The Switch from NF-YAl to NF-YAs Isoform Impairs Myotubes Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Proliferation Assay
2.2. Derivation of C2-YAl-KO Clones
2.3. Protein Extraction and Western Blot Analysis
2.4. Reverse Transcriptase PCR and Real-Time PCR
2.5. Flow Cytometry Analyses
2.6. Immunofluorescence
2.7. Overexpression and RNA Interference Experiments
2.8. Chromatin Immunoprecipitation Assay (ChIP)
3. Results
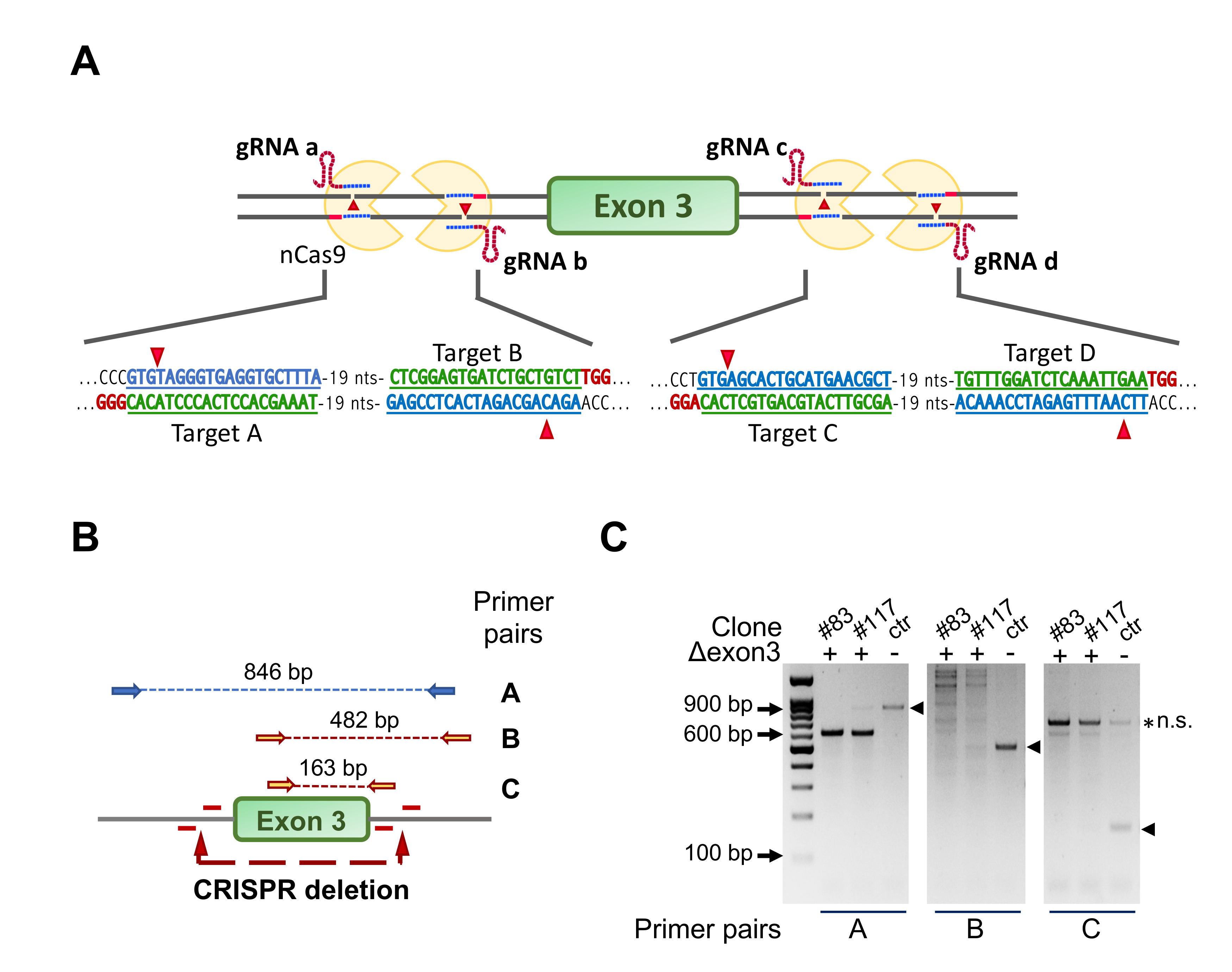
3.1. Ablation of NF-YA Exon 3 in C2C12 Cells by a Four Guides CRISPR/Cas9n Strategy
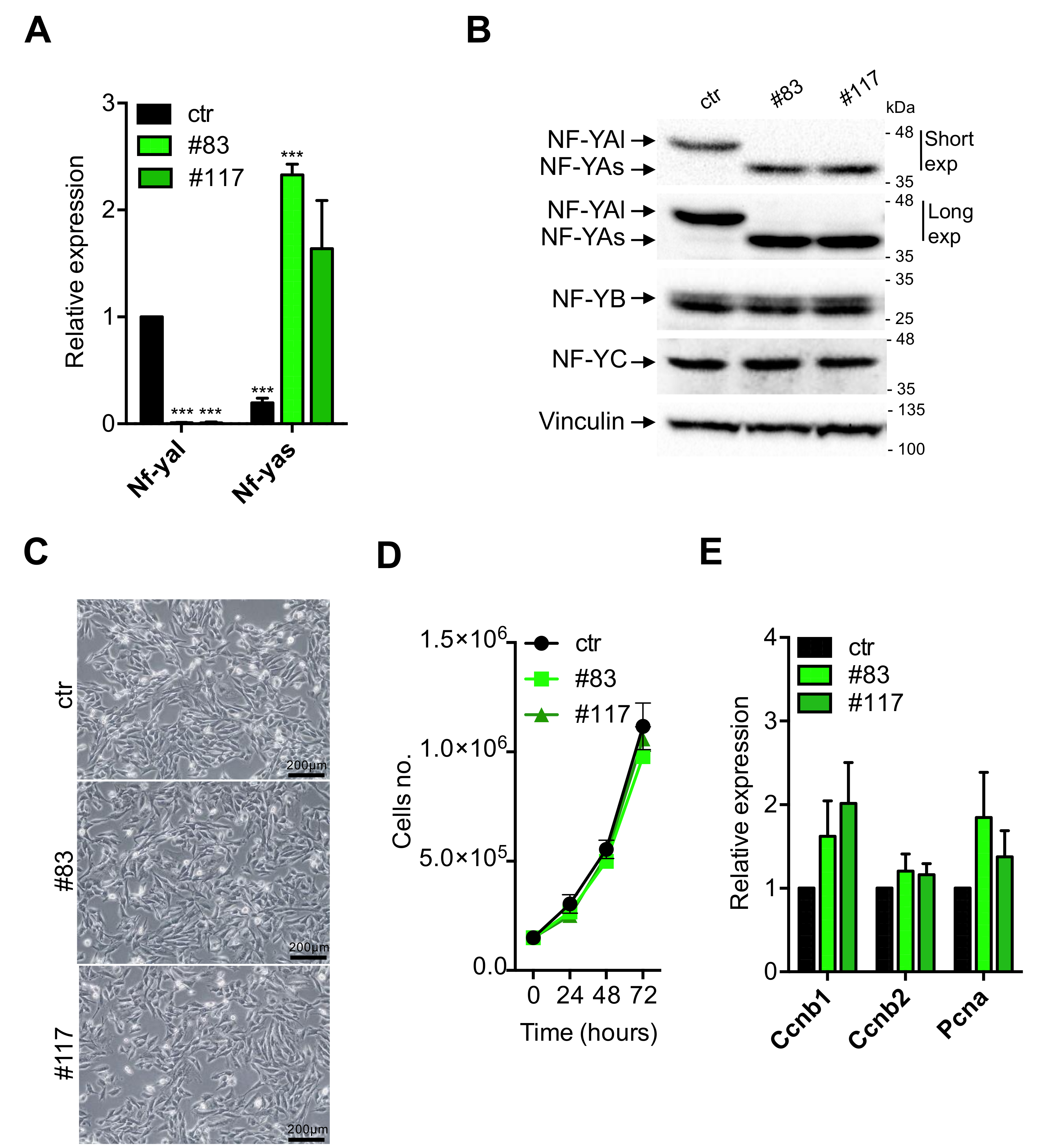
3.2. Characterization of C2-YAl-KO Cells
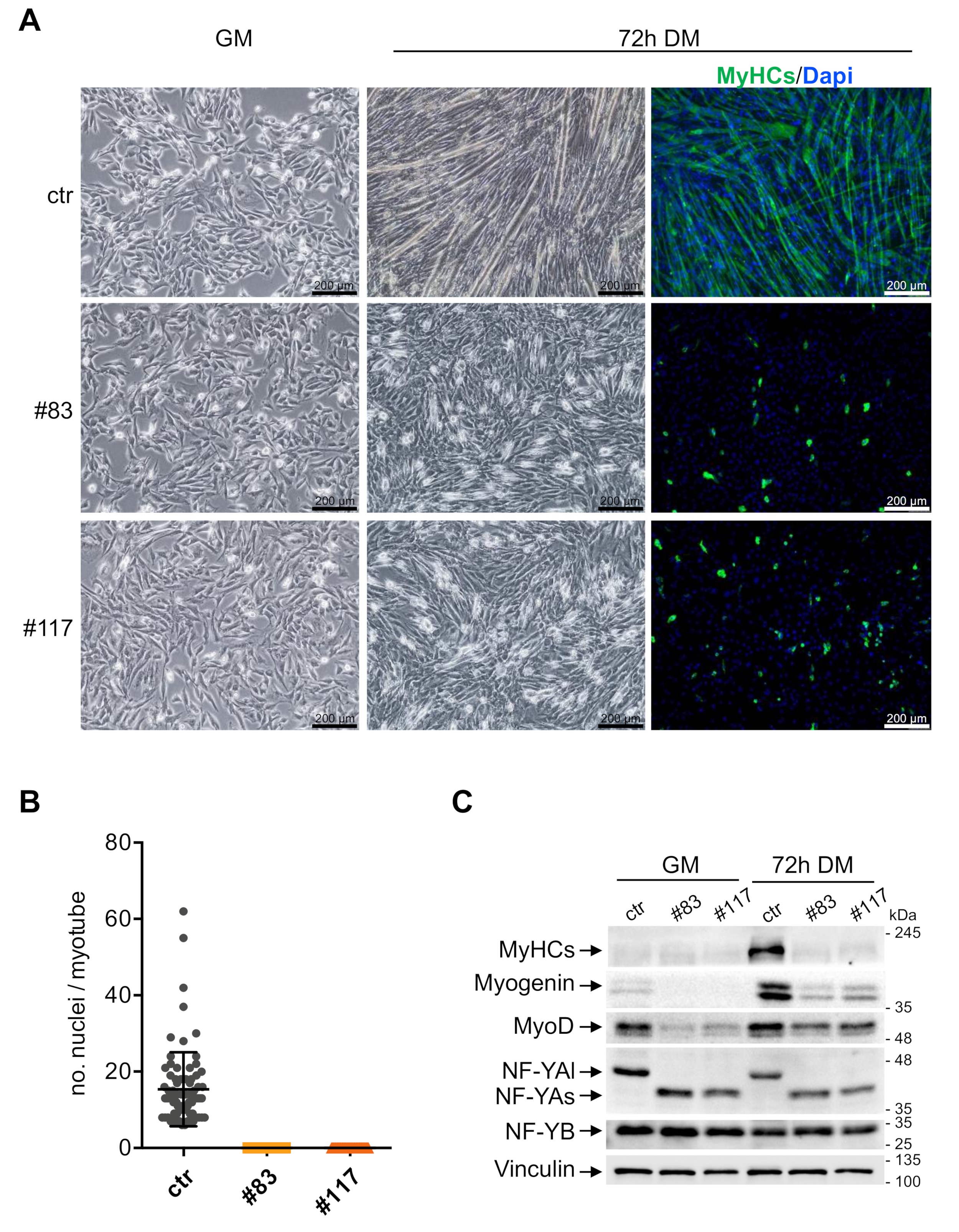
3.3. C2-YAl-KO Cells Fail to Differentiate and Fuse into Myotubes
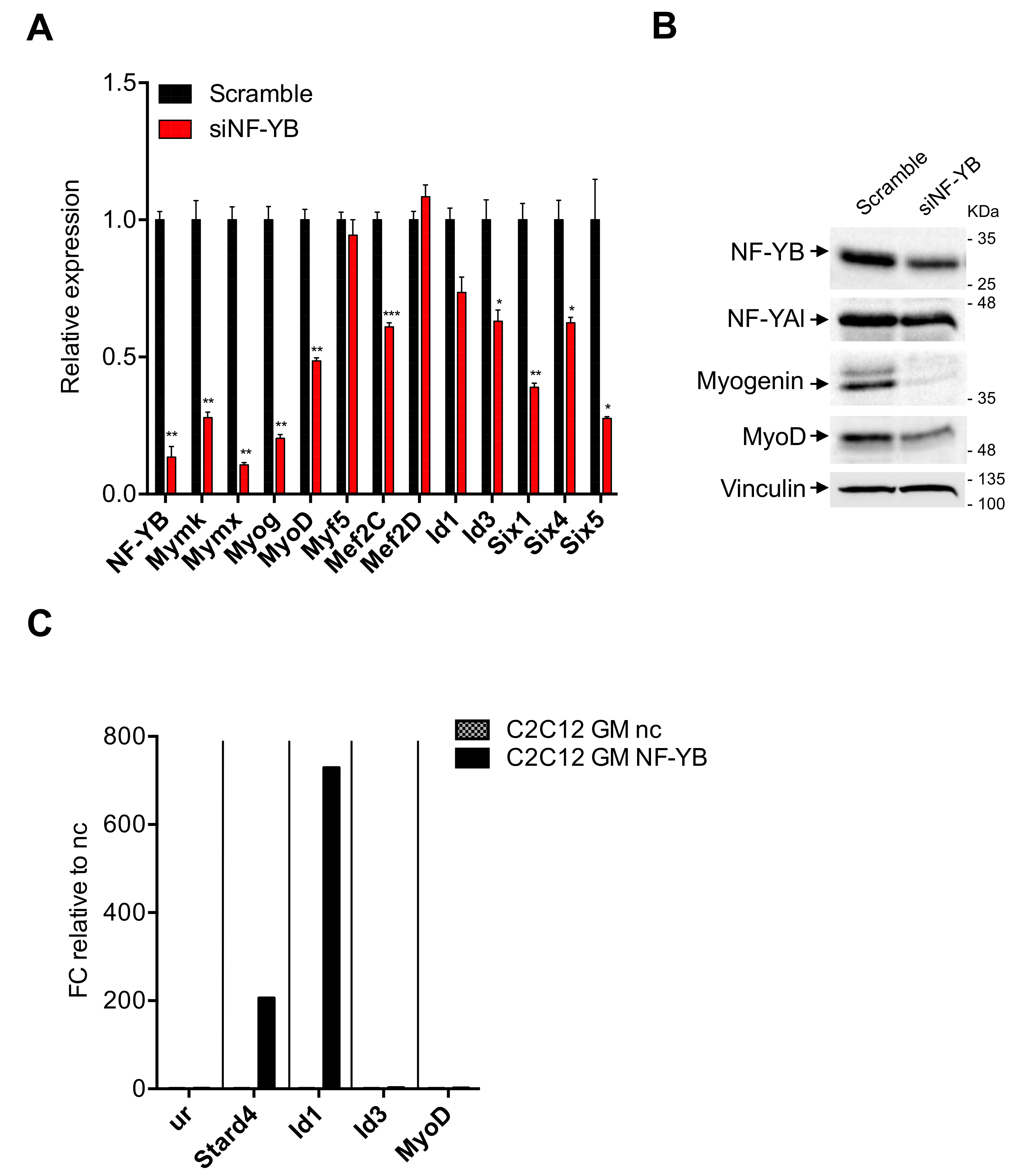
3.4. Expression of TFs in C2-YAl-KO
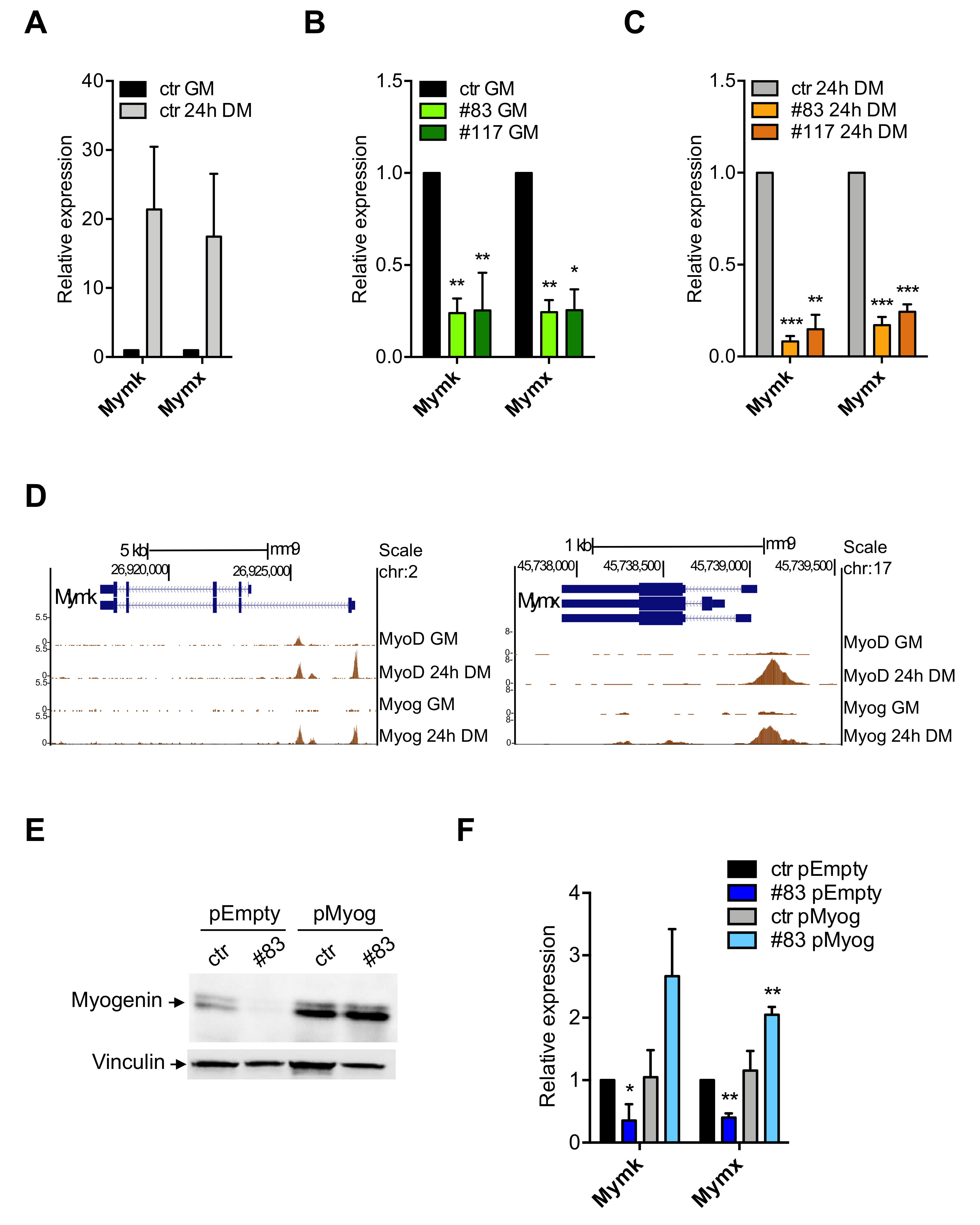
3.5. Expression of Myomaker and Myomixer Is Activated by Myogenin and It Is Impaired in C2-YAl-KO
3.6. Myogenin and MyoD Are—Indirectly—Regulated by NF-Y
4. Discussion
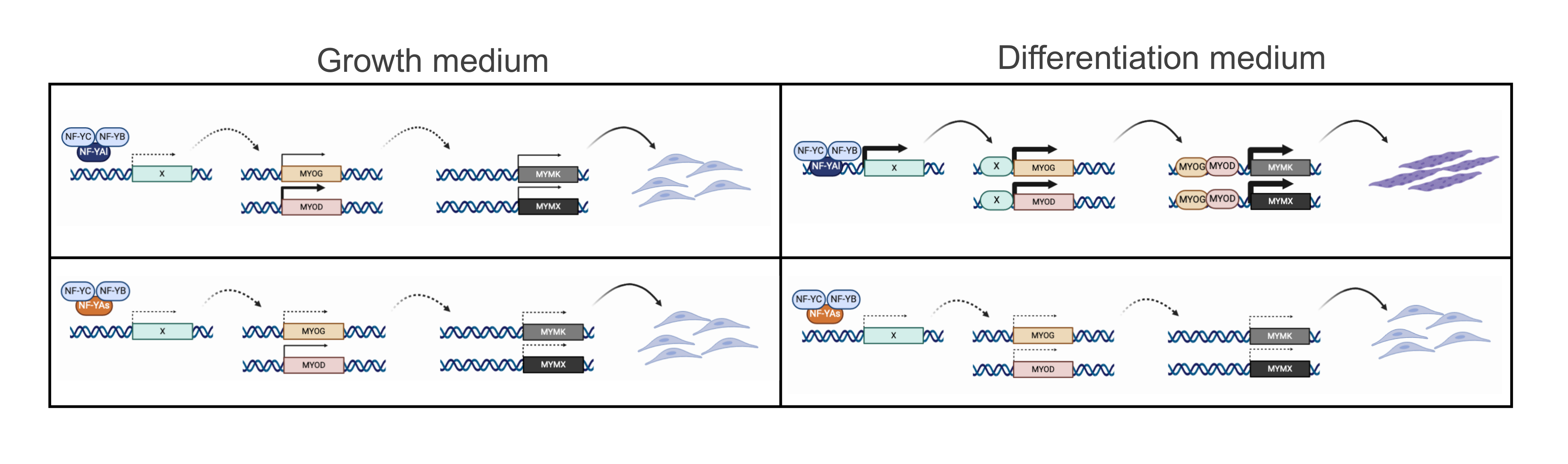
4.1. Role of NF-YA Alternative Splicing in Muscle Cells
4.2. NF-Y Does Not Target Directly Genes Involved in C2C12 Differentiation
4.3. NF-Y Regulates MRFs Expression Indirectly
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buckingham, M. Skeletal muscle formation in vertebrates. Curr. Opin. Genet. Dev. 2001, 11, 440–448. [Google Scholar] [CrossRef]
- Buckingham, M.; Rigby, P.W. Gene Regulatory Networks and Transcriptional Mechanisms that Control Myogenesis. Dev. Cell 2014, 28, 225–238. [Google Scholar] [CrossRef]
- Hernández-Hernández, J.M.; García-González, E.G.; Brun, C.E.; Rudnicki, M.A. The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Boil. 2017, 72, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Zammit, P.S. Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Boil. 2017, 72, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Boil. 2015, 44, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Black, B.L.; Olson, E.N. Transcriptional Control Of Muscle Development by Myocyte Enhancer Factor-2 (MEF2) Proteins. Annu. Rev. Cell Dev. Boil. 1998, 14, 167–196. [Google Scholar] [CrossRef]
- Taylor, M.V.; Hughes, S.M. Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Boil. 2017, 72, 33–44. [Google Scholar] [CrossRef]
- Kumar, D.; Shadrach, J.L.; Wagers, A.J.; Lassar, A.B. Id3 Is a Direct Transcriptional Target of Pax7 in Quiescent Satellite Cells. Mol. Boil. Cell 2009, 20, 3170–3177. [Google Scholar] [CrossRef]
- Wu, J.; Lim, R.W. Regulation of inhibitor of differentiation gene 3 (Id3) expression by Sp2-motif binding factor in myogenic C2C12 cells: Downregulation of DNA binding activity following skeletal muscle differentiation. Biochim. et Biophys. Acta (BBA) Gene Struct. Expr. 2005, 1731, 13–22. [Google Scholar] [CrossRef]
- Atherton, G.T.; Travers, H.; Deed, R.; Norton, J.D. Regulation of cell differentiation in C2C12 myoblasts by the Id3 helix-loop-helix protein. Cell Growth Differ. Mol. Boil. J. Am. Assoc. Cancer Res. 1996, 7, 1059–1066. [Google Scholar]
- Soleimani, V.D.; Yin, H.; Jahani-Asl, A.; Ming, H.; Kockx, C.; Van Ijcken, W.F.J.; Grosveld, F.; Rudnicki, M.A. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol. Cell 2012, 47, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Grifone, R.; Demignon, J.; Houbron, C.; Souil, E.; Niro, C.; Seller, M.J.; Hamard, G.; Maire, P. Six1 and Six4 homeoproteins are required for Pax3 and Mrf expression during myogenesis in the mouse embryo. Dev. 2005, 132, 2235–2249. [Google Scholar] [CrossRef] [PubMed]
- Yajima, H.; Motohashi, N.; Ono, Y.; Sato, S.; Ikeda, K.; Masuda, S.; Yada, E.; Kanesaki, H.; Miyagoe-Suzuki, Y.; Takeda, S.; et al. Six family genes control the proliferation and differentiation of muscle satellite cells. Exp. Cell Res. 2010, 316, 2932–2944. [Google Scholar] [CrossRef]
- Santolini, M.; Sakakibara, I.; Gauthier, M.; Aulinas, F.R.; Takahashi, H.; Sawasaki, T.; Mouly, V.; Concordet, J.-P.; Defossez, P.-A.; Hakim, V.; et al. MyoD reprogramming requires Six1 and Six4 homeoproteins: genome-wide cis-regulatory module analysis. Nucleic Acids Res. 2016, 44, 8621–8640. [Google Scholar] [CrossRef] [PubMed]
- Yajima, H.; Kawakami, K. Low Six4 and Six5 gene dosage improves dystrophic phenotype and prolongs life span of mdx mice. Dev. Growth Differ. 2016, 58, 546–561. [Google Scholar] [CrossRef]
- Guadagnin, E.; Mázala, D.; Chen, Y.-W. STAT3 in Skeletal Muscle Function and Disorders. Int. J. Mol. Sci. 2018, 19, 2265. [Google Scholar] [CrossRef] [PubMed]
- Messina, G.; Biressi, S.A.M.; Monteverde, S.; Magli, A.; Cassano, M.; Perani, L.; Roncaglia, E.; Tagliafico, E.; Starnes, L.; Campbell, C.E.; et al. Nfix Regulates Fetal-Specific Transcription in Developing Skeletal Muscle. Cell 2010, 140, 554–566. [Google Scholar] [CrossRef]
- Rossi, G.; Antonini, S.; Bonfanti, C.; Monteverde, S.; Vezzali, C.; Tajbakhsh, S.; Cossu, G.; Messina, G. Nfix Regulates Temporal Progression of Muscle Regeneration through Modulation of Myostatin Expression. Cell Rep. 2016, 14, 2238–2249. [Google Scholar] [CrossRef]
- Hayashi, S.; Manabe, I.; Suzuki, Y.; Relaix, F.; Oishi, Y. Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. eLife 2016, 5, 37798. [Google Scholar] [CrossRef]
- Sunadome, K.; Yamamoto, T.; Ebisuya, M.; Kondoh, K.; Sehara-Fujisawa, A.; Nishida, E. ERK5 Regulates Muscle Cell Fusion through Klf Transcription Factors. Dev. Cell 2011, 20, 192–205. [Google Scholar] [CrossRef]
- Potthoff, M.J.; Olson, E.N. MEF2: a central regulator of diverse developmental programs. Dev. 2007, 134, 4131–4140. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Kang, B.; Sun, X.H. Id proteins: small molecules, mighty regulators. Curr. Top Dev. Biol. 2014, 110, 189–216. [Google Scholar] [PubMed]
- Christensen, K.L.; Patrick, A.N.; McCoy, E.L.; Ford, H.L. Chapter 5 The Six Family of Homeobox Genes in Development and Cancer. Advances in Cancer Research 2008, 101, 93–126. [Google Scholar] [PubMed]
- Bialkowska, A.; Yang, V.W.; Mallipattu, S.K. Krüppel-like factors in mammalian stem cells and development. Dev. 2017, 144, 737–754. [Google Scholar] [CrossRef]
- Piper, M.; Gronostajski, R.; Messina, G. Nuclear Factor One X in Development and Disease. Trends Cell Boil. 2019, 29, 20–30. [Google Scholar] [CrossRef]
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Boil. 2011, 47, 29–49. [Google Scholar] [CrossRef]
- Fleming, J.D.; Pavesi, G.; Benatti, P.; Imbriano, C.; Mantovani, R.; Struhl, K. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013, 23, 1195–1209. [Google Scholar] [CrossRef]
- Sherwood, R.I.; Hashimoto, T.; O’Donnell, C.P.; Lewis, S.; A Barkal, A.; Van Hoff, J.P.; Karun, V.; Jaakkola, T.; Gifford, D.K. Discovery of directional and nondirectional pioneer transcription factors by modeling DNase profile magnitude and shape. Nat. Biotechnol. 2014, 32, 171–178. [Google Scholar] [CrossRef]
- Oldfield, A.; Yang, P.; Conway, A.E.; Cinghu, S.; Freudenberg, J.; Yellaboina, S.; Jothi, R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell 2014, 55, 708–722. [Google Scholar] [CrossRef]
- Oldfield, A.; Henriques, T.; Kumar, D.; Burkholder, A.B.; Cinghu, S.; Paulet, D.; Bennett, B.D.; Yang, P.; Scruggs, B.S.; Lavender, C.A.; et al. NF-Y controls fidelity of transcription initiation at gene promoters through maintenance of the nucleosome-depleted region. Nat. Commun. 2019, 10, 3072. [Google Scholar] [CrossRef]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Van Huijsduijnen, R.H.; Mantovani, R.; Benoist, C.; Mathis, D. Intron-exon organization of the NF-Y genes. Tissue-specific splicing modifies an activation domain. J. Boil. Chem. 1992, 267, 8984–8990. [Google Scholar]
- Ceribelli, M.; Benatti, P.; Imbriano, C.; Mantovani, R. NF-YC Complexity Is Generated by Dual Promoters and Alternative Splicing. J. Biol. Chem. 2009, 284, 34189–34200. [Google Scholar] [CrossRef] [PubMed]
- Benatti, P.; Dolfini, D.; Vigano, M.A.; Ravo, M.; Weisz, A.; Imbriano, C. Specific inhibition of NF-Y subunits triggers different cell proliferation defects. Nucleic Acids Res. 2011, 39, 5356–5368. [Google Scholar] [CrossRef] [PubMed]
- Farina, A.; Manni, I.; Fontemaggi, G.; Tiainen, M.; Cenciarelli, C.; Bellorini, M.; Mantovani, R.; Sacchi, A.; Piaggio, G. Down-regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene 1999, 18, 2818–2827. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gurtner, A.; Manni, I.; Fuschi, P.; Mantovani, R.; Guadagni, F.; Sacchi, A.; Piaggio, G. Requirement for Down-Regulation of the CCAAT-binding Activity of the NF-Y Transcription Factor during Skeletal Muscle Differentiation. Mol. Boil. Cell 2003, 14, 2706–2715. [Google Scholar] [CrossRef] [PubMed]
- Gurtner, A.; Fuschi, P.; Magi, F.; Colussi, C.; Gaetano, C.; Dobbelstein, M.; Sacchi, A.; Piaggio, G. NF-Y Dependent Epigenetic Modifications Discriminate between Proliferating and Postmitotic Tissue. PLOS ONE 2008, 3, e2047. [Google Scholar] [CrossRef]
- Goeman, F.; Manni, I.; Artuso, S.; Ramachandran, B.; Toietta, G.; Bossi, G.; Rando, G.; Cencioni, C.; Germoni, S.; Straino, S.; et al. Molecular imaging of nuclear factor-Y transcriptional activity maps proliferation sites in live animals. Mol. Boil. Cell 2012, 23, 1467–1474. [Google Scholar] [CrossRef]
- Basile, V.; Baruffaldi, F.; Dolfini, D.; Belluti, S.; Benatti, P.; Ricci, L.; Artusi, V.; Tagliafico, E.; Mantovani, R.; Molinari, S.; et al. NF-YA splice variants have different roles on muscle differentiation. Biochim. et Biophys. Acta (BBA) - Gene Regul. Mech. 2016, 1859, 627–638. [Google Scholar] [CrossRef]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Cell Boil. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Maity, S.N. NF-Y (CBF) regulation in specific cell types and mouse models. Biochim. et Biophys. Acta (BBA) Bioenerg. 2016, 1860, 598–603. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, T.; Nishikawa, A.; Kume, S.; Chayama, K.; Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014, 4, 5400. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Minuzzo, M.; Pavesi, G.; Mantovani, R. The Short Isoform of NF-YA Belongs to the Embryonic Stem Cell Transcription Factor Circuitry. STEM CELLS 2012, 30, 2450–2459. [Google Scholar] [CrossRef] [PubMed]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.-L.; Bassel-Duby, R.; Olson, E.N. CRISPR Correction of Duchenne Muscular Dystrophy. Annu. Rev. Med. 2018, 70, 239–255. [Google Scholar] [CrossRef] [PubMed]
- Bungartz, G.; Land, H.; Scadden, D.T.; Emerson, S.G. NF-Y is necessary for hematopoietic stem cell proliferation and survival. Blood 2012, 119, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Belluti, S.; Semeghini, V.; Basile, V.; Rigillo, G.; Salsi, V.; Genovese, F.; Dolfini, D.; Imbriano, C. An autoregulatory loop controls the expression of the transcription factor NF-Y. Biochim. et Biophys. Acta (BBA) Bioenerg. 2018, 1861, 509–518. [Google Scholar] [CrossRef]
- Moran, J.; Li, Y.; Hill, A.A.; Mounts, W.M.; Miller, C.P. Gene expression changes during mouse skeletal myoblast differentiation revealed by transcriptional profiling. Physiol. Genom. 2002, 10, 103–111. [Google Scholar] [CrossRef]
- Clever, J.L.; Sakai, Y.; Wang, R.A.; Schneider, D.B. Inefficient skeletal muscle repair in inhibitor of differentiation knockout mice suggests a crucial role for BMP signaling during adult muscle regeneration. Am. J. Physiol. Physiol. 2010, 298, C1087–C1099. [Google Scholar] [CrossRef]
- Salizzato, V.; Zanin, S.; Borgo, C.; Lidron, E.; Salvi, M.; Rizzuto, R.; Pallafacchina, G.; Donella-Deana, A. Protein kinase CK2 subunits exert specific and coordinated functions in skeletal muscle differentiation and fusogenic activity. FASEB J. 2019, 33, 10648–10667. [Google Scholar] [CrossRef]
- Millay, D.P.; Gamage, D.G.; Quinn, M.E.; Min, Y.-L.; Mitani, Y.; Bassel-Duby, R.; Olson, E.N. Structure–function analysis of myomaker domains required for myoblast fusion. Proc. Natl. Acad. Sci. 2016, 113, 2116–2121. [Google Scholar] [CrossRef] [PubMed]
- Petrany, M.J.; Millay, D.P. Cell Fusion: Merging Membranes and Making Muscle. Trends Cell Boil. 2019, 29, 964–973. [Google Scholar] [CrossRef] [PubMed]
- Ganassi, M.; Badodi, S.; Quiroga, H.P.O.; Zammit, P.S.; Hinits, Y.; Hughes, S.M. Myogenin promotes myocyte fusion to balance fibre number and size. Nat. Commun. 2018, 9, 4232. [Google Scholar] [CrossRef] [PubMed]
- Pedraza-Alva, G.; Zingg, J.M.; Jost, J.P. AP-1 binds to a putative cAMP response element of the MyoD1 promoter and negatively modulates MyoD1 expression in dividing myoblasts. J. Boil. Chem. 1994, 269, 6978–6985. [Google Scholar]
- Murray-Zmijewski, F.; Lane, D.P.; Bourdon, J.C. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006, 13, 962–972. [Google Scholar] [CrossRef]
- Imbriano, C.; Molinari, S. Alternative Splicing of Transcription Factors Genes in Muscle Physiology and Pathology. Genes 2018, 9, 107. [Google Scholar] [CrossRef]
- Nakka, K.; Ghigna, C.; Gabellini, D.; Dilworth, F.J. Diversification of the muscle proteome through alternative splicing. Skelet. Muscle 2018, 8, 8. [Google Scholar] [CrossRef]
- Zhang, M.; Zhu, B.; Davie, J. Alternative Splicing of MEF2C pre-mRNA Controls Its Activity in Normal Myogenesis and Promotes Tumorigenicity in Rhabdomyosarcoma Cells*. J. Boil. Chem. 2014, 290, 310–324. [Google Scholar] [CrossRef]
- Sebastian, S.; Faralli, H.; Yao, Z.; Rakopoulos, P.; Palii, C.; Cao, Y.; Singh, K.; Liu, Q.-C.; Chu, A.; Aziz, A.; et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genome Res. 2013, 27, 1247–1259. [Google Scholar] [CrossRef]
- Vogan, K.; Underhill, D.A.; Gros, P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol. Cell. Boil. 1996, 16, 6677–6686. [Google Scholar] [CrossRef]
- Barber, T.D.; Barber, M.C.; Cloutier, T.E.; Friedman, T.B. PAX3 gene structure, alternative splicing and evolution. Gene 1999, 237, 311–319. [Google Scholar] [CrossRef]
- Pritchard, C.; Grosveld, G.; Hollenbach, A.D. Alternative splicing of Pax3 produces a transcriptionally inactive protein. Gene 2003, 305, 61–69. [Google Scholar] [CrossRef]
- Charytonowicz, E.; Matushansky, I.; Castillo-Martin, M.; Hricik, T.; Cordon-Cardo, C.; Ziman, M. Alternate PAX3 and PAX7 C-terminal isoforms in myogenic differentiation and sarcomagenesis. Clin. Transl. Oncol. 2011, 13, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Vorobyov, E.; Horst, J. Expression of two protein isoforms of PAX7 is controlled by competing cleavage-polyadenylation and splicing. Gene 2004, 342, 107–112. [Google Scholar] [CrossRef] [PubMed]
- LiBetti, D.; Bernardini, A.; Chiaramonte, M.L.; Minuzzo, M.; Gnesutta, N.; Messina, G.; Dolfini, D.; Mantovani, R. NF-YA enters cells through cell penetrating peptides. Biochim. et Biophys. Acta (BBA) Bioenerg. 2019, 1866, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Cappabianca, L.; Farina, A.R.; Di Marcotullio, L.; Infante, P.; De Simone, D.; Sebastiano, M.; Mackay, A. Discovery, characterization and potential roles of a novel NF-YAx splice variant in human neuroblastoma. J. Exp. Clin. Cancer Res. 2019, 38, 1–25. [Google Scholar] [CrossRef]
- Wang, J.; Zhuang, J.; Iyer, S.; Lin, X.; Whitfield, T.W.; Greven, M.C.; Pierce, B.G.; Dong, X.; Kundaje, A.; Cheng, Y.; et al. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012, 22, 1798–1812. [Google Scholar] [CrossRef]
- Zambelli, F.; Pavesi, G. Genome wide features, distribution and correlations of NF-Y binding sites. Biochim. et Biophys. Acta (BBA) Bioenerg. 2017, 1860, 581–589. [Google Scholar] [CrossRef]
- Welch, R.D.; Guo, C.; Sengupta, M.; Carpenter, K.J.; Stephens, N.A.; Arnett, S.A.; Meyers, M.J.; Sparks, L.M.; Smith, S.R.; Zhang, J.; et al. Rev-Erb co-regulates muscle regeneration via tethered interaction with the NF-Y cistrome. Mol. Metab. 2017, 6, 703–714. [Google Scholar] [CrossRef]
- Van Wageningen, S.; Ridder, M.C.B.-D.; Nigten, J.; Nikoloski, G.; Erpelinck-Verschueren, C.A.J.; Löwenberg, B.; De Witte, T.; Tenen, D.G.; Van Der Reijden, B.A.; Jansen, J.H. Gene transactivation without direct DNA binding defines a novel gain-of-function for PML-RARα. Blood 2008, 111, 1634–1643. [Google Scholar] [CrossRef]
- Moeinvaziri, F.; Shahhosseini, M. Epigenetic role of CCAAT box-binding transcription factor NF-Y onIDgene family in human embryonic carcinoma cells. IUBMB Life 2015, 67, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Dolfini, D.; Mantovani, R.; Zambelli, F.; Pavesi, G. A perspective of promoter architecture from the CCAAT box. Cell Cycle 2009, 8, 4127–4137. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Dilworth, F.J. Differential modulation of cell cycle progression distinguishes members of the myogenic regulatory factor family of transcription factors. FEBS J. 2013, 280, 3991–4003. [Google Scholar] [CrossRef]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Libetti, D.; Bernardini, A.; Sertic, S.; Messina, G.; Dolfini, D.; Mantovani, R. The Switch from NF-YAl to NF-YAs Isoform Impairs Myotubes Formation. Cells 2020, 9, 789. https://doi.org/10.3390/cells9030789
Libetti D, Bernardini A, Sertic S, Messina G, Dolfini D, Mantovani R. The Switch from NF-YAl to NF-YAs Isoform Impairs Myotubes Formation. Cells. 2020; 9(3):789. https://doi.org/10.3390/cells9030789
Chicago/Turabian StyleLibetti, Debora, Andrea Bernardini, Sarah Sertic, Graziella Messina, Diletta Dolfini, and Roberto Mantovani. 2020. "The Switch from NF-YAl to NF-YAs Isoform Impairs Myotubes Formation" Cells 9, no. 3: 789. https://doi.org/10.3390/cells9030789
APA StyleLibetti, D., Bernardini, A., Sertic, S., Messina, G., Dolfini, D., & Mantovani, R. (2020). The Switch from NF-YAl to NF-YAs Isoform Impairs Myotubes Formation. Cells, 9(3), 789. https://doi.org/10.3390/cells9030789






