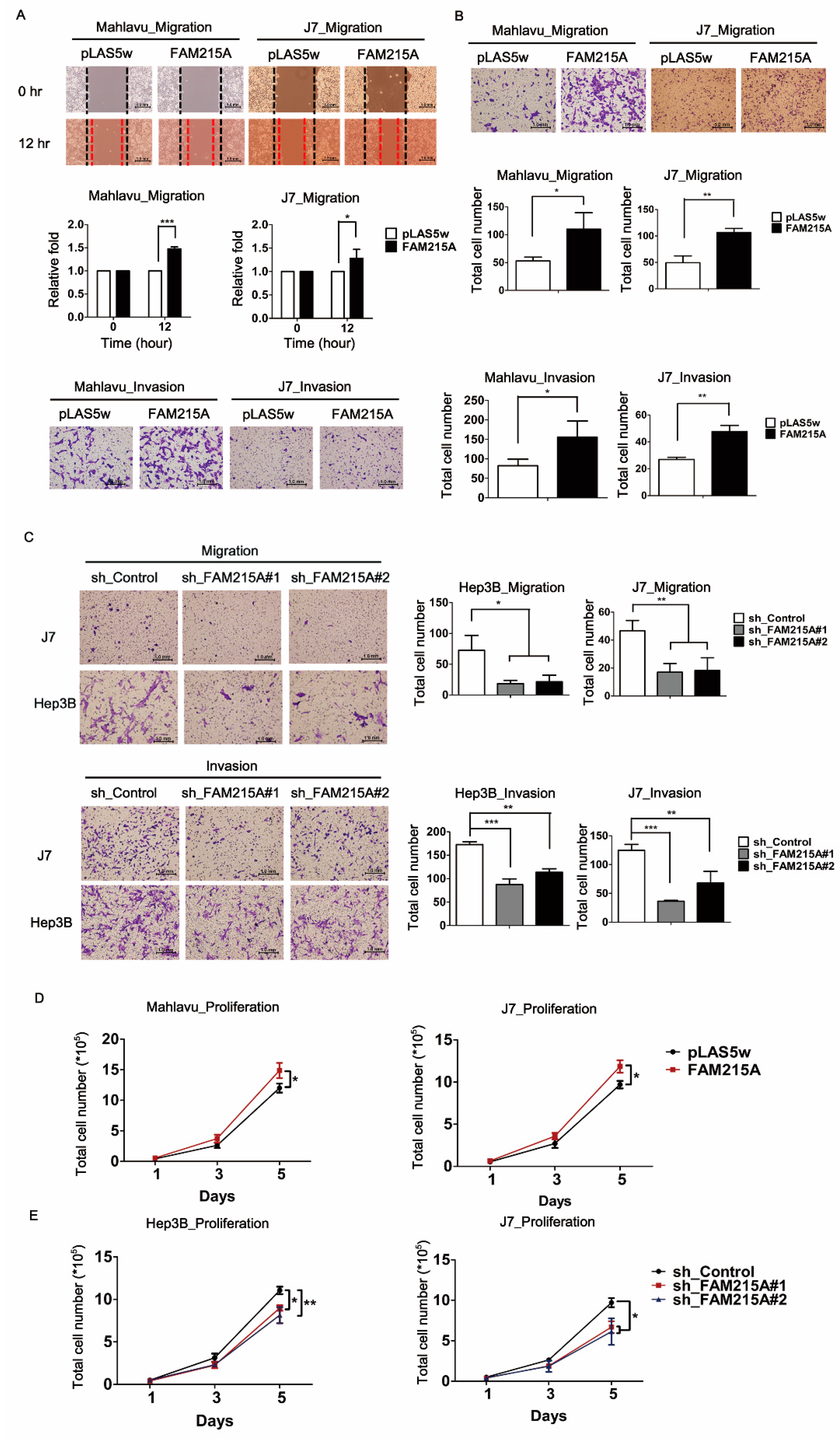
3.2. FAM215A Promotes HCC Cell Metastasis and Proliferation
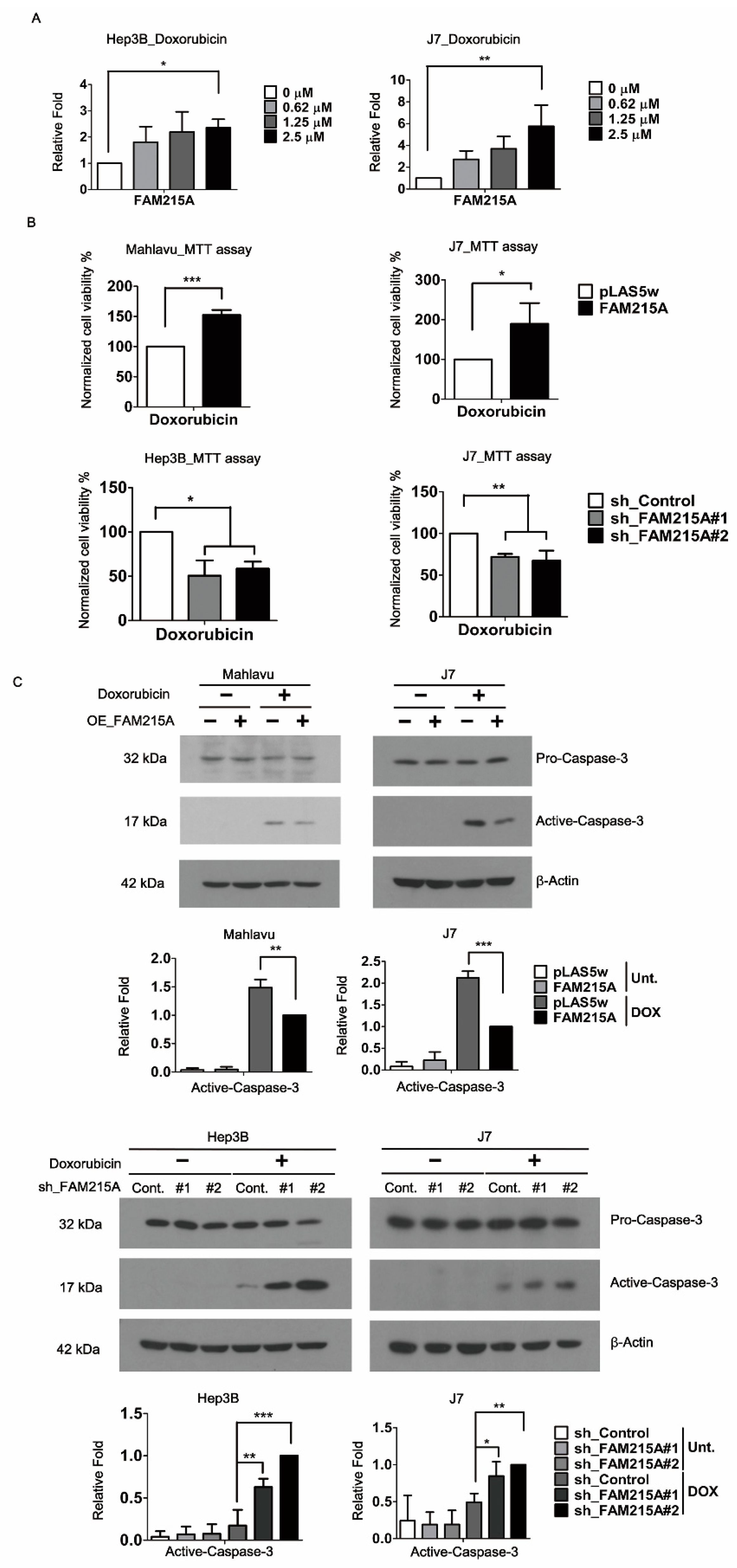
3.3. FAM215A Promotes Doxorubicin Resistance and Is Highly Expressed in Doxorubicin-Resistant HCC Cells
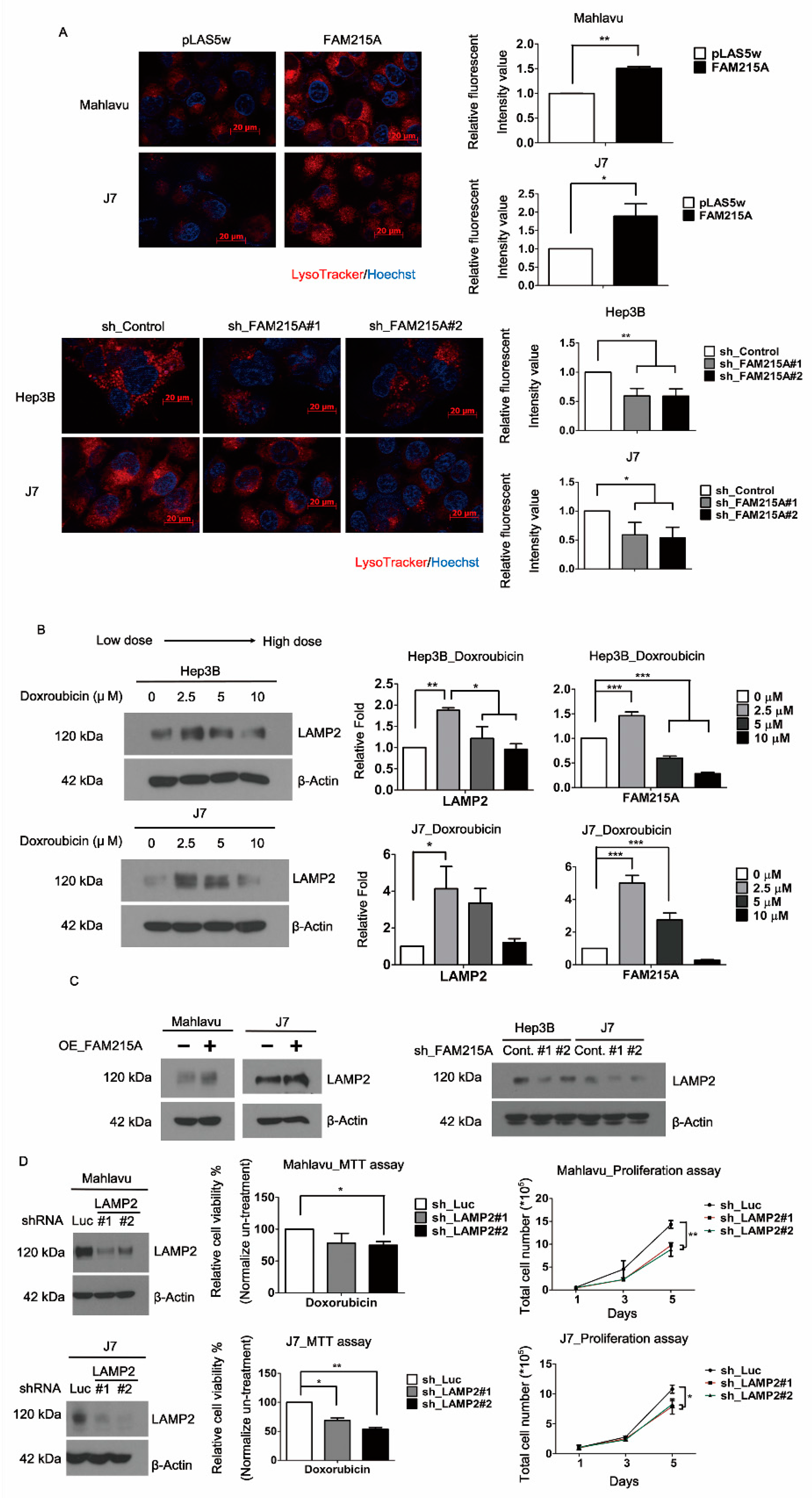
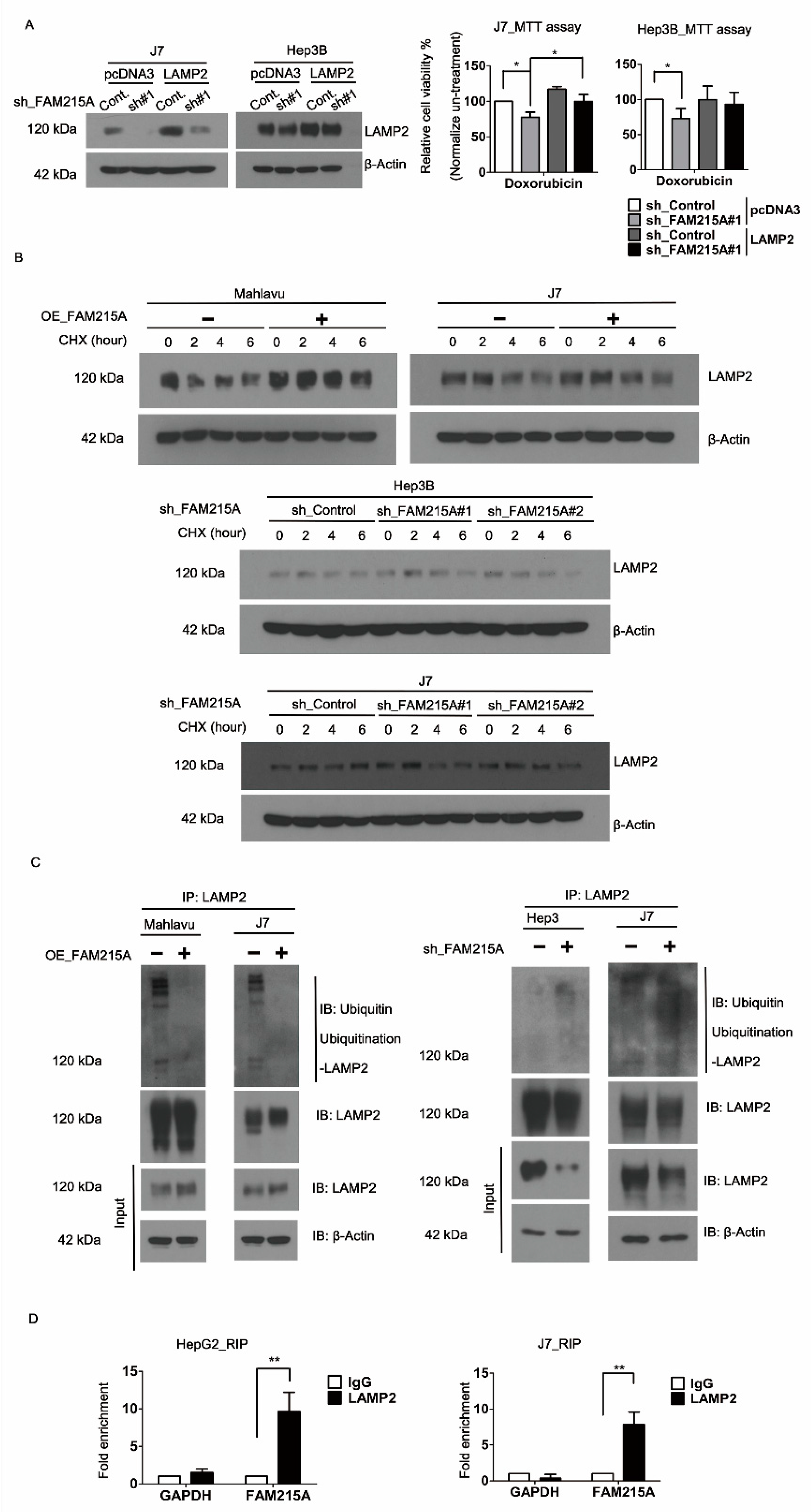
3.4. FAM215A Increases HCC Progression and Drug Resistance through LAMP2
3.5. FAM215A Interacts with and Stabilizes LAMP2
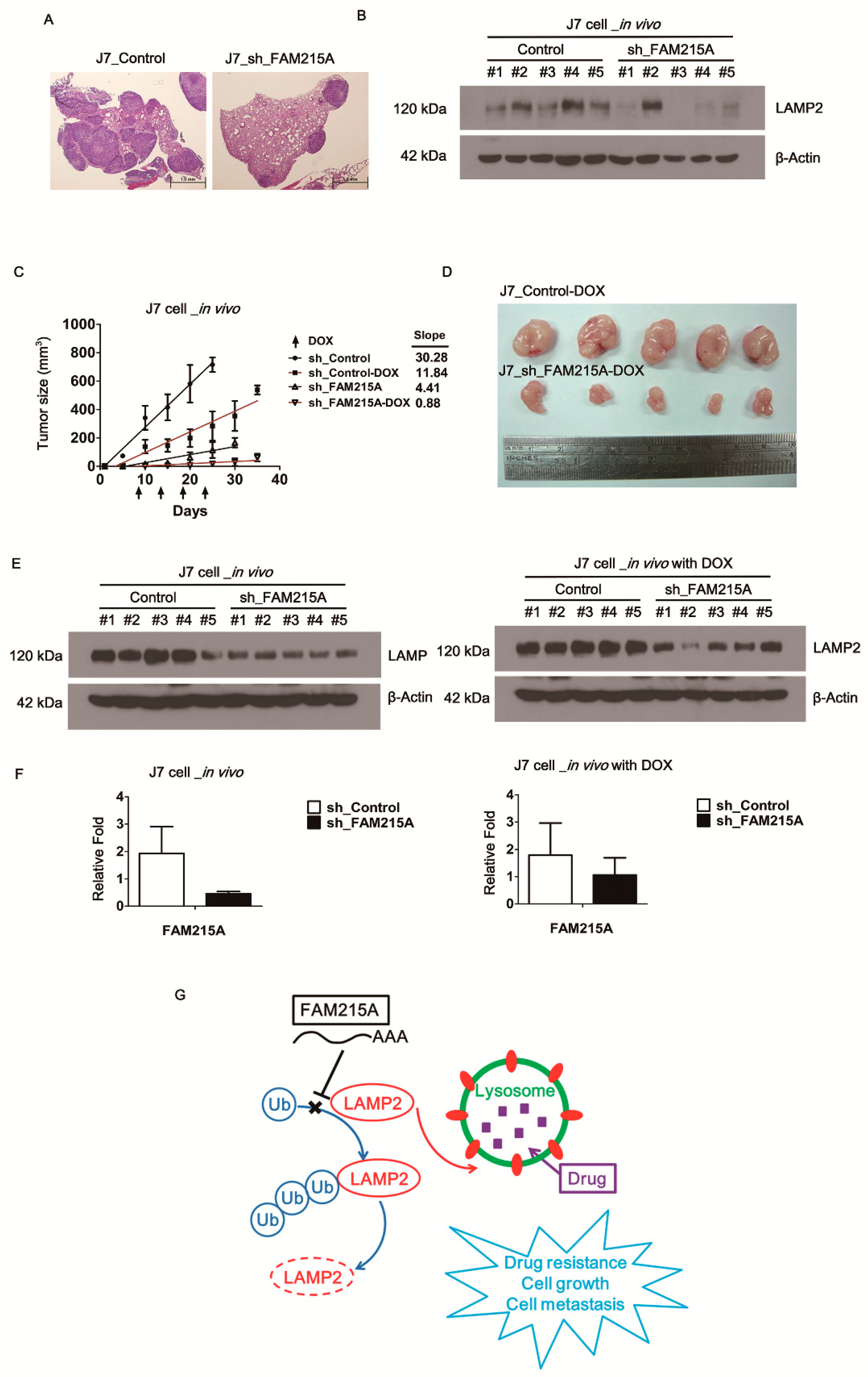
3.6. FAM215A Increases HCC Progression and Drug Resistance In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levrero, M. Viral hepatitis and liver cancer: The case of hepatitis C. Oncogene 2006, 25, 3834–3847. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Strathearn, L.S.; Stepanov, A.I.; Font-Burgada, J. Inflammation in Primary and Metastatic Liver Tumorigenesis-Under the Influence of Alcohol and High-Fat Diets. Nutrients 2020, 12, 933. [Google Scholar] [CrossRef] [PubMed]
- Charriere, B.; Maulat, C.; Suc, B.; Muscari, F. Contribution of alpha-fetoprotein in liver transplantation for hepatocellular carcinoma. hepatology 2016, 8, 881–890. [Google Scholar] [CrossRef]
- Wu, S.M.; Lin, S.L.; Lee, K.Y.; Chuang, H.C.; Feng, P.H.; Cheng, W.L.; Liao, C.J.; Chi, H.C.; Lin, Y.H.; Tsai, C.Y.; et al. Hepatoma cell functions modulated by NEK2 are associated with liver cancer progression. Cancer 2017, 140, 1581–1596. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. cell 2011, 43, 904–914. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nature reviews. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef]
- He, Y.; Meng, X.M.; Huang, C.; Wu, B.M.; Zhang, L.; Lv, X.W.; Li, J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014, 344, 20–27. [Google Scholar] [CrossRef]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhou, L.; Wu, L.M.; Lai, M.C.; Xie, H.Y.; Zhang, F.; Zheng, S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol. 2011, 18, 1243–1250. [Google Scholar] [CrossRef] [PubMed]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011, 71, 6320–6326. [Google Scholar] [CrossRef] [PubMed]
- Ghidini, M.; Braconi, C. Non-Coding RNAs in Primary Liver Cancer. Front. Med. 2015, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Kallunki, T.; Olsen, O.D.; Jaattela, M. Cancer-associated lysosomal changes: Friends or foes? Oncogene 2013, 32, 1995–2004. [Google Scholar] [CrossRef] [PubMed]
- Alessandrini, F.; Pezze, L.; Ciribilli, Y. LAMPs: Shedding light on cancer biology. Semin. Oncol. 2017, 44, 239–253. [Google Scholar] [CrossRef]
- Krishnan, V.; Bane, S.M.; Kawle, P.D.; Naresh, K.N.; Kalraiya, R.D. Altered melanoma cell surface glycosylation mediates organ specific adhesion and metastasis via lectin receptors on the lung vascular endothelium. Clin. Exp. Meta. 2005, 22, 11–24. [Google Scholar] [CrossRef]
- Saitoh, O.; Wang, W.C.; Lotan, R.; Fukuda, M. Differential glycosylation and cell surface expression of lysosomal membrane glycoproteins in sublines of a human colon cancer exhibiting distinct metastatic potentials. Biolo. Chem. 1992, 267, 5700–5711. [Google Scholar]
- Ding, Z.B.; Fu, X.T.; Shi, Y.H.; Zhou, J.; Peng, Y.F.; Liu, W.R.; Shi, G.M.; Gao, Q.; Wang, X.Y.; Song, K.; et al. Lamp2a is required for tumor growth and promotes tumor recurrence of hepatocellular carcinoma. Oncology 2016, 49, 2367–2376. [Google Scholar] [CrossRef]
- Huang, P.S.; Chung, I.H.; Lin, Y.H.; Lin, T.K.; Chen, W.J.; Lin, K.H. The Long Non-Coding RNA MIR503HG Enhances Proliferation of Human ALK-Negative Anaplastic Large-Cell Lymphoma. Mol. Sci. 2018, 19, 1463. [Google Scholar] [CrossRef]
- Lin, Y.H.; Wu, M.H.; Huang, Y.H.; Yeh, C.T.; Cheng, M.L.; Chi, H.C.; Tsai, C.Y.; Chung, I.H.; Chen, C.Y.; Lin, K.H. Taurine up-regulated gene 1 functions as a master regulator to coordinate glycolysis and metastasis in hepatocellular carcinoma. Hepatology 2018, 67, 188–203. [Google Scholar] [CrossRef]
- Lagarde, J.; Uszczynska-Ratajczak, B.; Santoyo-Lopez, J.; Gonzalez, J.M.; Tapanari, E.; Mudge, J.M.; Steward, C.A.; Wilming, L.; Tanzer, A.; Howald, C.; et al. Extension of human lncRNA transcripts by RACE coupled with long-read high-throughput sequencing (RACE-Seq). Nat. Commun. 2016, 7, 12339. [Google Scholar] [CrossRef]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. Clin. Invest. 2009, 119, 1429–1437. [Google Scholar] [CrossRef]
- Garg, M. Epithelial-mesenchymal transition activating transcription factors multifunctional regulators in cancer. Stem cells 2013, 5, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, W.Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. Recep. Signal Trans. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, J.; Wu, T.; Xu, X.; Cao, G.; Li, H.; Chen, X. Histone deacetylase 2 regulates the doxorubicin (Dox) resistance of hepatocarcinoma cells and transcription of ABCB1. Life Sci. 2019, 216, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.; Lo, C.M.; Guo, D.Y.; Qi, X.; Li, C.X.; Geng, W.; Liu, X.B.; Ling, C.C.; Ma, Y.Y.; Yeung, W.H.; et al. Identification of transmembrane protein 98 as a novel chemoresistance-conferring gene in hepatocellular carcinoma. Mol. Cancer Thera. 2014, 13, 1285–1297. [Google Scholar] [CrossRef]
- Ueno, M.; Kakinuma, Y.; Yuhki, K.; Murakoshi, N.; Iemitsu, M.; Miyauchi, T.; Yamaguchi, I. Doxorubicin induces apoptosis by activation of caspase-3 in cultured cardiomyocytes in vitro and rat cardiac ventricles in vivo. Pharmacol. Sci. 2006, 101, 151–158. [Google Scholar] [CrossRef]
- Sheng, J.; Qin, H.; Zhang, K.; Li, B.; Zhang, X. Targeting autophagy in chemotherapy-resistant of hepatocellular carcinoma. Cancer Res. 2018, 8, 354–365. [Google Scholar]
- Piao, S.; Amaravadi, R.K. Targeting the lysosome in cancer. Sciences 2016, 1371, 45–54. [Google Scholar] [CrossRef]
- Eskelinen, E.L.; Illert, A.L.; Tanaka, Y.; Schwarzmann, G.; Blanz, J.; Von Figura, K.; Saftig, P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol. Biol. Cell 2002, 13, 3355–3368. [Google Scholar] [CrossRef]
- Yan, K.; Fu, Y.; Zhu, N.; Wang, Z.; Hong, J.L.; Li, Y.; Li, W.J.; Zhang, H.B.; Song, J.H. Repression of lncRNA NEAT1 enhances the antitumor activity of CD8(+)T cells against hepatocellular carcinoma via regulating miR-155/Tim-3. Biochem. Cell Biol. 2019, 110, 1–8. [Google Scholar] [CrossRef]
- Chen, F.; Li, Y.; Li, M.; Wang, L. Long noncoding RNA GAS5 inhibits metastasis by targeting miR-182/ANGPTL1 in hepatocellular carcinoma. Cancer Res. 2019, 9, 108–121. [Google Scholar]
- Fu, Y.; Biglia, N.; Wang, Z.; Shen, Y.; Risch, H.A.; Lu, L.; Canuto, E.M.; Jia, W.; Katsaros, D.; Yu, H. Long non-coding RNAs, ASAP1-IT1, FAM215A, and LINC00472, in epithelial ovarian cancer. Gynecol. Oncol. 2016, 143, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Dis. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- van Zijl, F.; Zulehner, G.; Petz, M.; Schneller, D.; Kornauth, C.; Hau, M.; Machat, G.; Grubinger, M.; Huber, H.; Mikulits, W. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol. 2009, 5, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Han, L.L.; Yin, X.R.; Zhang, S.Q. miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. oncology 2018, 53, 2433–2444. [Google Scholar] [CrossRef]
- Bae, Y.K.; Choi, J.E.; Kang, S.H.; Lee, S.J. Epithelial-Mesenchymal Transition Phenotype Is Associated with Clinicopathological Factors That Indicate Aggressive Biological Behavior and Poor Clinical Outcomes in Invasive Breast Cancer. breast cancer 2015, 18, 256–263. [Google Scholar] [CrossRef]
- Guegan, J.P.; Fremin, C.; Baffet, G. The MAPK MEK1/2-ERK1/2 Pathway and Its Implication in Hepatocyte Cell Cycle Control. Int. J. Hepatol. 2012, 2012, 328372. [Google Scholar] [CrossRef]
- Moreno, M.; Lombardi, A.; Silvestri, E.; Senese, R.; Cioffi, F.; Goglia, F.; Lanni, A.; de Lange, P. PPARs: Nuclear Receptors Controlled by, and Controlling, Nutrient Handling through Nuclear and Cytosolic Signaling. PPAR research 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Valenzuela, R.; Videla, L.A. Impact of the Co-Administration of N-3 Fatty Acids and Olive Oil Components in Preclinical Nonalcoholic Fatty Liver Disease Models: A Mechanistic View. Nutrients 2020, 12, 499. [Google Scholar] [CrossRef] [PubMed]
- Corona, J.C.; Duchen, M.R. PPARgamma as a therapeutic target to rescue mitochondrial function in neurological disease. Free Radic. Biol. Med. 2016, 100, 153–163. [Google Scholar] [CrossRef]
- Ye, P.; Xing, H.; Lou, F.; Wang, K.; Pan, Q.; Zhou, X.; Gong, L.; Li, D. Histone deacetylase 2 regulates doxorubicin (Dox) sensitivity of colorectal cancer cells by targeting ABCB1 transcription. Cancer Chemother. Pharmacol. 2016, 77, 613–621. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogen. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, Q.; Qi, H.; Wang, C.; Wang, C.; Zhang, J.; Dong, L. Doxorubicin-Induced Systemic Inflammation Is Driven by Upregulation of Toll-Like Receptor TLR4 and Endotoxin Leakage. Cancer Res. 2016, 76, 6631–6642. [Google Scholar] [CrossRef] [PubMed]
- Fallahi-Sichani, M.; Becker, V.; Izar, B.; Baker, G.J.; Lin, J.R.; Boswell, S.A.; Shah, P.; Rotem, A.; Garraway, L.A.; Sorger, P.K. Adaptive resistance of melanoma cells to RAF inhibition via reversible induction of a slowly dividing de-differentiated state. Mol. Syst. Biol. 2017, 13, 905. [Google Scholar] [CrossRef]
- Wang, H.; Fang, L.; Jiang, J.; Kuang, Y.; Wang, B.; Shang, X.; Han, P.; Li, Y.; Liu, M.; Zhang, Z.; et al. The cisplatin-induced lncRNA PANDAR dictates the chemoresistance of ovarian cancer via regulating SFRS2-mediated p53 phosphorylation. Cell Death Dis. 2018, 9, 1103. [Google Scholar] [CrossRef]
- Gavini, J.; Dommann, N.; Jakob, M.O.; Keogh, A.; Bouchez, L.C.; Karkampouna, S.; Julio, M.K.; Medova, M.; Zimmer, Y.; Schlafli, A.M.; et al. Verteporfin-induced lysosomal compartment dysregulation potentiates the effect of sorafenib in hepatocellular carcinoma. Cell Death Dis. 2019, 10, 749. [Google Scholar] [CrossRef]
- Towers, C.G.; Thorburn, A. Targeting the Lysosome for Cancer Therapy. Cancer Dis. 2017, 7, 1218–1220. [Google Scholar] [CrossRef]
- Colombo, F.; Trombetta, E.; Cetrangolo, P.; Maggioni, M.; Razini, P.; De Santis, F.; Torrente, Y.; Prati, D.; Torresani, E.; Porretti, L. Giant Lysosomes as a Chemotherapy Resistance Mechanism in Hepatocellular Carcinoma Cells. PLoS ONE 2014, 9, e114787. [Google Scholar] [CrossRef][Green Version]
- Fennelly, C.; Amaravadi, R.K. Lysosomal Biology in Cancer. Methods Mol. Biol. 2017, 1594, 293–308. [Google Scholar]
- Eskelinen, E.L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Molecular Aspects Med. 2006, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Dubois, A.; Furstoss, N.; Calleja, A.; Zerhouni, M.; Cluzeau, T.; Savy, C.; Marchetti, S.; Hamouda, M.A.; Boulakirba, S.; Orange, F.; et al. LAMP2 expression dictates azacytidine response and prognosis in MDS/AML. Leukemia 2019, 33, 1501–1513. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Zhang, M.; Zhang, S.; Wang, J.; Zhou, X.; Guo, G.; Wang, L.; Wang, M.; Peng, Z.; Guo, C.; et al. Characterisation of Lamp2-deficient rats for potential new animal model of Danon disease. Sci. Rep. 2018, 8, 6932. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Yasuda, S.; Fujita, T.; Hamasaki, M.; Murakami, A.; Kawawaki, J.; Iwai, K.; Saeki, Y.; Yoshimori, T.; Matsuda, N.; et al. Ubiquitination of exposed glycoproteins by SCF(FBXO27) directs damaged lysosomes for autophagy. Proc. Acad. Sci. USA 2017, 114, 8574–8579. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yang, L.; Li, X.; Li, B.; Li, Y.; Zhang, X.; Ma, Y.; Peng, X.; Jin, H.; Li, H. New insights into autophagy in hepatocellular carcinoma: Mechanisms and therapeutic strategies. Cancer Res. 2019, 9, 1329–1353. [Google Scholar]
- Sun, T.; Liu, H.; Ming, L. Multiple Roles of Autophagy in the Sorafenib Resistance of Hepatocellular Carcinoma. Cell. Physiol. Biochem.: Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 44, 716–727. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, P.-S.; Lin, Y.-H.; Chi, H.-C.; Tseng, Y.-H.; Chen, C.Y.; Lin, T.-K.; Yeh, C.-T.; Lin, K.-H. Dysregulated FAM215A Stimulates LAMP2 Expression to Confer Drug-Resistant and Malignant in Human Liver Cancer. Cells 2020, 9, 961. https://doi.org/10.3390/cells9040961
Huang P-S, Lin Y-H, Chi H-C, Tseng Y-H, Chen CY, Lin T-K, Yeh C-T, Lin K-H. Dysregulated FAM215A Stimulates LAMP2 Expression to Confer Drug-Resistant and Malignant in Human Liver Cancer. Cells. 2020; 9(4):961. https://doi.org/10.3390/cells9040961
Chicago/Turabian StyleHuang, Po-Shuan, Yang-Hsiang Lin, Hsiang-Cheng Chi, Yi-Hsin Tseng, Cheng Yi Chen, Tzu-Kang Lin, Chau-Ting Yeh, and Kwang-Huei Lin. 2020. "Dysregulated FAM215A Stimulates LAMP2 Expression to Confer Drug-Resistant and Malignant in Human Liver Cancer" Cells 9, no. 4: 961. https://doi.org/10.3390/cells9040961
APA StyleHuang, P.-S., Lin, Y.-H., Chi, H.-C., Tseng, Y.-H., Chen, C. Y., Lin, T.-K., Yeh, C.-T., & Lin, K.-H. (2020). Dysregulated FAM215A Stimulates LAMP2 Expression to Confer Drug-Resistant and Malignant in Human Liver Cancer. Cells, 9(4), 961. https://doi.org/10.3390/cells9040961




