Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Cells
2.3. Microspectrofluorometry
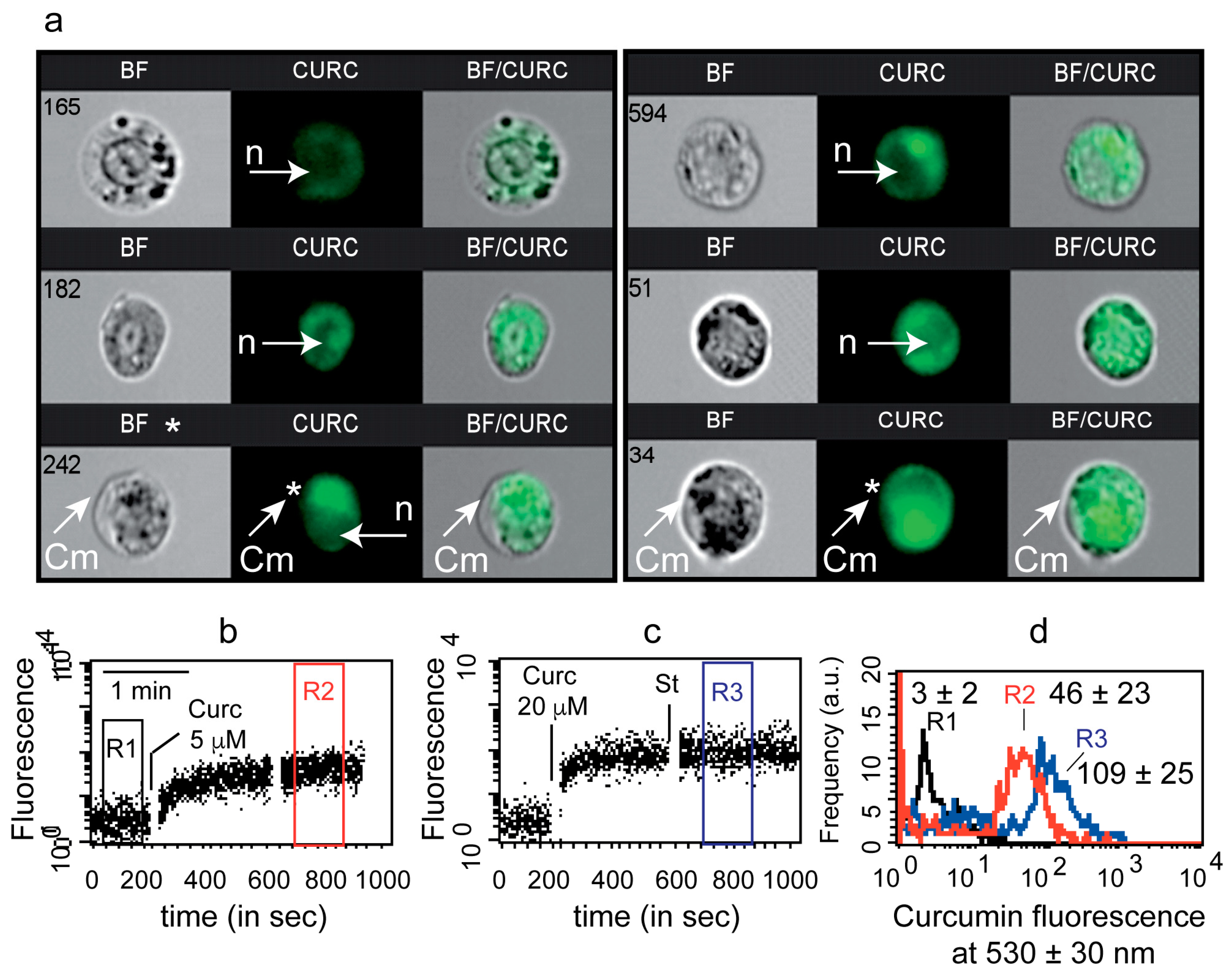
2.4. Imaging Flow Cytometry
2.5. Determination of Mitochondrial Membrane Potential (∆Ψm), Reactive Oxygen Species, and Cytosolic Ca2+ Levels
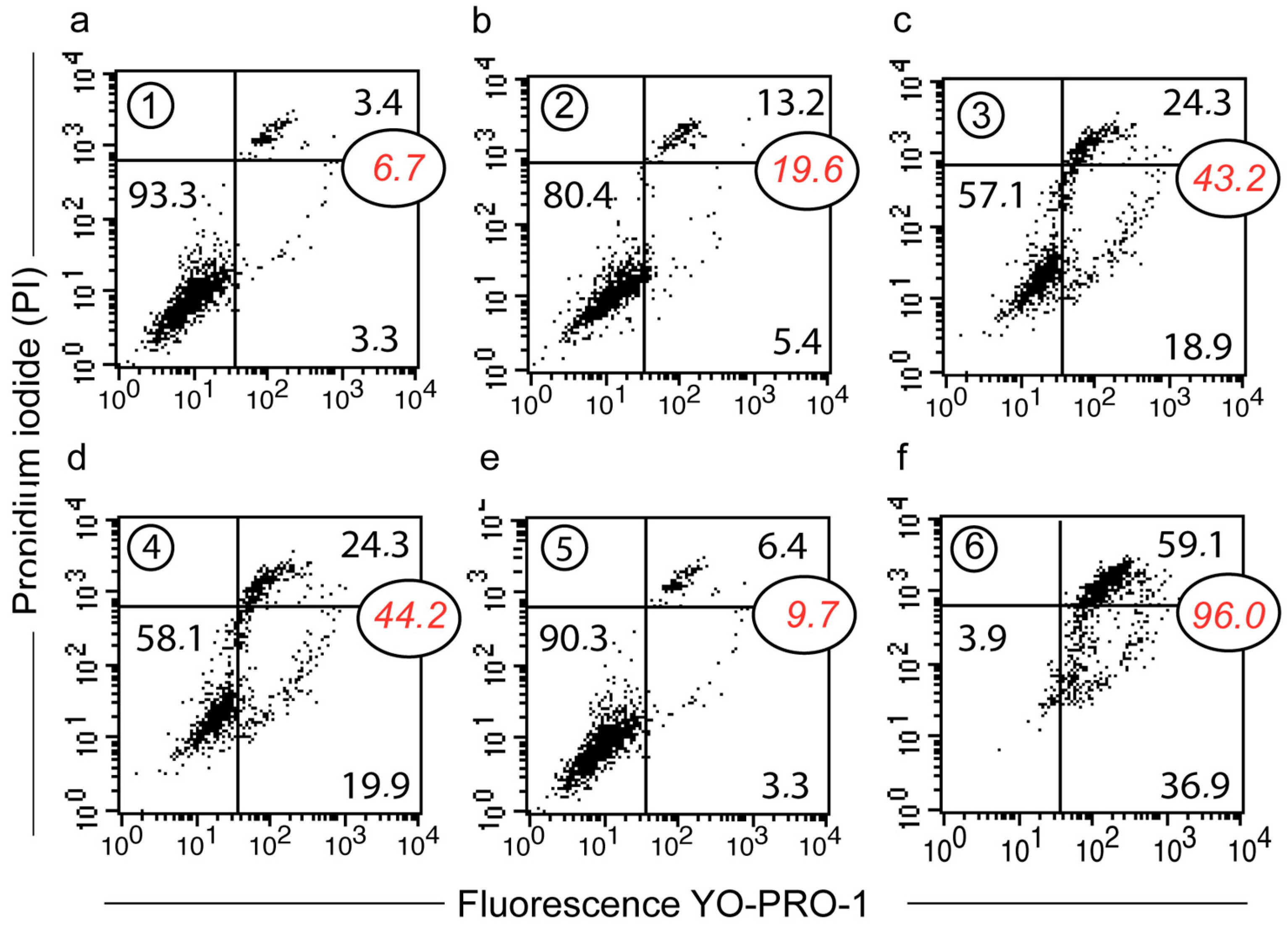
2.6. Cell Cycle Analysis by Flow Cytometry
2.7. Control of ROS Production by Antioxidants
2.8. Control of Calcium Efflux by Calcium Chelators or Inhibition of the Mitochondrial Calcium Uniport
2.9. Analysis of Cathepsin and Calpain Activities by Flow Cytometry
2.10. Analysis of CHOP and GRP78 Activities by Flow Cytometry
2.11. Caspase Activation, Fluorimetric Assays
2.12. Impedancemetry with xCELLigence for the Measurement of Cell Proliferation
2.13. Measurement of Ca2+-ATPase Activity
2.14. Electron Microscopy
3. Results
3.1. Uptake of Curcumin and Invalidation of Its Putative Cytoplasmic Membrane/Nuclei Localization
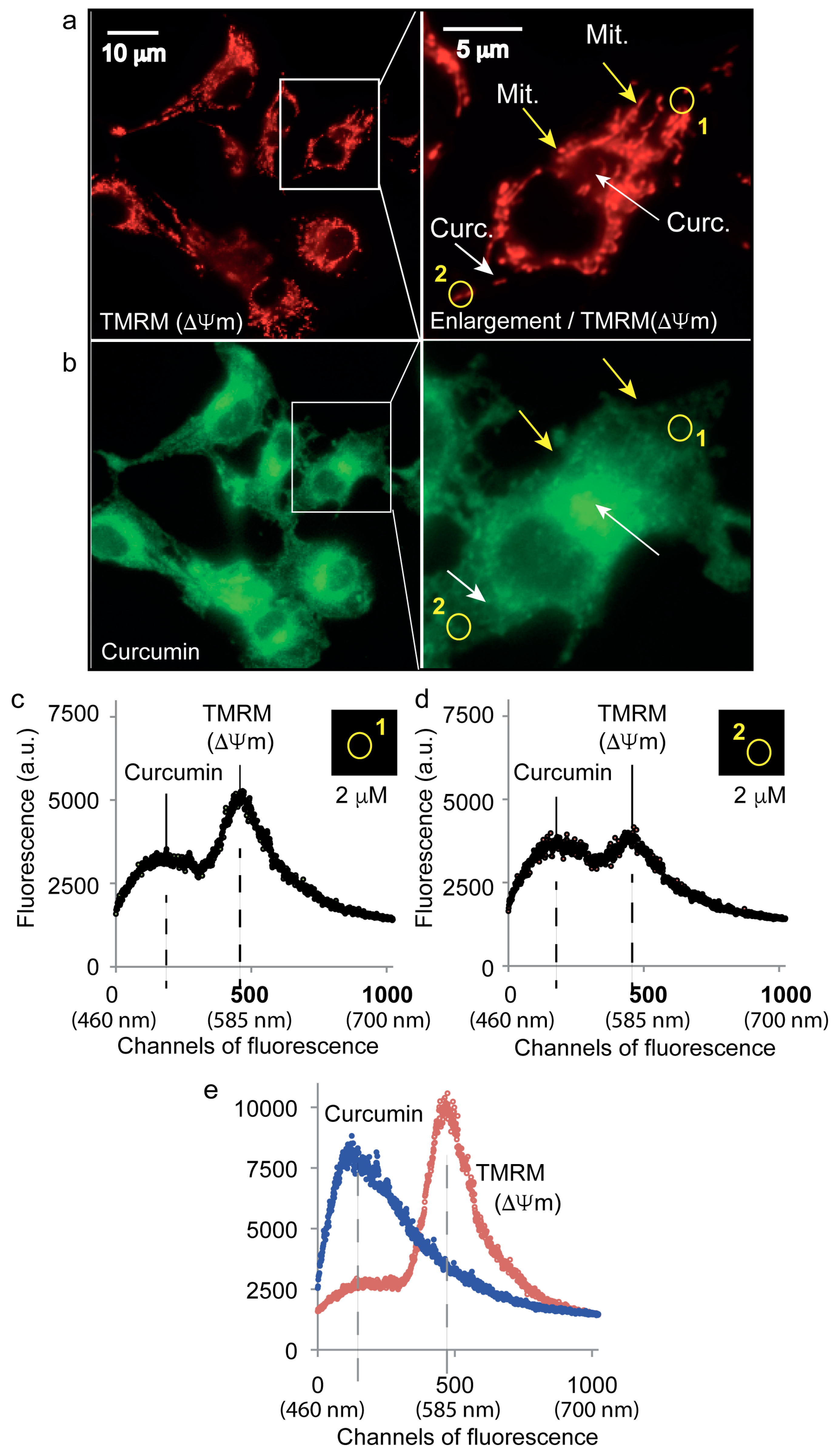
3.2. Curcumin Is Not Localized on the Mitochondrial Network
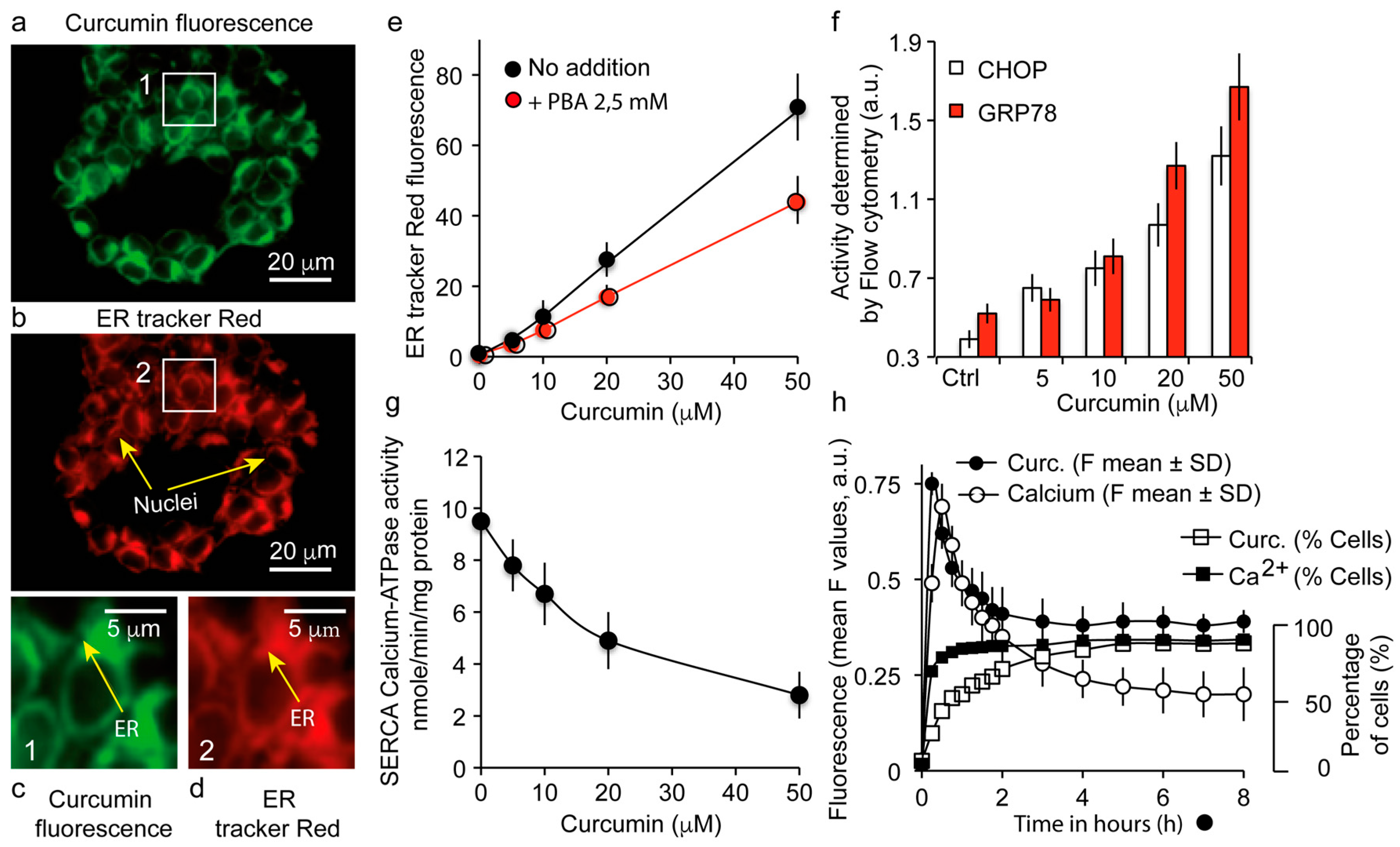
3.3. The Major Fraction of Curcumin Locates to the Endoplasmic Reticulum
3.4. Curcumin Localization at the Endoplasmic Reticulum Causes a UPR Response Together with a Calcium Status Change
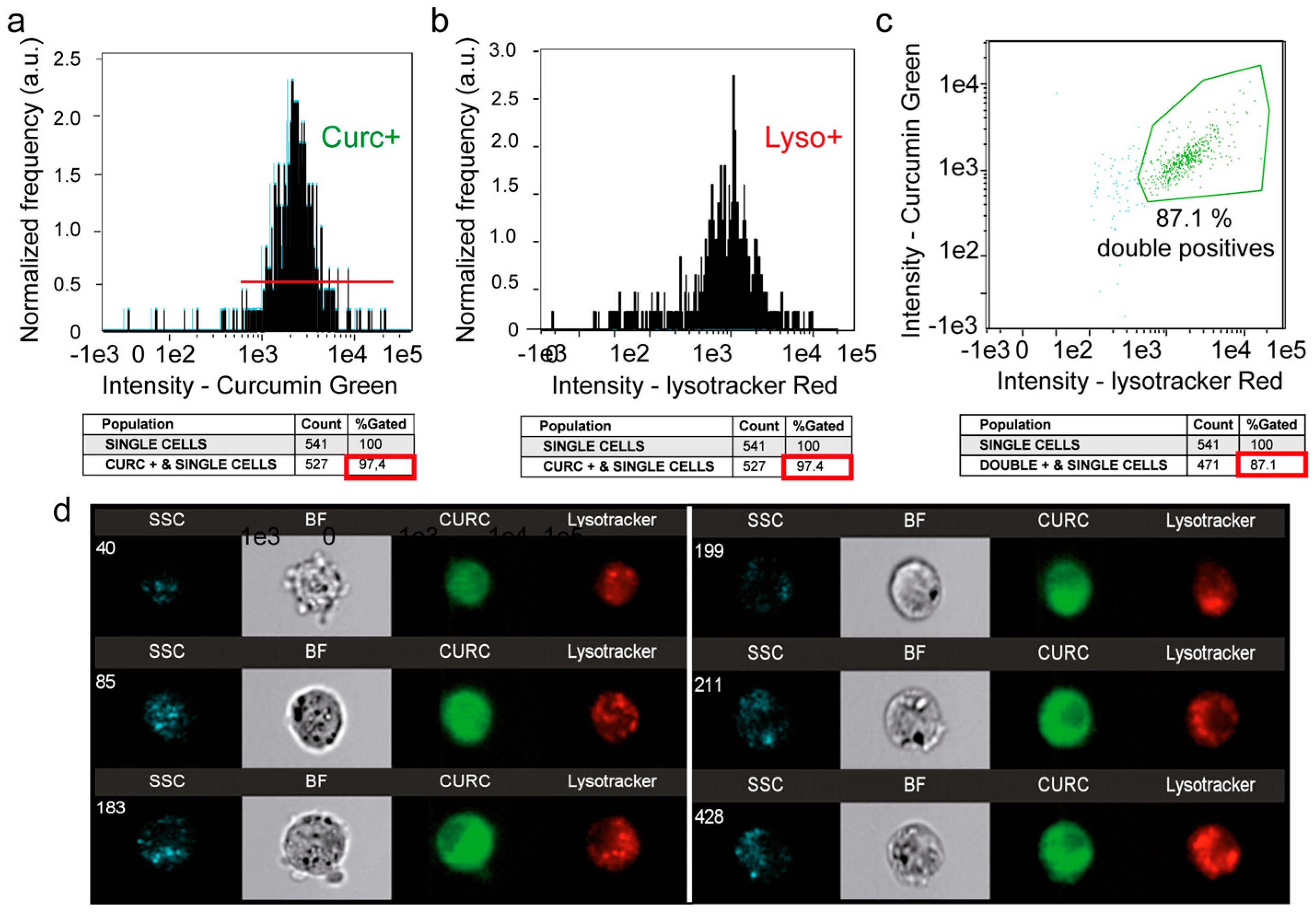
3.5. A Second Fraction of Curcumin Is Lysosomal
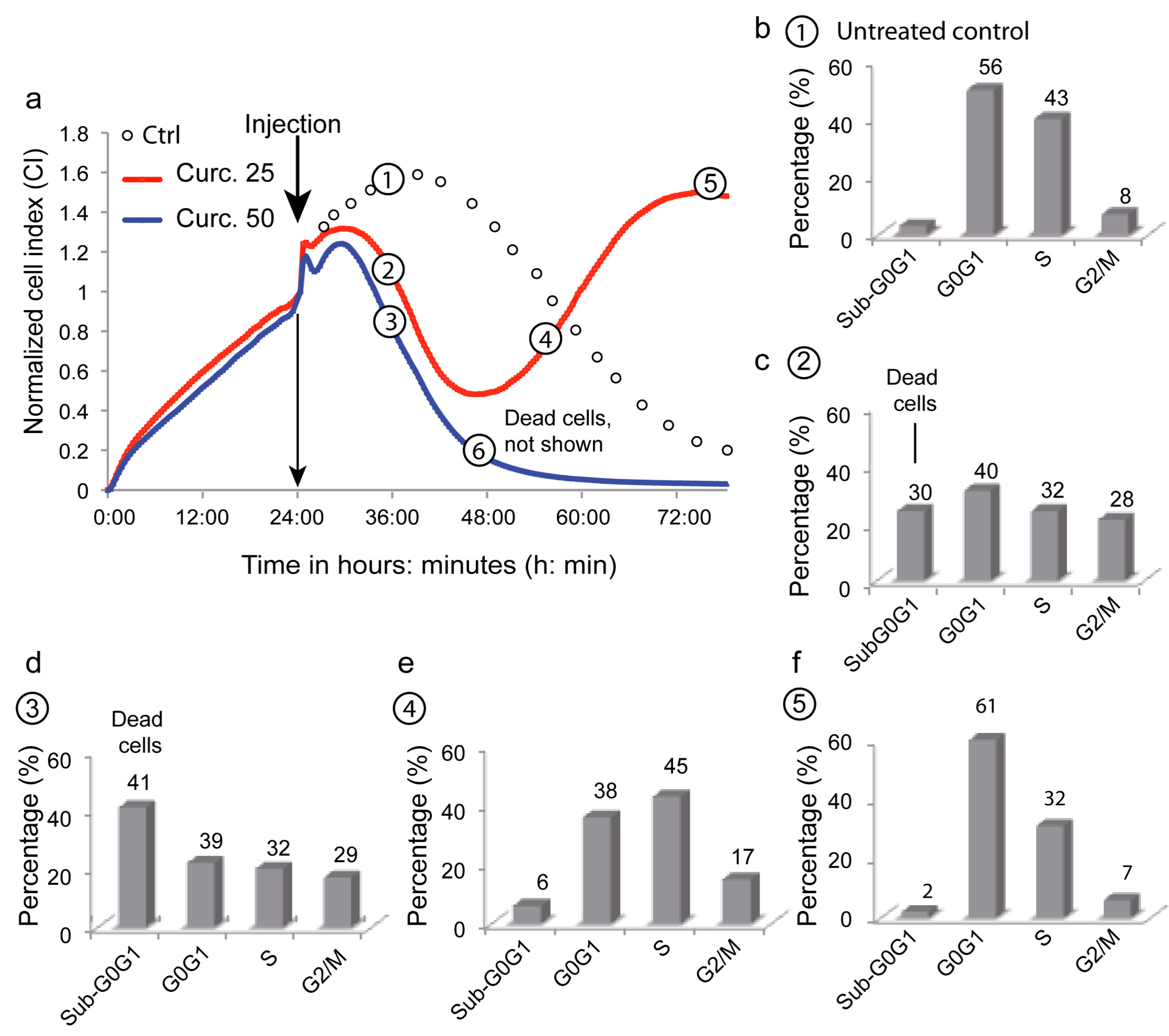
3.6. Depicting Curcumin-Induced Autophagy and Apoptosis by Impedancemetry Combined with Flow Cytometry
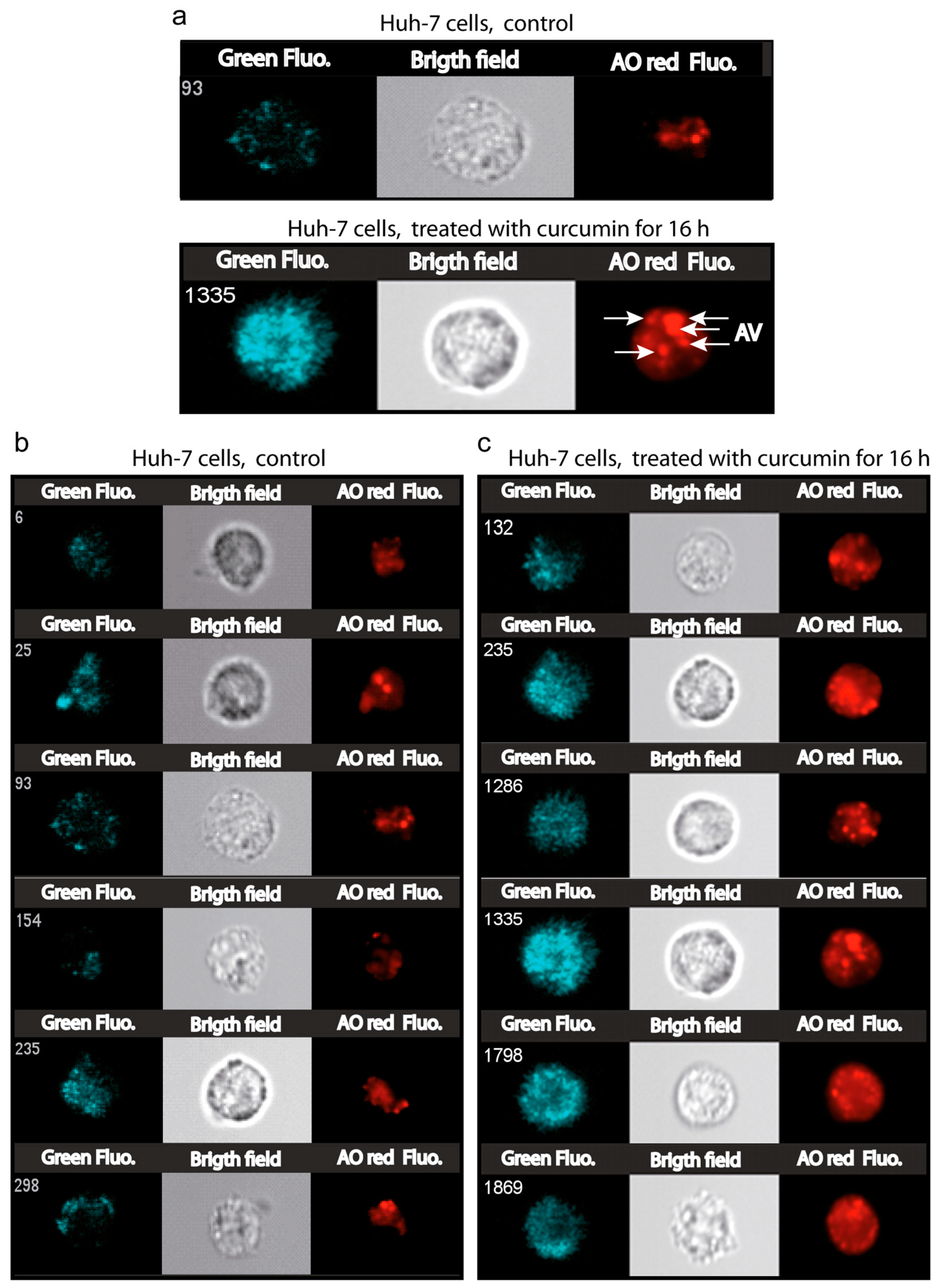
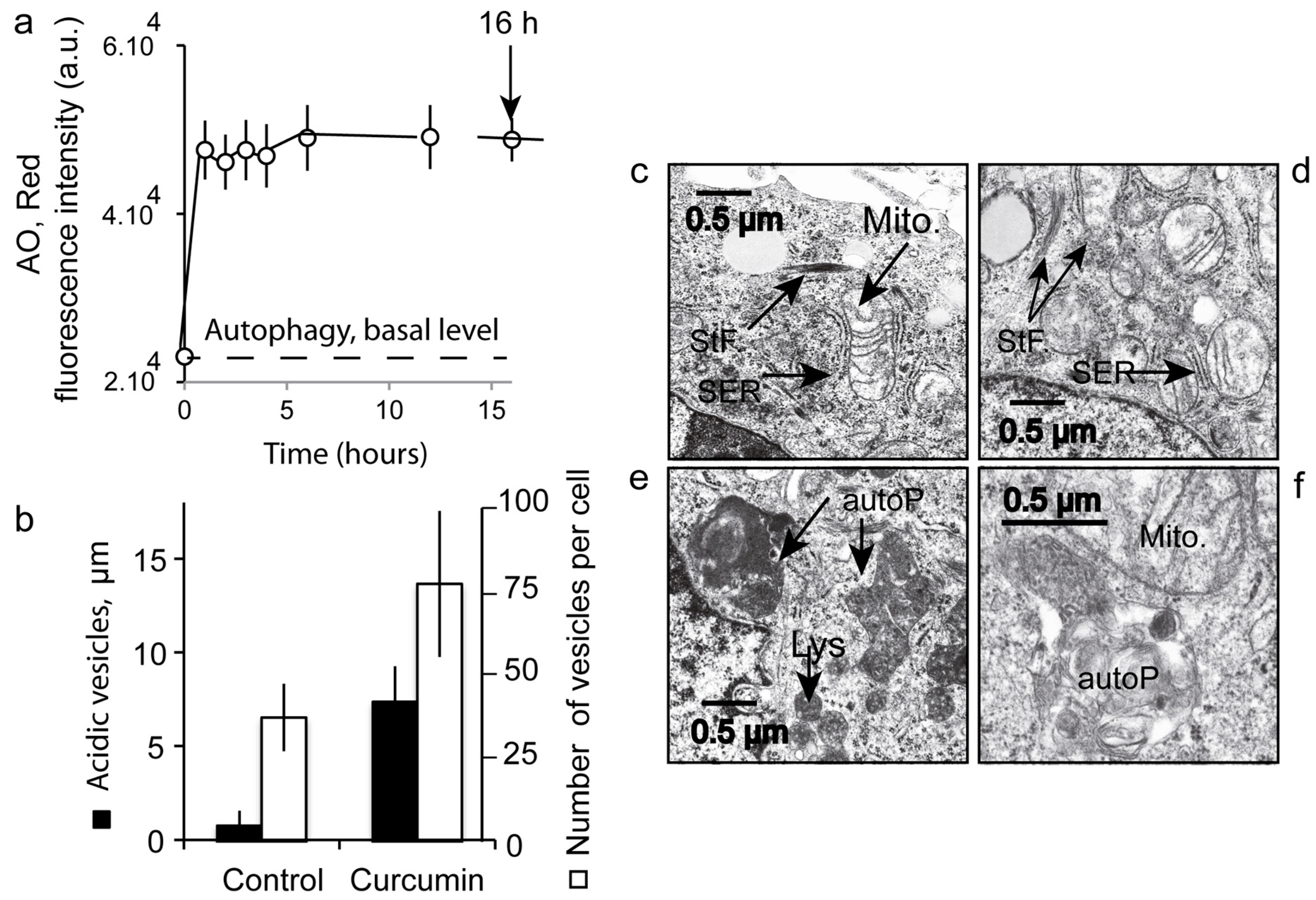
3.7. Number and Size of the Acidic Compartment as Evidence of Autolysosome Formation
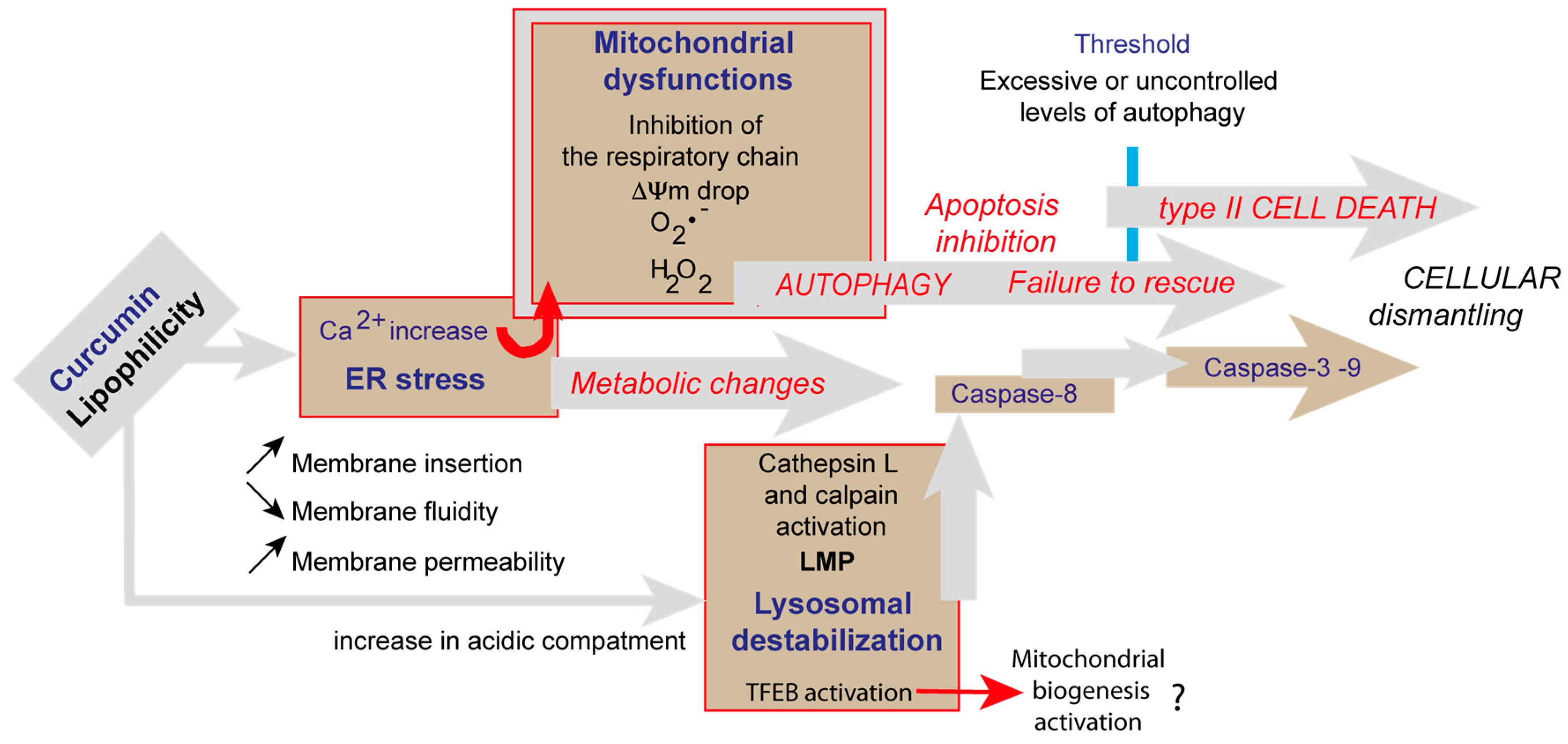
4. Discussion
4.1. Curcumin Does Not Localize at the Mitochondria
4.2. Curcumin-Induced Endoplasmic Reticulum Alterations and Its Consequences
4.3. Curcumin and Secondary Lysosomes Stained
4.4. Successful Curcumin-Induced Autophagy Detected by Impedancemetry Coupled to Multiparametric Flow Cytometry Analysis
4.5. Curcumin at Low Concentration Exacerbates Autophagy (Mitophagy)
Author Contributions
Funding
Conflicts of Interest
References
- Gupta, S.C.; Patchva, S.; Aggarwal, B.B. Therapeutic roles of curcumin: Lessons learned from clinical trials. AAPS J. 2013, 15, 195–218. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Sundaram, C.; Jhurani, S.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin and cancer: An “old-age” disease with an “age-old” solution. Cancer Lett. 2008, 267, 133–164. [Google Scholar] [CrossRef] [PubMed]
- Marchiani, A.; Rozzo, C.; Fadda, A.; Delogu, G.; Ruzza, P. Curcumin and curcumin-like molecules: From spice to drugs. Curr. Med. Chem. 2014, 21, 204–222. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsini, K.I. The chemistry of curcumin: From extraction to therapeutic agent. Molecules 2014, 19, 20091–20112. [Google Scholar] [CrossRef] [PubMed]
- Sharma, O.P. Antioxidant activity of curcumin and related compounds. Biochem. Pharm. 1976, 25, 1811–1812. [Google Scholar] [CrossRef]
- Lee, K.J.; Kim, Y.S.; Ma, J.Y. Separation and identification of curcuminoids from Asian turmeric (Curcuma longa L.) using RP-HPLC and LC-MS. Asian J. Chem. 2013, 25, 909. [Google Scholar] [CrossRef]
- Ali, I.; Haque, A.; Saleem, K. Separation and identification of curcuminoids in turmeric powder by HPLC using phenyl column. Anal. Methods 2014, 6, 2526–2536. [Google Scholar] [CrossRef]
- Moustapha, A.; Peretout, P.A.; Rainey, N.E.; Sureau, F.; Geze, M.; Petit, J.M.; Dewailly, E.; Slomianny, C.; Petit, P.X. Curcumin induces crosstalk between autophagy and apoptosis mediated by calcium release from the endoplasmic reticulum, lysosomal destabilization and mitochondrial events. Cell Death Discov. 2015, 1, 15017. [Google Scholar] [CrossRef]
- Szebeni, G.J.; Balazs, A.; Madarasz, I.; Pocz, G.; Ayaydin, F.; Kanizsai, I.; Fajka-Boja, R.; Alfoldi, R.; Hackler, L., Jr.; Puskas, L.G. Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells. Int. J. Mol. Sci. 2017, 18, 2105. [Google Scholar] [CrossRef]
- Mohammadi, K.; Thompson, K.H.; Patrick, B.O.; Storr, T.; Martins, C.; Polishchuk, E.; Yuen, V.G.; McNeill, J.H.; Orvig, C. Synthesis and characterization of dual function vanadyl, gallium and indium curcumin complexes for medicinal applications. J. Inorg. Biochem. 2005, 99, 2217–2225. [Google Scholar] [CrossRef]
- Barik, A.; Mishra, B.; Shen, L.; Mohan, H.; Kadam, R.M.; Dutta, S.; Zhang, H.Y.; Priyadarsini, K.I. Evaluation of a new copper(II)-curcumin complex as superoxide dismutase mimic and its free radical reactions. Free Radic. Biol. Med. 2005, 39, 811–822. [Google Scholar] [CrossRef] [PubMed]
- Karewicz, A.; Bielska, D.; Gzyl-Malcher, B.; Kepczynski, M.; Lach, R.; Nowakowska, M. Interaction of curcumin with lipid monolayers and liposomal bilayers. Colloids Surf. B Biointerfaces 2011, 88, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Chen, Y.; Yang, N.; Zhu, X.; Sun, L.; Li, G. Interaction between curcumin and mimetic biomembrane. Sci. China Life Sci. 2012, 55, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lee, C.C.; Hung, W.C.; Chen, F.Y.; Lee, M.T.; Huang, H.W. The bound states of amphipathic drugs in lipid bilayers: Study of curcumin. Biophys. J. 2008, 95, 2318–2324. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.C.; Chen, F.Y.; Lee, C.C.; Sun, Y.; Lee, M.T.; Huang, H.W. Membrane-thinning effect of curcumin. Biophys. J. 2008, 94, 4331–4338. [Google Scholar] [CrossRef] [PubMed]
- Ingolfsson, H.I.; Koeppe, R.E.; Andersen, O.S. Curcumin is a modulator of bilayer material properties. Biochemistry 2007, 46, 10384–10391. [Google Scholar] [CrossRef]
- Koiram, P.R.; Veerapur, V.P.; Kunwar, A.; Mishra, B.; Barik, A.; Priyadarsini, I.K.; Mazhuvancherry, U.K. Effect of curcumin and curcumin copper complex (1:1) on radiation-induced changes of anti-oxidant enzymes levels in the livers of Swiss albino mice. J. Radiat. Res. 2007, 48, 241–245. [Google Scholar] [CrossRef]
- Rainey, N.; Motte, L.; Aggarwal, B.B.; Petit, P.X. Curcumin hormesis mediates a cross-talk between autophagy and cell death. Cell Death Dis. 2015, 6, e2003. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Gupta, S.C.; Sung, B. Curcumin: An orally bioavailable blocker of TNF and other pro-inflammatory biomarkers. Br. J. Pharmacol. 2013, 169, 1672–1692. [Google Scholar] [CrossRef]
- Kunnumakkara, A.B.; Anand, P.; Aggarwal, B.B. Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett. 2008, 269, 199–225. [Google Scholar] [CrossRef]
- Gupta, S.C.; Prasad, S.; Kim, J.H.; Patchva, S.; Webb, L.J.; Priyadarsini, I.K.; Aggarwal, B.B. Multitargeting by curcumin as revealed by molecular interaction studies. Nat. Prod. Rep. 2011, 28, 1937–1955. [Google Scholar] [CrossRef] [PubMed]
- Sureau, F.; Chinsky, L.; Duquesne, M.; Laigle, A.; Turpin, P.Y.; Amirand, C.; Ballini, J.P.; Vigny, P. Microspectrofluorimetric study of the kinetics of cellular uptake and metabolization of benzo(a)pyrene in human T 47D mammary tumor cells: Evidence for cytochrome P1450 induction. Eur. Biophys. J. 1990, 18, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Basiji, D.A.; Ortyn, W.E.; Liang, L.; Venkatachalam, V.; Morrissey, P. Cellular image analysis and imaging by flow cytometry. Clin. Lab. Med. 2007, 27, 653–670. [Google Scholar] [CrossRef] [PubMed]
- Zamzami, N.; Marchetti, P.; Castedo, M.; Decaudin, D.; Macho, A.; Hirsch, T.; Susin, S.A.; Petit, P.X.; Mignotte, B.; Kroemer, G. Sequential reduction of mitochondrial transmembrane potential and generation of reactive oxygen species in early programmed cell death. J. Exp. Med. 1995, 182, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Gendron, M.C.; Schrantz, N.; Metivier, D.; Kroemer, G.; Maciorowska, Z.; Sureau, F.; Koester, S.; Petit, P.X. Oxidation of pyridine nucleotides during Fas- and ceramide-induced apoptosis in Jurkat cells: Correlation with changes in mitochondria, glutathione depletion, intracellular acidification and caspase 3 activation. Biochem. J. 2001, 353, 357–367. [Google Scholar] [PubMed]
- Petit, P.X.; Lecoeur, H.; Zorn, E.; Dauguet, C.; Mignotte, B.; Gougeon, M.L. Alterations of mitochondrial structure and function are early events of dexamethasone-induced thymocyte apoptosis. J. Cell. Biol. 1995, 130, 157–167. [Google Scholar] [CrossRef]
- Hanneman, W.H.; Legare, M.E.; Tiffany-Castiglioni, E.; Safe, S.H. The need for cellular, biochemical, and mechanistic studies. Neurotoxicol. Teratol. 1996, 18, 247–250. [Google Scholar] [CrossRef]
- Solly, K.; Wang, X.; Xu, X.; Strulovici, B.; Zheng, W. Application of real-time cell electronic sensing (RT-CES) technology to cell-based assays. Assay. Drug. Dev. Technol. 2004, 2, 363–372. [Google Scholar] [CrossRef]
- Ke, N.; Wang, X.; Xu, X.; Abassi, Y.A. The xCELLigence system for real-time and label-free monitoring of cell viability. Methods Mol. Biol. 2011, 740, 33–43. [Google Scholar]
- Zamzami, N.; Marchetti, P.; Castedo, M.; Zanin, C.; Vayssière, J.L.; Petit, P.X.; Kroemer, G. Reduction in mitochondrial potential constitutes an early irrseversible step of programmed lymphocyte death in vivo. J. Exp. Med. 1995, 181, 1661–1672. [Google Scholar] [CrossRef]
- Thome, M.P.; Filippi-Chiela, E.C.; Villodre, E.S.; Migliavaca, C.B.; Onzi, G.R.; Felipe, K.B.; Lenz, G. Ratiometric analysis of Acridine Orange staining in the study of acidic organelles and autophagy. J. Cell. Sci. 2016, 129, 4622–4632. [Google Scholar] [CrossRef] [PubMed]
- Kuttikrishnan, S.; Siveen, K.S.; Prabhu, K.S.; Khan, A.Q.; Ahmed, E.I.; Akhtar, S.; Ali, T.A.; Merhi, M.; Dermime, S.; Steinhoff, M.; et al. Curcumin Induces Apoptotic Cell Death via Inhibition of PI3-Kinase/AKT Pathway in B-Precursor Acute Lymphoblastic Leukemia. Front. Oncol. 2019, 9, 484. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, M.R.; Jardim, F.R.; Setzer, W.N.; Nabavi, S.M.; Nabavi, S.F. Curcumin, mitochondrial biogenesis, and mitophagy: Exploring recent data and indicating future needs. Biotechnol. Adv. 2016, 34, 813–826. [Google Scholar] [CrossRef]
- Yang, C.; Ma, X.; Wang, Z.; Zeng, X.; Hu, Z.; Ye, Z.; Shen, G. Curcumin induces apoptosis and protective autophagy in castration-resistant prostate cancer cells through iron chelation. Drug Des. Dev. Ther. 2017, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Csordas, G.; Weaver, D.; Hajnoczky, G. Endoplasmic Reticulum-Mitochondrial Contactology: Structure and Signaling Functions. Trends Cell Biol. 2018, 28, 523–540. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Min, K.T. The Interface between ER and Mitochondria: Molecular Compositions and Functions. Mol. Cells 2018, 41, 1000–1007. [Google Scholar]
- Stacchiotti, A.; Favero, G.; Lavazza, A.; Garcia-Gomez, R.; Monsalve, M.; Rezzani, R. Perspective: Mitochondria-ER Contacts in Metabolic Cellular Stress Assessed by Microscopy. Cells 2018, 8, 5. [Google Scholar] [CrossRef]
- Reddy, C.A.; Somepalli, V.; Golakoti, T.; Kanugula, A.K.; Karnewar, S.; Rajendiran, K.; Vasagiri, N.; Prabhakar, S.; Kuppusamy, P.; Kotamraju, S.; et al. Mitochondrial-targeted curcuminoids: A strategy to enhance bioavailability and anticancer efficacy of curcumin. PLoS ONE 2014, 9, e89351. [Google Scholar] [CrossRef]
- Li, X.; Xu, Z.; Wang, S.; Guo, H.; Dong, S.; Wang, T.; Zhang, L.; Jiang, Z. Emodin ameliorates hepatic steatosis through endoplasmic reticulum-stress sterol regulatory element-binding protein 1c pathway in liquid fructose-feeding rats. Hepatol. Res. 2016, 46, E105–E117. [Google Scholar] [CrossRef]
- Zhang, L.; Su, F.; Kong, X.; Lee, F.; Day, K.; Gao, W.; Vecera, M.E.; Sohr, J.M.; Buizer, S.; Tian, Y.; et al. Ratiometric fluorescent pH-sensitive polymers for high-throughput monitoring of extracellular pH. RSC Adv. 2016, 6, 46134–46142. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The Unfolded Protein Response: From Stress Pathway to Homeostatic Regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Bilmen, J.G.; Wootton, L.L.; Godfrey, R.E.; Smart, O.S.; Michelangeli, F. Inhibition of SERCA Ca2+ pumps by 2-aminoethoxydiphenyl borate (2-APB). 2-APB reduces both Ca2+ binding and phosphoryl transfer from ATP, by interfering with the pathway leading to the Ca2+-binding sites. Eur. J. Biochem. 2002, 269, 3678–3687. [Google Scholar] [CrossRef] [PubMed]
- Bilmen, J.G.; Khan, S.Z.; Javed, M.H.; Michelangeli, F. Inhibition of the SERCA Ca2+ pumps by curcumin. Curcumin putatively stabilizes the interaction between the nucleotide-binding and phosphorylation domains in the absence of ATP. Eur. J. Biochem. 2001, 268, 6318–6327. [Google Scholar] [CrossRef] [PubMed]
- Nishitoh, H. CHOP is a multifunctional transcription factor in the ER stress response. J. Biochem. 2012, 151, 217–219. [Google Scholar] [CrossRef]
- Roczniak-Ferguson, A.; Petit, C.S.; Froehlich, F.; Qian, S.; Ky, J.; Angarola, B.; Walther, T.C.; Ferguson, S.M. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal 2012, 5, ra42. [Google Scholar] [CrossRef]
- Settembre, C.; Zoncu, R.; Medina, D.L.; Vetrini, F.; Erdin, S.; Huynh, T.; Ferron, M.; Karsenty, G.; Vellard, M.C.; Facchinetti, V.; et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012, 31, 1095–1108. [Google Scholar] [CrossRef]
- Li, J.; Ni, M.; Lee, B.; Barron, E.; Hinton, D.R.; Lee, A.S. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008, 15, 1460–1471. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Xu, J.; Lu, Y.; Jiang, J.; Wang, L.; Shen, H.M.; Xia, D. Curcumin targets the TFEB-lysosome pathway for induction of autophagy. Oncotarget 2016, 7, 75659–75671. [Google Scholar] [CrossRef]
- Choudhuri, T.; Pal, S.; Agwarwal, M.L.; Das, T.; Sa, G. Curcumin induces apoptosis in human breast cancer cells through p53-dependent Bax induction. FEBS Lett. 2002, 512, 334–340. [Google Scholar] [CrossRef]
- Sahu, R.P.; Batra, S.; Srivastava, S.K. Activation of ATM/Chk1 by curcumin causes cell cycle arrest and apoptosis in human pancreatic cancer cells. Br. J. Cancer 2009, 100, 1425–1433. [Google Scholar] [CrossRef]
- Kroemer, G.; Petit, P.X.; Zamzami, N.; Vayssière, J.-L.; Mignotte, B. The biochemistry of apoptosis. FASEB J. 1995, 9, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.; Allais, D.P.; Duroux, J.L.; Basly, J.P.; Durand-Fontanier, S.; Delage, C. Inhibitory effect of curcuminoids on MCF-7 cell proliferation and structure-activity relationships. Cancer Lett. 1998, 129, 111–116. [Google Scholar] [CrossRef]
- Rieder, C.L.; Maiato, H. Stuck in division or passing through: What happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 2004, 7, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Singh, P.; Panda, D. Curcumin suppresses the dynamic instability of microtubules, activates the mitotic checkpoint and induces apoptosis in MCF-7 cells. FEBS J. 2010, 277, 3437–3448. [Google Scholar] [CrossRef]
- Palmieri, M.; Pal, R.; Nelvagal, H.R.; Lotfi, P.; Stinnett, G.R.; Seymour, M.L.; Chaudhury, A.; Bajaj, L.; Bondar, V.V.; Bremner, L.; et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 2017, 8, 14338. [Google Scholar] [CrossRef]
- Zhao, F.; Gong, Y.; Hu, Y.; Lu, M.; Wang, J.; Dong, J.; Chen, D.; Chen, L.; Fu, F.; Qiu, F. Curcumin and its major metabolites inhibit the inflammatory response induced by lipopolysaccharide: Translocation of nuclear factor-kappaB as potential target. Mol. Med. Rep. 2015, 11, 3087–3093. [Google Scholar] [CrossRef]
- Miranda, J.; Lasa, A.; Aguirre, L.; Fernandez-Quintela, A.; Milton, I.; Portillo, M.P. Potential application of non-flavonoid phenolics in diabetes: Antiinflammatory effects. Curr. Med. Chem. 2015, 22, 112–131. [Google Scholar] [CrossRef]
- Li, X.N.; Ma, L.Y.; Ji, H.; Qin, Y.H.; Jin, S.S.; Xu, L.X. Resveratrol protects against oxidative stress by activating the Keap-1/Nrf2 antioxidant defense system in obese-asthmatic rats. Exp. Ther. Med. 2018, 16, 4339–4348. [Google Scholar] [CrossRef]
- Millot, C.; Millot, J.M.; Morjani, H.; Desplaces, A.; Manfait, M. Characterization of acidic vesicles in multidrug-resistant and sensitive cancer cells by acridine orange staining and confocal microspectrofluorometry. J. Histochem. Cytochem. 1997, 45, 1255–1264. [Google Scholar] [CrossRef]
- Settembre, C.; Di Malta, C.; Polito, V.A.; Garcia Arencibia, M.; Vetrini, F.; Erdin, S.; Erdin, S.U.; Huynh, T.; Medina, D.; Colella, P.; et al. TFEB links autophagy to lysosomal biogenesis. Science 2011, 332, 1429–1433. [Google Scholar] [CrossRef]
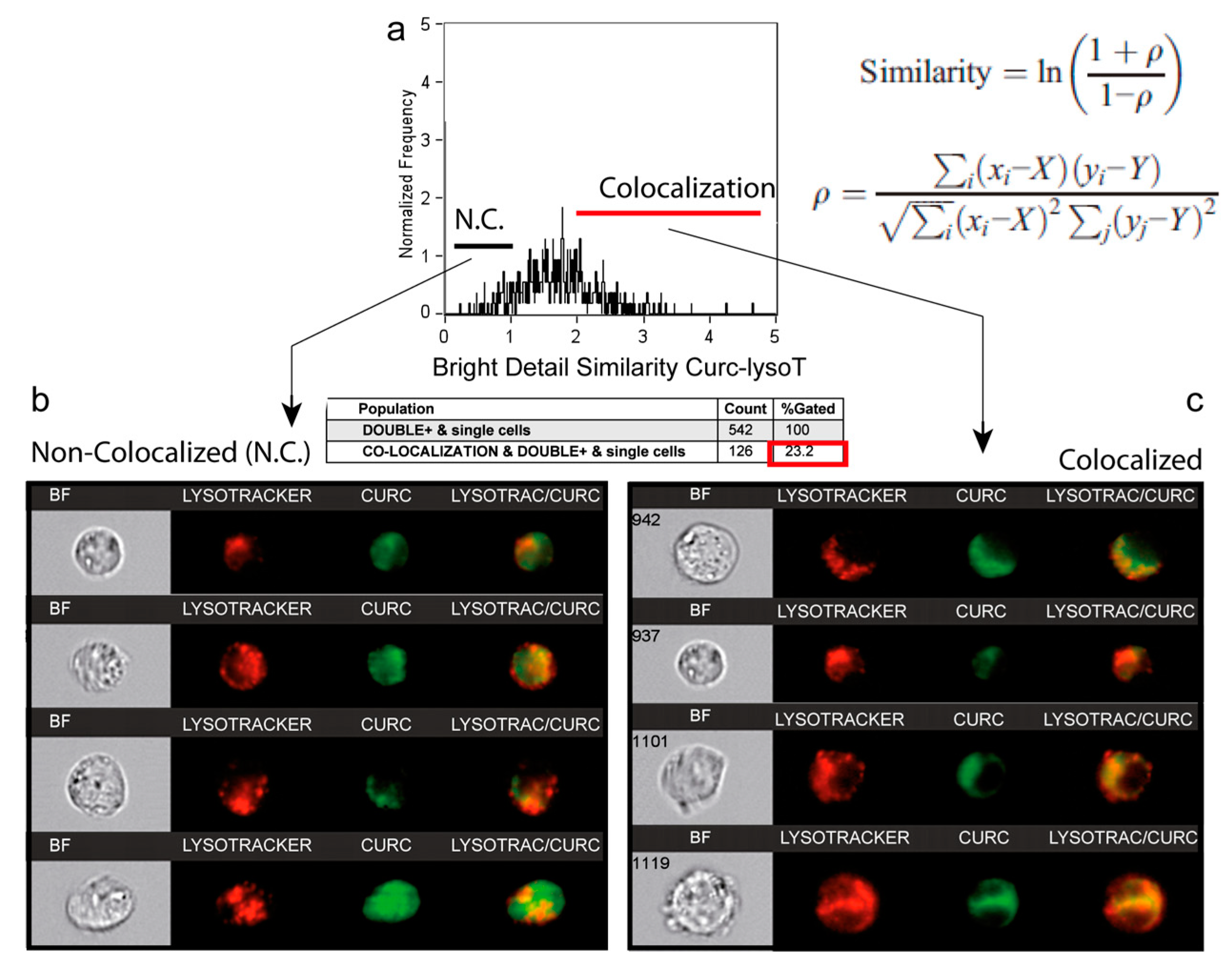
| Curcumin Incubation 3 h | 5 μM | 20 μM | 50 μM |
| True colocalisation of curcumin and lysotracker red cells (% of cells) | 1.95 | 14.8 | 23.4 |
| Double curcumin and lysotracker red Staining (% of cells) | 63.6 | 66.7 | 95.1 |
| Curcumin Incubation 24 h | 5 μM | 20 μM | 50 μM |
| True colocalisation of curcumin and lysotracker red cells (% of cells) | 67.8 | 87.6 | 97.8 |
| Double curcumin and lysotracker red Staining (% of cells) | 92.2 | 95.3 | 96.8 |
| Metabolic Events (% of Cells) | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| ΔΨm high | 95 ± 2 | 58 ± 4 | 50 ± 5 | 62 ± 5 | 97 ± 2 | 4 ± 1 |
| Superoxide anions | 4 ± 3 | 36 ± 6 | 49 ± 5 | 12 ± 2 | 2 ± 2 | NA |
| Peroxidase activity | 3 ± 4 | 35 ± 7 | 48 ± 6 | 10 ± 3 | 3 ± 3 | NA |
| Calcium rise | 1 ± 1 | 30 ± 8 | 45 ± 7 | 5 ± 3 | 1 ± 1 | NA |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sala de Oyanguren, F.J.; Rainey, N.E.; Moustapha, A.; Saric, A.; Sureau, F.; O’Connor, J.-E.; Petit, P.X. Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization. Cells 2020, 9, 361. https://doi.org/10.3390/cells9020361
Sala de Oyanguren FJ, Rainey NE, Moustapha A, Saric A, Sureau F, O’Connor J-E, Petit PX. Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization. Cells. 2020; 9(2):361. https://doi.org/10.3390/cells9020361
Chicago/Turabian StyleSala de Oyanguren, Francisco J., Nathan E. Rainey, Aoula Moustapha, Ana Saric, Franck Sureau, José-Enrique O’Connor, and Patrice X. Petit. 2020. "Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization" Cells 9, no. 2: 361. https://doi.org/10.3390/cells9020361
APA StyleSala de Oyanguren, F. J., Rainey, N. E., Moustapha, A., Saric, A., Sureau, F., O’Connor, J.-E., & Petit, P. X. (2020). Highlighting Curcumin-Induced Crosstalk between Autophagy and Apoptosis as Supported by Its Specific Subcellular Localization. Cells, 9(2), 361. https://doi.org/10.3390/cells9020361






