High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
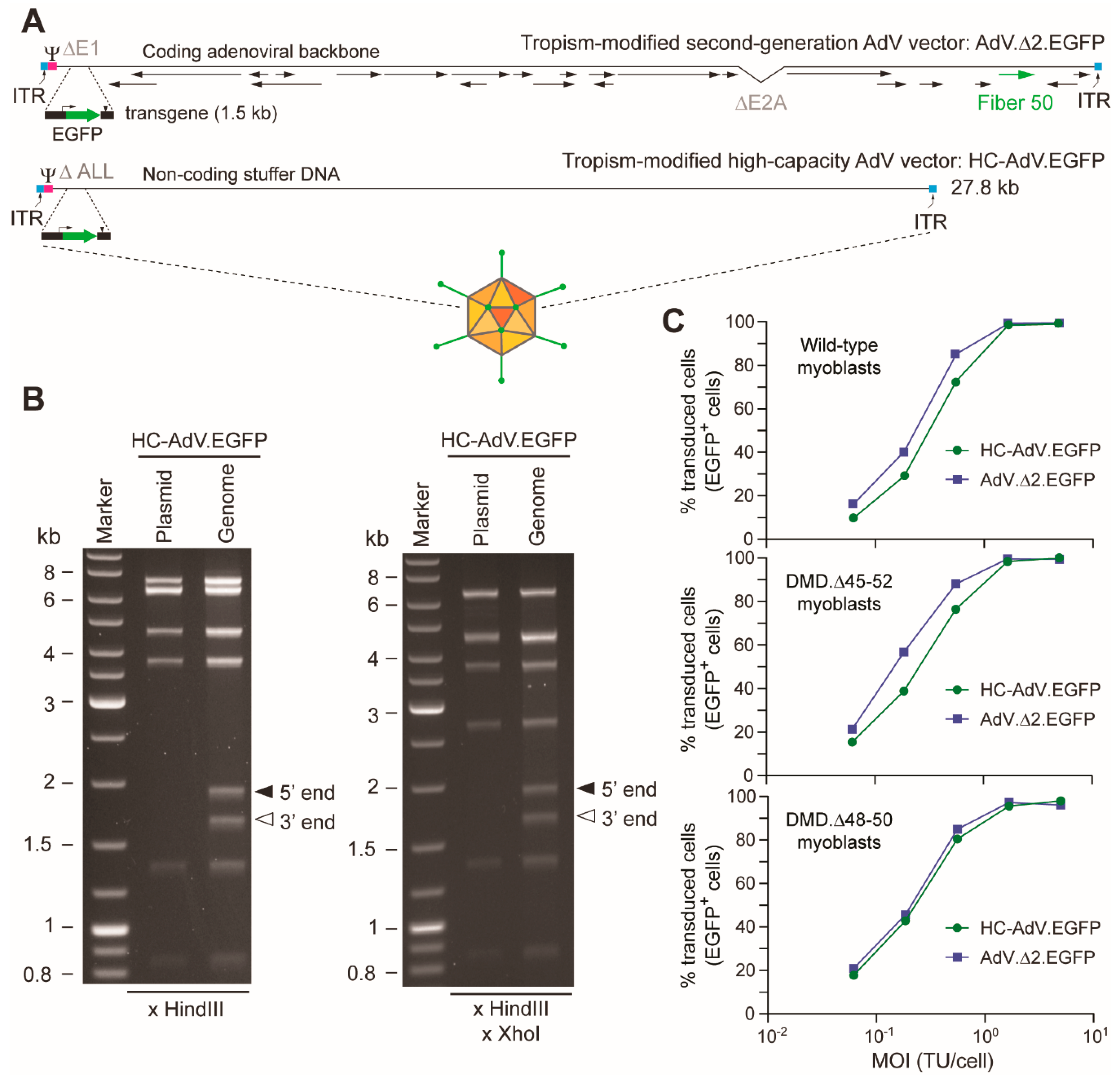
2.2. Production and Characterization of Adenoviral Vectors
2.3. Transduction Experiments
2.4. Cell Viability Assays
2.5. Live-Cell Light Microscopy
2.6. Flow Cytometry
2.7. qPCR Assays
2.8. Immunofluorescence Microscopy Analyses
2.9. Western Blot Analysis
2.10. Statistical Analyses
3. Results
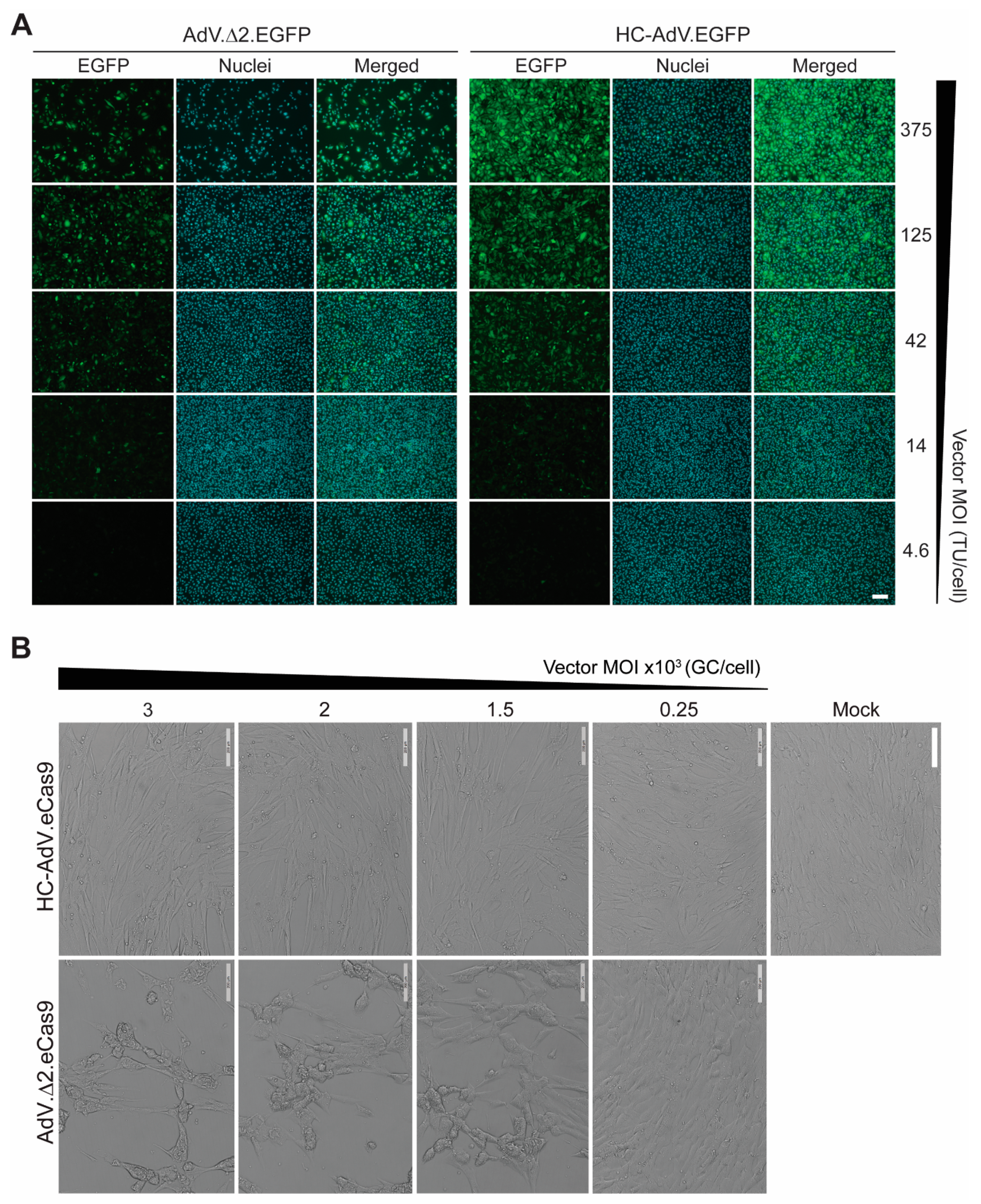
3.1. At High Doses HC-AdVs Are Significantly Less Cytotoxic Than Second-Generation AdVs
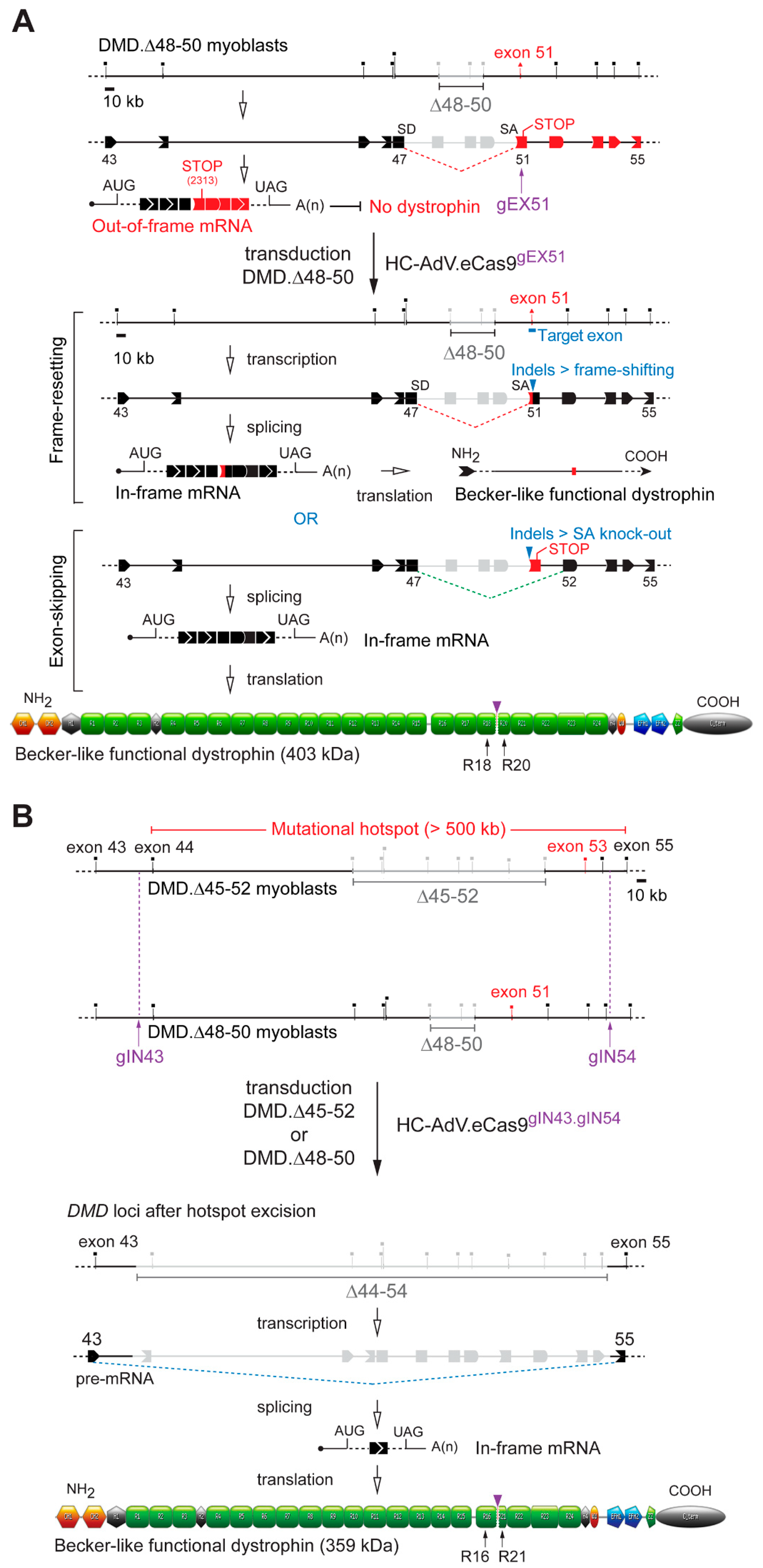
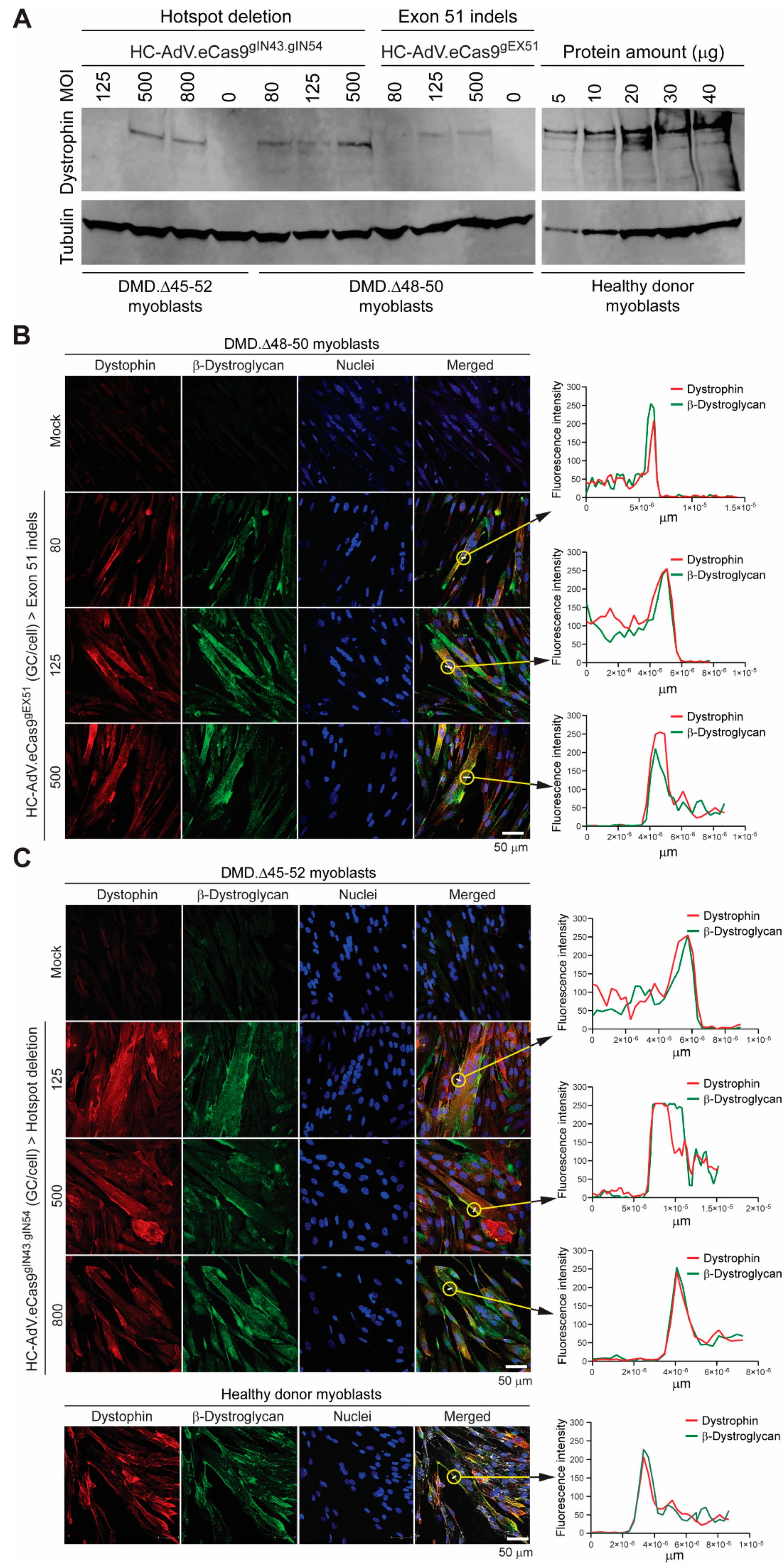
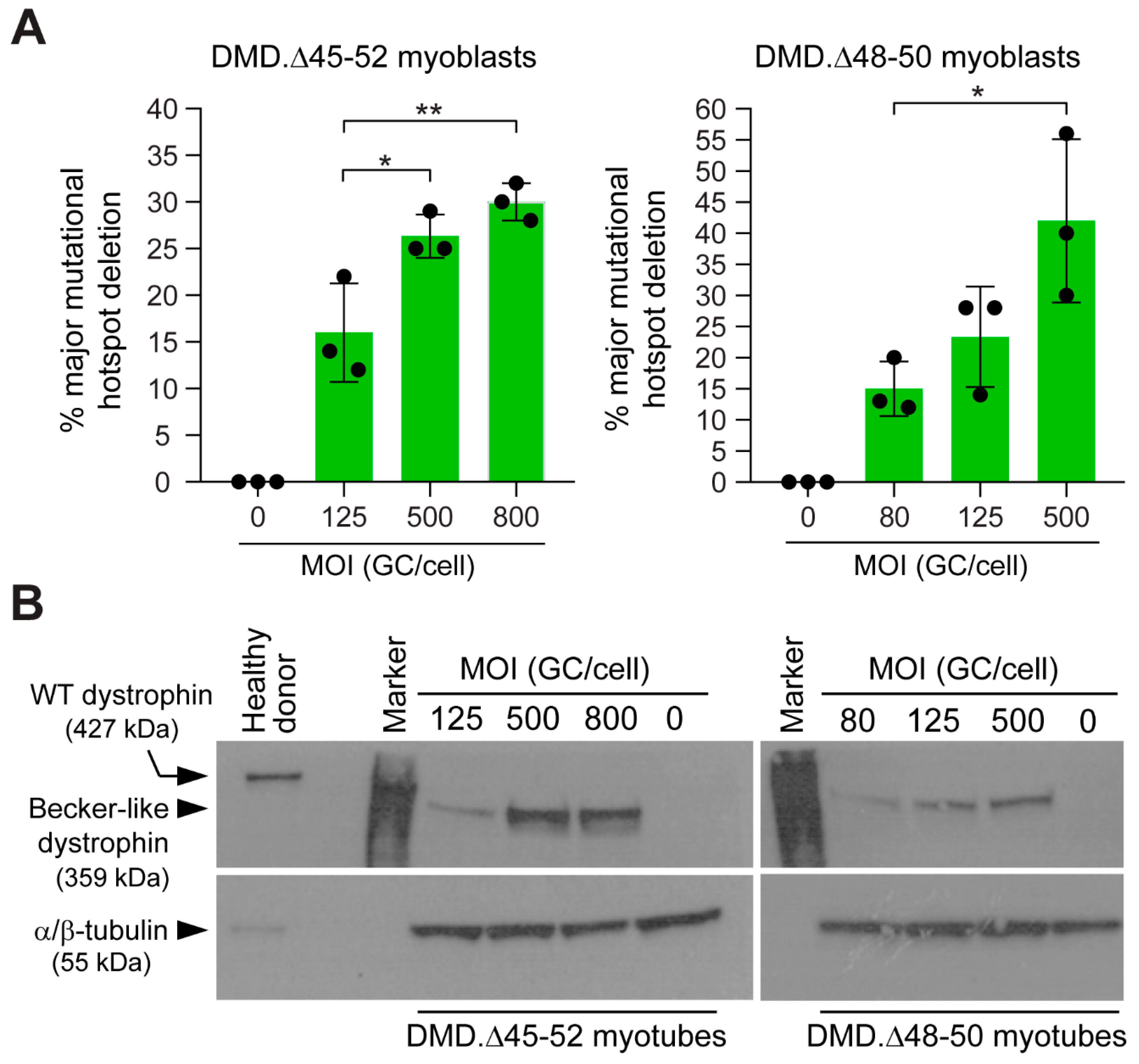
3.2. HC-AdVs Encoding Single or Dual RGNs Induce Robust Dystrophin Synthesis in Unselected DMD Muscle Cell Populations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mah, J.K.; Korngut, L.; Fiest, K.M.; Dykeman, J.; Day, L.J.; Pringsheim, T.; Jette, N. A systematic review and meta-analysis on the epidemiology of the muscular dystrophies. Can. J. Neurol. Sci. 2016, 43, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, E.P.; Brown Jr, R.H.; Kunkel, L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell 1987, 51, 919–928. [Google Scholar] [CrossRef]
- Hoffman, E.P.; Fischbeck, K.H.; Brown, R.H.; Johnson, M.; Medori, R.; Loire, J.D.; Harris, J.B.; Waterston, R.; Brooke, M.; Specht, L. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N. Engl. J. Med. 1988, 318, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Ahn, A.H.; Kunkel, L.M. The structural and functional diversity of dystrophin. Nat. Gen. 1993, 3, 283–291. [Google Scholar] [CrossRef]
- Guiraud, S.; Aartsma-Rus, A.; Vieira, N.M.; Davies, K.E.; van Ommen, G.-J.B.; Kunkel, L.M. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Gen. 2015, 16, 281–308. [Google Scholar] [CrossRef]
- Bladen, C.L.; Salgado, D.; Monges, S.; Foncuberta, M.E.; Kekou, K.; Kosma, K.; Dawkins, H.; Lamont, L.; Roy, A.J.; Chamova, T.; et al. The TREAT-NMD DMD Global Database: Analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015, 36, 395–402. [Google Scholar] [CrossRef]
- Koenig, M.; Beggs, A.H.; Moyer, M.; Scherpf, S.; Heindrich, K.; Bettecken, T.; Meng, G.; Müller, C.R.; Lindlöf, M.; Kaariainen, H.; et al. The molecular basis for Duchenne versus Becker muscular dystrophy: Correlation of severity with type of deletion. Am. J. Hum. Genet. 1989, 45, 498–506. [Google Scholar]
- Verhaart, I.E.; Aartsma-Rus, A. Therapeutic developments for Duchenne muscular dystrophy. Nat. Rev. Neurol. 2019, 15, 373–386. [Google Scholar] [CrossRef]
- Nelson, C.E.; Robinson-Hamm, J.N.; Gersbach, C.A. Genome engineering: A new approach to gene therapy for neuromuscular disorders. Nat. Rev. Neurol. 2017, 13, 647–661. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gonçalves, M.A. DNA, RNA and Protein Tools for Editing the Genetic Information in Human Cells. iScience 2018, 6, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Gonçalves, M.A. Genome editing at the crossroads of delivery, specificity, and fidelity. Trends Biotechnol. 2015, 33, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Ousterout, D.G.; Perez-Pinera, P.; Thakore, P.I.; Kabadi, A.M.; Brown, M.T.; Qin, X.; Fedrigo, O.; Mouly, V.; Tremblay, J.P.; Gersbach, C.A. Reading frame correction by targeted genome editing restores dystrophin expression in cells from Duchenne muscular dystrophy patients. Mol. Ther. 2013, 21, 1718–1726. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports 2015, 4, 143–154. [Google Scholar] [CrossRef]
- Ousterout, D.G.; Kabadi, A.M.; Thakore, P.I.; Majoros, W.H.; Reddy, T.E.; Gersbach, C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015, 6, 6244. [Google Scholar] [CrossRef]
- Ousterout, D.G.; Kabadi, A.M.; Thakore, P.I.; Perez-Pinera, P.; Brown, M.T.; Majoros, W.H.; Reddy, T.E.; Gersbach, C.A. Correction of dystrophin expression in cells from Duchenne muscular dystrophy patients through genomic excision of exon 51 by zinc finger nucleases. Mol. Ther. 2015, 23, 523–532. [Google Scholar] [CrossRef]
- Maggio, I.; Chen, X.; Gonçalves, M.A. The emerging role of viral vectors as vehicles for DMD gene editing. Genome Med. 2016, 8, 59. [Google Scholar] [CrossRef]
- Gonçalves, M.A.; de Vries, A.A. Adenovirus: From foe to friend. Rev. Medical Virol. 2006, 16, 167–186. [Google Scholar] [CrossRef]
- Gonçalves, M.A. Adeno-associated virus: From defective virus to effective vector. Virol. J. 2005, 2, 43. [Google Scholar] [CrossRef]
- Maggio, I.; Stefanucci, L.; Janssen, J.M.; Liu, J.; Chen, X.; Mouly, V.; Gonçalves, M.A. Selection-free gene repair after adenoviral vector transduction of designer nucleases: Rescue of dystrophin synthesis in DMD muscle cell populations. Nucleic Acids Res. 2016, 44, 1449–1470. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Liu, J.; Janssen, J.M.; Chen, X.; Gonçalves, M.A. Adenoviral vectors encoding CRISPR/Cas9 multiplexes rescue dystrophin synthesis in unselected populations of DMD muscle cells. Sci. Rep. 2016, 6, 37051. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Park, K.H.; Zhao, L.; Xu, J.; El Refaey, M.; Gao, Y.; Zhu, H.; Ma, J.; Han, R. CRISPR-mediated genome editing restores dystrophin expression and function in mdx mice. Mol. Ther. 2016, 24, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Long, C.; McAnally, J.R.; Shelton, J.M.; Mireault, A.A.; Bassel-Duby, R.; Olson, E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9–mediated editing of germline DNA. Science 2014, 345, 1184–1188. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Rivera, R.M.C.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Miller, D.G.; Rutledge, E.A.; Russell, D.W. Chromosomal effects of adeno-associated virus vector integration. Nature Genet. 2002, 30, 147–148. [Google Scholar] [CrossRef]
- Porteus, M.H.; Cathomen, T.; Weitzman, M.D.; Baltimore, D. Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol. Cell. Biol. 2003, 23, 3558–3565. [Google Scholar] [CrossRef]
- Miller, D.G.; Petek, L.M.; Russell, D.W. Adeno-associated virus vectors integrate at chromosome breakage sites. Nat. Genet. 2004, 36, 767–773. [Google Scholar] [CrossRef]
- Li, H.; Haurigot, V.; Doyon, Y.; Li, T.; Wong, S.Y.; Bhagwat, A.S.; Malani, N.; Anguela, X.M.; Sharma, R.; Ivanciu, L.; et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 2011, 475, 217–221. [Google Scholar] [CrossRef]
- Anguela, X.M.; Sharma, R.; Doyon, Y.; Miller, J.C.; Li, H.; Haurigot, V.; Rohde, M.E.; Wong, S.Y.; Davidson, R.J.; Zhou, S.; et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood 2013, 122, 3283–3287. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Rivera, R.M.C.; Collier, J.H.; et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, K.S.; Kleinstiver, B.P.; Garcia, S.P.; Zaborowski, M.P.; Volak, A.; Spirig, S.E.; Muller, A.; Sousa, A.A.; Tsai, S.Q.; Bengtsson, N.E.; et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun. 2019, 10, 4439. [Google Scholar] [CrossRef] [PubMed]
- Bergelson, J.M.; Cunningham, J.A.; Droguett, G.; Kurt-Jones, E.A.; Krithivas, A.; Hong, J.S.; Horwitz, M.S.; Crowell, R.L.; Finberg, R.W. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 1997, 275, 1320–1323. [Google Scholar] [CrossRef]
- Knaän-Shanzer, S.; van de Watering, M.J.M.; van der Velde, I.; Gonçalves, M.A.F.V.; Valerio, D.; de Vries, A.A.F. Endowing human adenovirus serotype 5 vectors with fiber domains of species B greatly enhances gene transfer into human mesenchymal stem cells. Stem Cells 2005, 23, 1598–1607. [Google Scholar] [CrossRef]
- Gonçalves, M.A.F.V.; de Vries, A.A.F.; Holkers, M.; van de Watering, M.J.M.; van der Velde, I.; van Nierop, G.P.; Valerio, D.; Knaän-Shanzer, S. Human mesenchymal stem cells ectopically expressing full-length dystrophin can complement Duchenne muscular dystrophy myotubes by cell fusion. Hum. Mol. Genet. 2005, 15, 213–221. [Google Scholar] [CrossRef]
- Gonçalves, M.A.F.V.; Holkers, M.; Cudré-Mauroux, C.; Van Nierop, G.P.; Knaän-Shanzer, S.; Van Der Velde, I.; Valerio, D.; De Vries, A.A. Transduction of myogenic cells by retargeted dual high-capacity hybrid viral vectors: Robust dystrophin synthesis in duchenne muscular dystrophy muscle cells. Mol. Ther. 2006, 13, 976–986. [Google Scholar] [CrossRef]
- Gaggar, A.; Shayakhmetov, D.M.; Lieber, A. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 2003, 9, 1408–1412. [Google Scholar] [CrossRef]
- Shayakhmetov, D.M.; Papayannopoulou, T.; Stamatoyannopoulos, G.; Lieber, A. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 2000, 74, 2567–2583. [Google Scholar] [CrossRef]
- Knaän-Shanzer, S.; Van Der Velde, I.; Havenga, M.J.E.; Lemckert, A.A.C.; De Vries, A.A.F.; Valerio, D. Highly efficient targeted transduction of undifferentiated human hematopoietic cells by adenoviral vectors displaying fiber knobs of subgroup B. Hum. Gene Ther. 2001, 12, 1989–2005. [Google Scholar] [CrossRef]
- Morsy, M.A.; Gu, M.; Motzel, S.; Zhao, J.; Lin, J.; Su, Q.; Allen, H.; Franlin, L.; Parks, R.J.; Graham, F.L. An adenoviral vector deleted for all viral coding sequences results in enhanced safety and extended expression of a leptin transgene. Proc. Natl. Acad. Sci. USA 1998, 95, 7866–7871. [Google Scholar] [CrossRef] [PubMed]
- Schiedner, G.; Morral, N.; Parks, R.J.; Wu, Y.; Koopmans, S.C.; Langston, C.; Graham, F.L.; Beaudet, A.L.; Kochanek, S. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 1998, 18, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Morral, N.; O’Neal, W.; Rice, K.; Leland, M.; Kaplan, J.; Piedra, P.A.; Zhou, H.; Parks, R.J.; Velji, R.; Aguilar-Córdova, E.; et al. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. USA 1999, 96, 12816–12821. [Google Scholar] [CrossRef] [PubMed]
- Ehrke-Schulz, E.; Schiwon, M.; Leitner, T.; Dávid, S.; Bergmann, T.; Liu, J.; Ehrhardt, A. CRISPR/Cas9 delivery with one single adenoviral vector devoid of all viral genes. Sci. Rep. 2017, 7, 17113. [Google Scholar] [CrossRef] [PubMed]
- Schiwon, M.; Ehrke-Schulz, E.; Oswald, A.; Bergmann, T.; Michler, T.; Protzer, U.; Ehrhardt, A. One-vector system for multiplexed CRISPR/Cas9 against hepatitis B virus cccDNA utilizing high-capacity adenoviral vectors. Mol. Ther. Nucleic Acids 2018, 12, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Mamchaoui, K.; Trollet, C.; Bigot, A.; Negroni, E.; Chaouch, S.; Wolff, A.; Kandalla, P.K.; Marie, S.; Di Santo, J.; St Guily, J.L.; et al. Immortalized pathological human myoblasts: Towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle 2011, 1, 34. [Google Scholar] [CrossRef]
- Fallaux, F.J.; Bout, A.; van der Velde, I.; van den Wollenberg, D.J.M.; Hehir, K.M.; Keegan, J.; Auger, C.; Cramer, S.J.; van Ormondt, H.; van der Eb, A.J.; et al. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 1998, 9, 1909–1917. [Google Scholar] [CrossRef]
- Ensinger, M.J.; Ginsberg, H.S. Selection and preliminary characterization of temperature-sensitive mutants of type 5 adenovirus. J. Virol. 1972, 10, 328–339. [Google Scholar] [CrossRef]
- Van Der Vliet, P.C.; Levine, A.J.; Ensinger, M.J.; Ginsberg, H.S. Thermolabile DNA binding proteins from cells infected with a temperature-sensitive mutant of adenovrius defective in viral DNA synthesis. J. Virol. 1975, 15, 348–354. [Google Scholar] [CrossRef]
- Havenga, M.; Holterman, L.; Melis, I.; Smits, S.; Kaspers, J.; Heemskerk, E.; Vlugt, R.v.d.; Koldijk, M.; Schouten, G.; Hateboer, G. Serum-free transient protein production system based on adenoviral vector and PER. C6 technology: High yield and preserved bioactivity. Biotechnol. Bioeng. 2008, 100, 273–283. [Google Scholar] [CrossRef]
- Holkers, M.; Maggio, I.; Henriques, S.F.; Janssen, J.M.; Cathomen, T.; Gonçalves, M.A. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods 2014, 11, 1051–1057. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Jia, G.; Choi, J.; Ma, H.; Anaya, E.; Ye, C.; Shankar, P.; Wu, H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015, 16, 280. [Google Scholar] [CrossRef] [PubMed]
- Maggio, I.; Zittersteijn, H.A.; Wang, Q.; Liu, J.; Janssen, J.M.; Ojeda, I.T.; van der Maarel, S.M.; Lankester, A.C.; Hoeben, R.C.; Gonçalves, M.A.F.V. Integrating gene delivery and gene-editing technologies by adenoviral vector transfer of optimized CRISPR-Cas9 components. Gene Ther. 2020. [Google Scholar] [CrossRef] [PubMed]
- Janssen, J.M.; Liu, J.; Skokan, J.; Gonçalves, M.A.; de Vries, A.A. Development of an AdEasy-based system to produce first-and second-generation adenoviral vectors with tropism for CAR-or CD46-positive cells. J. Gene Med. 2013, 15, 1–11. [Google Scholar] [CrossRef]
- Holkers, M.; Cathomen, T.; Gonçalves, M.A.F.V. Construction and characterization of adenoviral vectors for the delivery of TALENs into human cells. Methods 2014, 69, 179–187. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef]
- Matsumura, K.; Campbell, K.P. Deficiency of dystrophin-associated proteins: A common mechanism leading to muscle cell necrosis in severe childhood muscular dystrophies. Neuromuscul. Disord. 1993, 3, 109–118. [Google Scholar] [CrossRef]
- Wasala, N.B.; Hakim, C.H.; Chen, S.-J.; Yang, N.N.; Duan, D. Questions Answered and Unanswered by the First CRISPR Editing Study in a Canine Model of Duchenne Muscular Dystrophy. Hum. Gene Ther. 2019, 30, 535–543. [Google Scholar] [CrossRef]
- Sun, C.; Serra, C.; Lee, G.; Wagner, K.R. Stem cell-based therapies for Duchenne muscular dystrophy. Exp. Neurol. 2020, 323, 113086. [Google Scholar] [CrossRef]
- Chen, X.; Rinsma, M.; Janssen, J.M.; Liu, J.; Maggio, I.; Gonçalves, M.A.F.V. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016, 44, 6482–6492. [Google Scholar] [CrossRef]
- Daer, R.M.; Cutts, J.P.; Brafman, D.A.; Haynes, K.A. The impact of chromatin dynamics on Cas9-mediated genome editing in human cells. ACS Synt. Biol. 2016, 6, 428–438. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Lucchetti-Miganeh, C.; Yaou, R.B.; Kaplan, J.-C.; Chelly, J.; Leturcq, F.; Barloy-Hubler, F.; Le Rumeur, E. Assessment of the structural and functional impact of in-frame mutations of the DMD gene, using the tools included in the eDystrophin online database. Orphanet J. Rare Dis. 2012, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, A.; Raguénès-Nicol, C.; Ben Yaou, R.; Ameziane-Le Hir, S.; Chéron, A.; Vié, V.; Claustres, M.; Leturcq, F.; Delalande, O.; Hubert, J.-F.; et al. Becker muscular dystrophy severity is linked to the structure of dystrophin. Hum. Mol. Genet. 2014, 24, 1267–1279. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brescia, M.; Janssen, J.M.; Liu, J.; Gonçalves, M.A.F.V. High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells. Cells 2020, 9, 869. https://doi.org/10.3390/cells9040869
Brescia M, Janssen JM, Liu J, Gonçalves MAFV. High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells. Cells. 2020; 9(4):869. https://doi.org/10.3390/cells9040869
Chicago/Turabian StyleBrescia, Marcella, Josephine M. Janssen, Jin Liu, and Manuel A. F. V. Gonçalves. 2020. "High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells" Cells 9, no. 4: 869. https://doi.org/10.3390/cells9040869
APA StyleBrescia, M., Janssen, J. M., Liu, J., & Gonçalves, M. A. F. V. (2020). High-Capacity Adenoviral Vectors Permit Robust and Versatile Testing of DMD Gene Repair Tools and Strategies in Human Cells. Cells, 9(4), 869. https://doi.org/10.3390/cells9040869




