Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient’s Cohort
2.2. Immunohistochemistry
2.3. Digital Microscopy
2.4. Statistical Analysis
2.5. Cell Cultures
2.6. Neutrophil Isolation and Transwell Migration Assay
2.7. Quantitative RT-PCR (qRT-PCR)
2.8. STAT3 Silencing
2.9. Data Preprocessing and Statistical Analysis of the TCGA, GSE32894 and GSE124305 Datasets
3. Results
3.1. Patient Characteristics
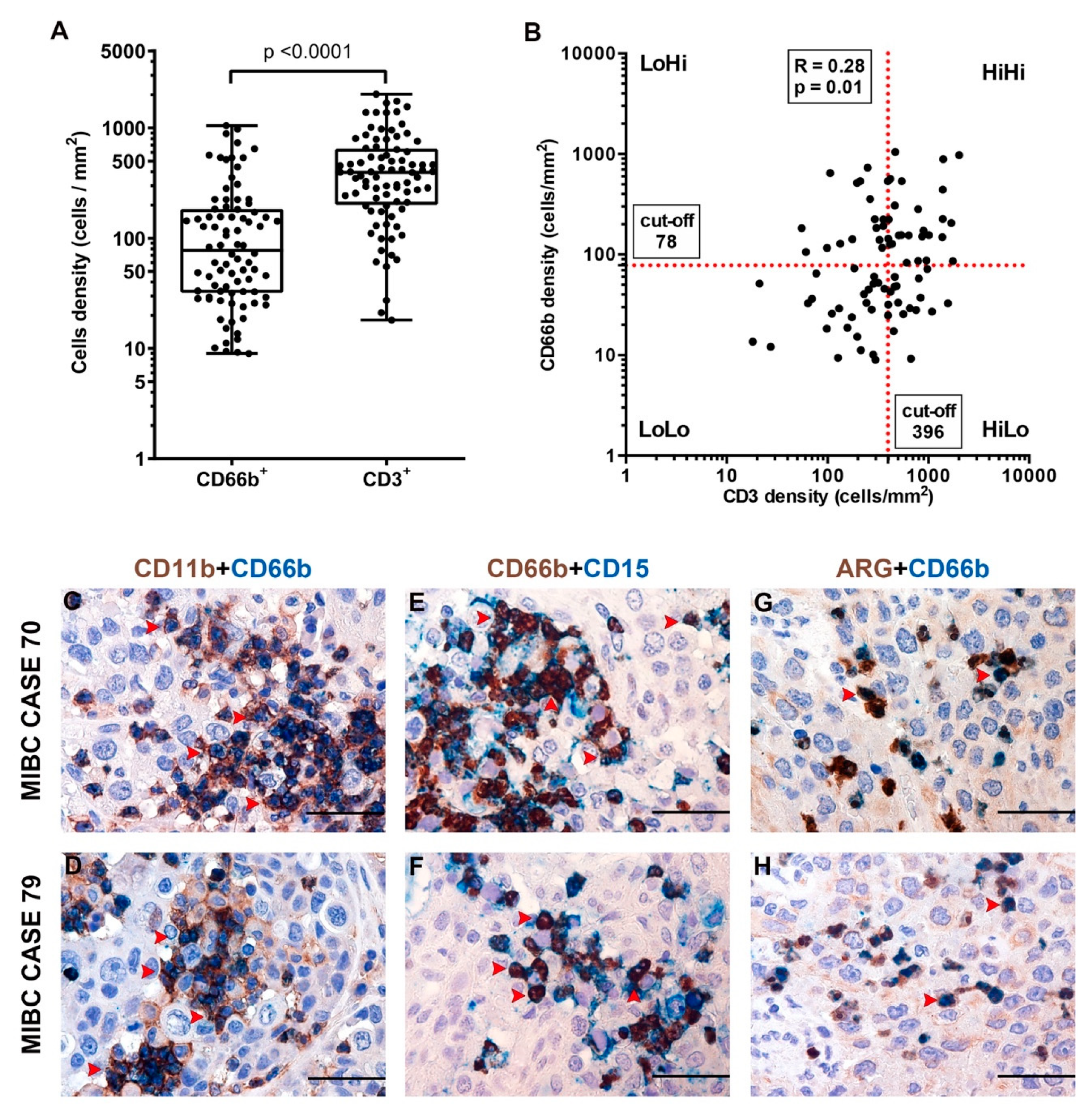
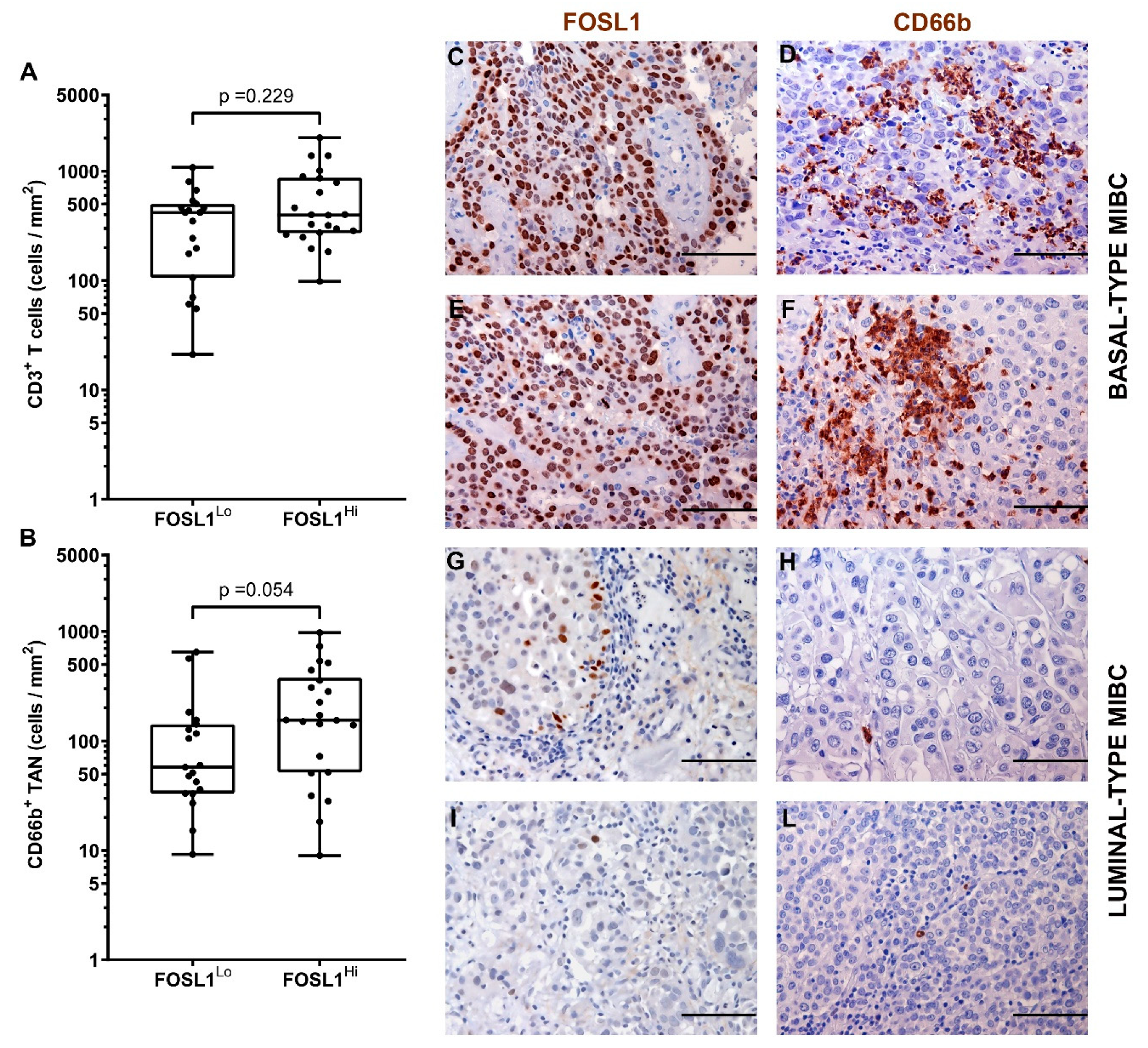
3.2. CD66b+ TAN and CD3+ T Cell Immune-Contexture in Muscle Invasive UBC
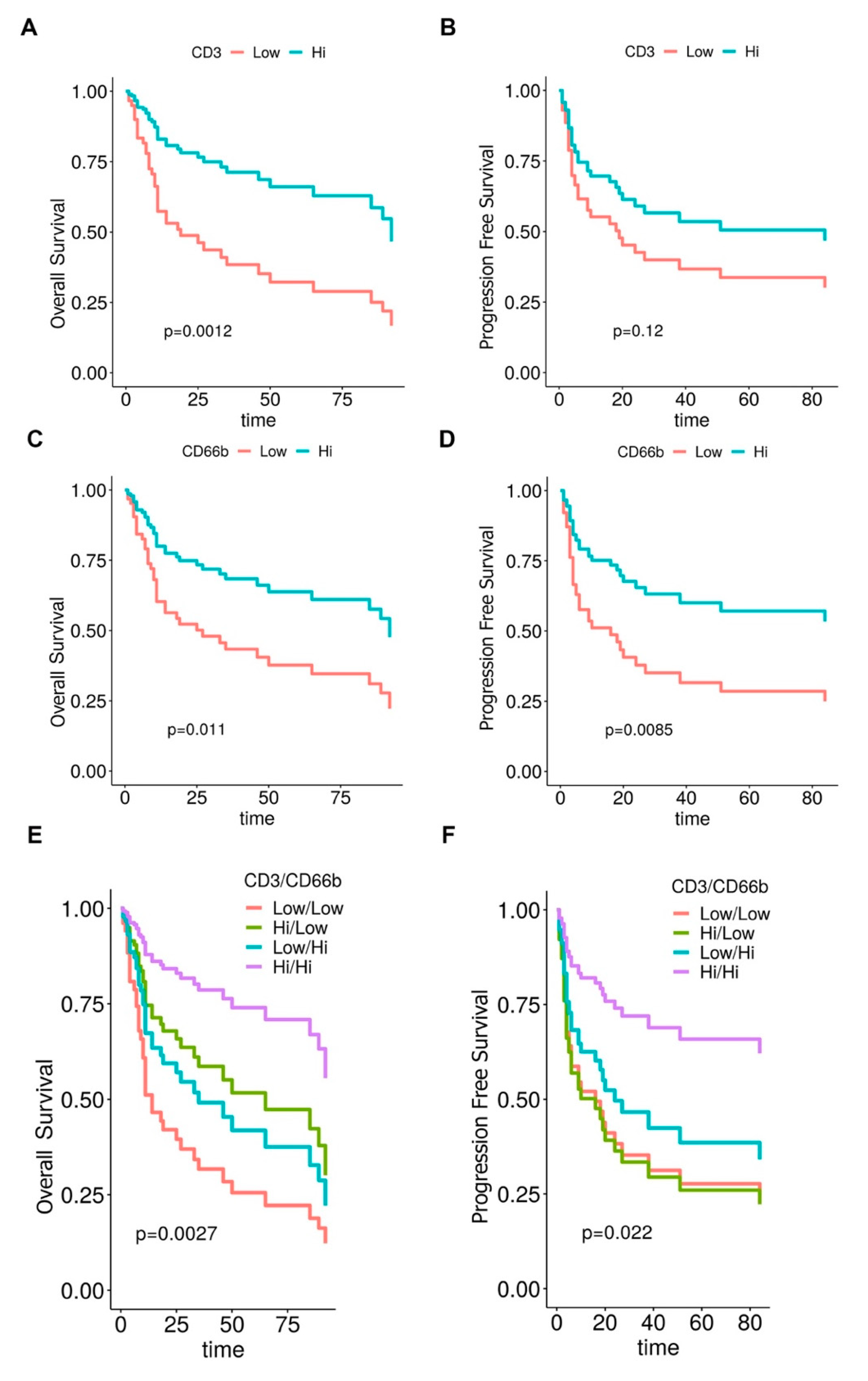
3.3. Clinical Significance of CD66b+ TAN and CD3+ T Cell Immune-Contexture in MIBC
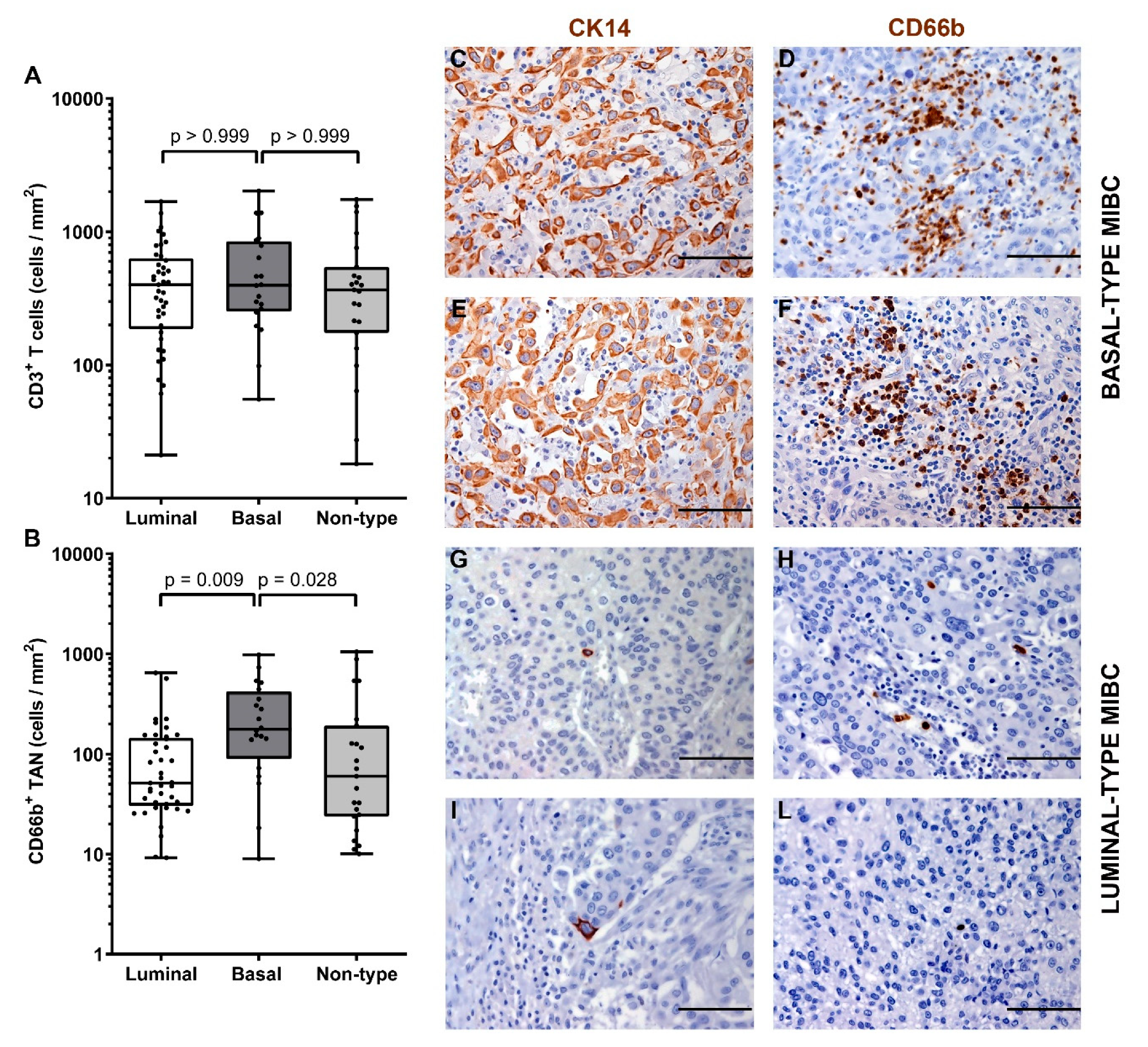
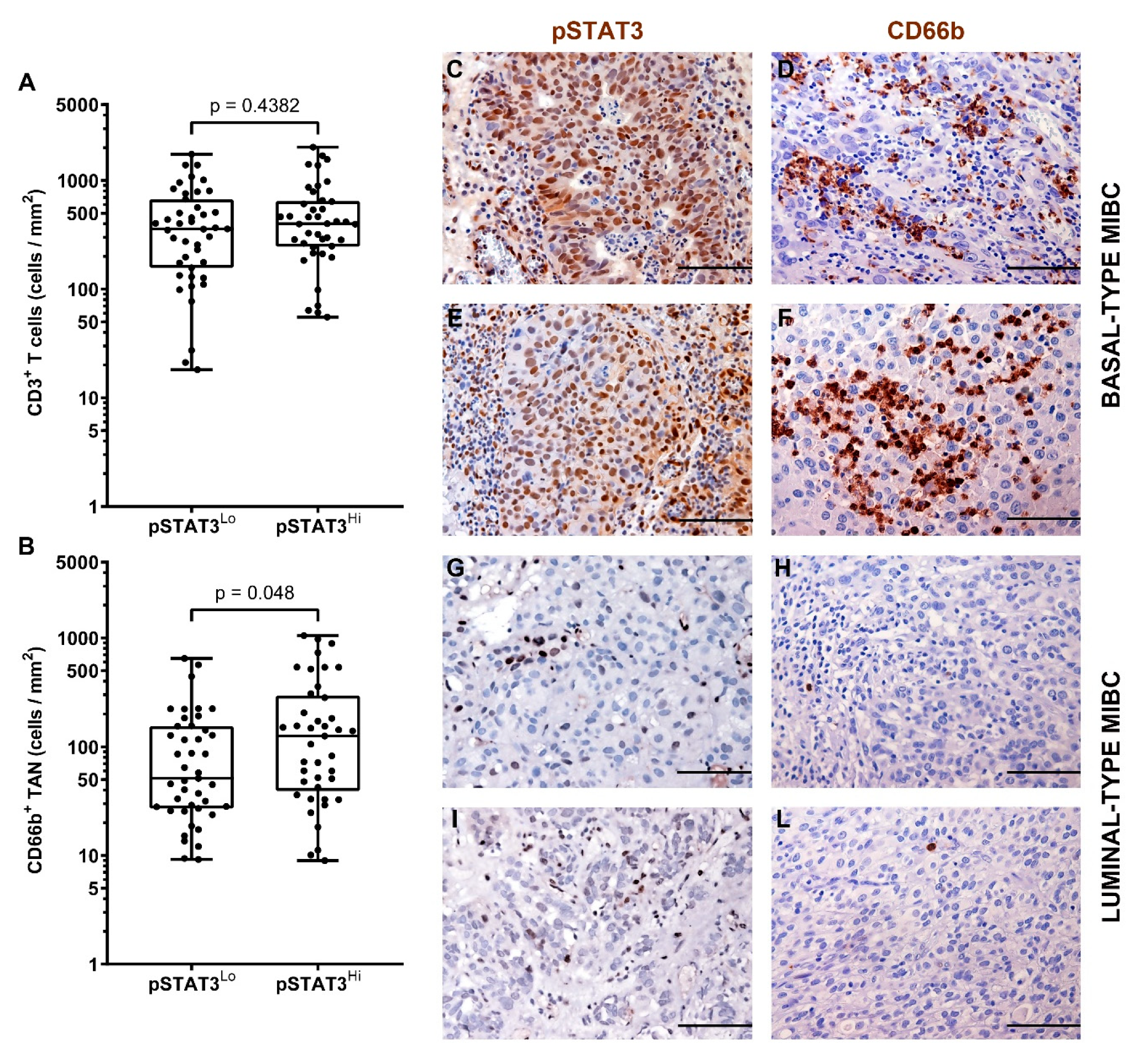
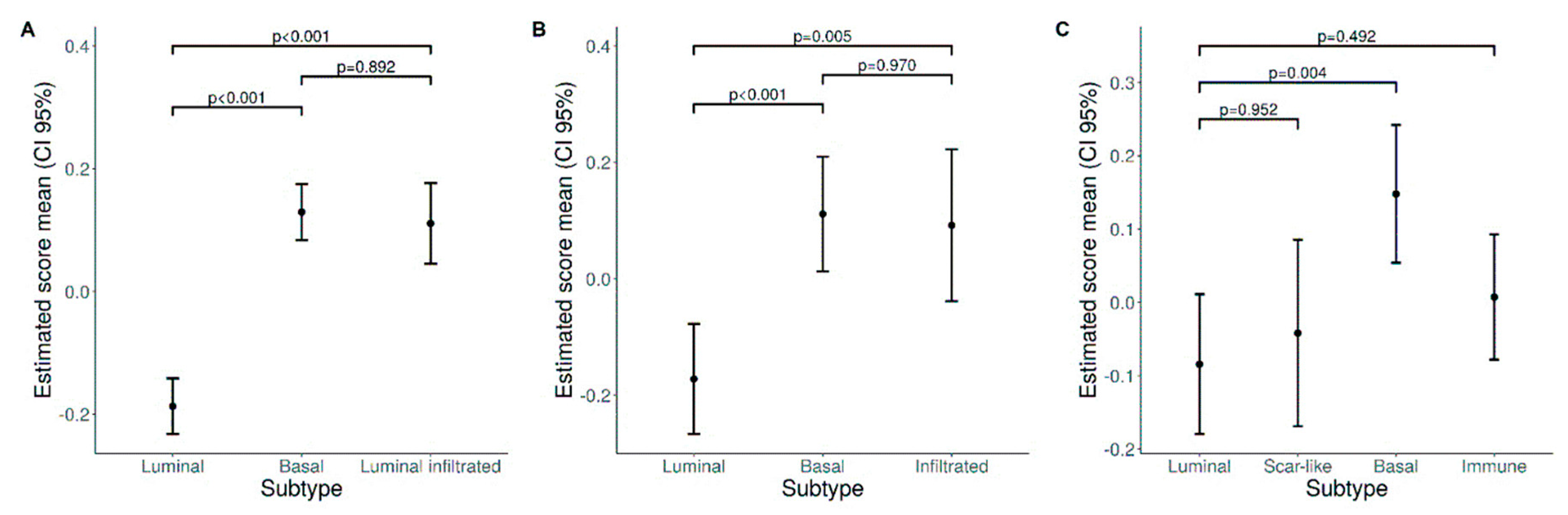
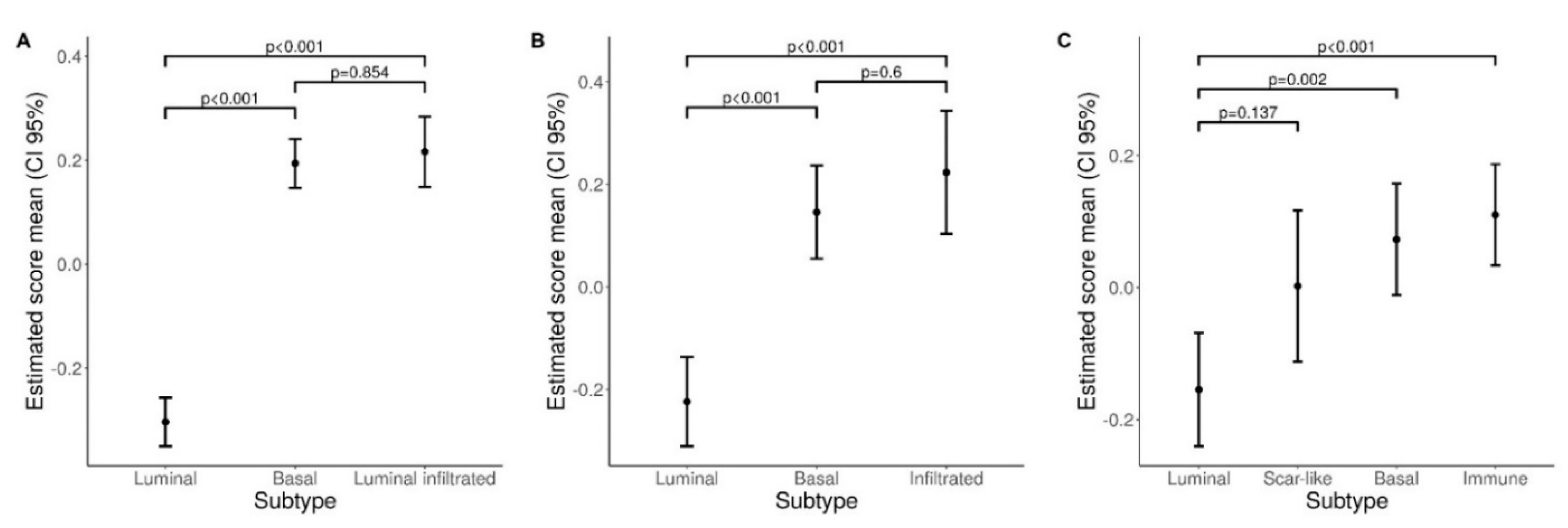
3.4. A Higher Density of CD66b+ TAN Is Restricted to Basal Type MIBC Expressing the Transcription Factor STAT3
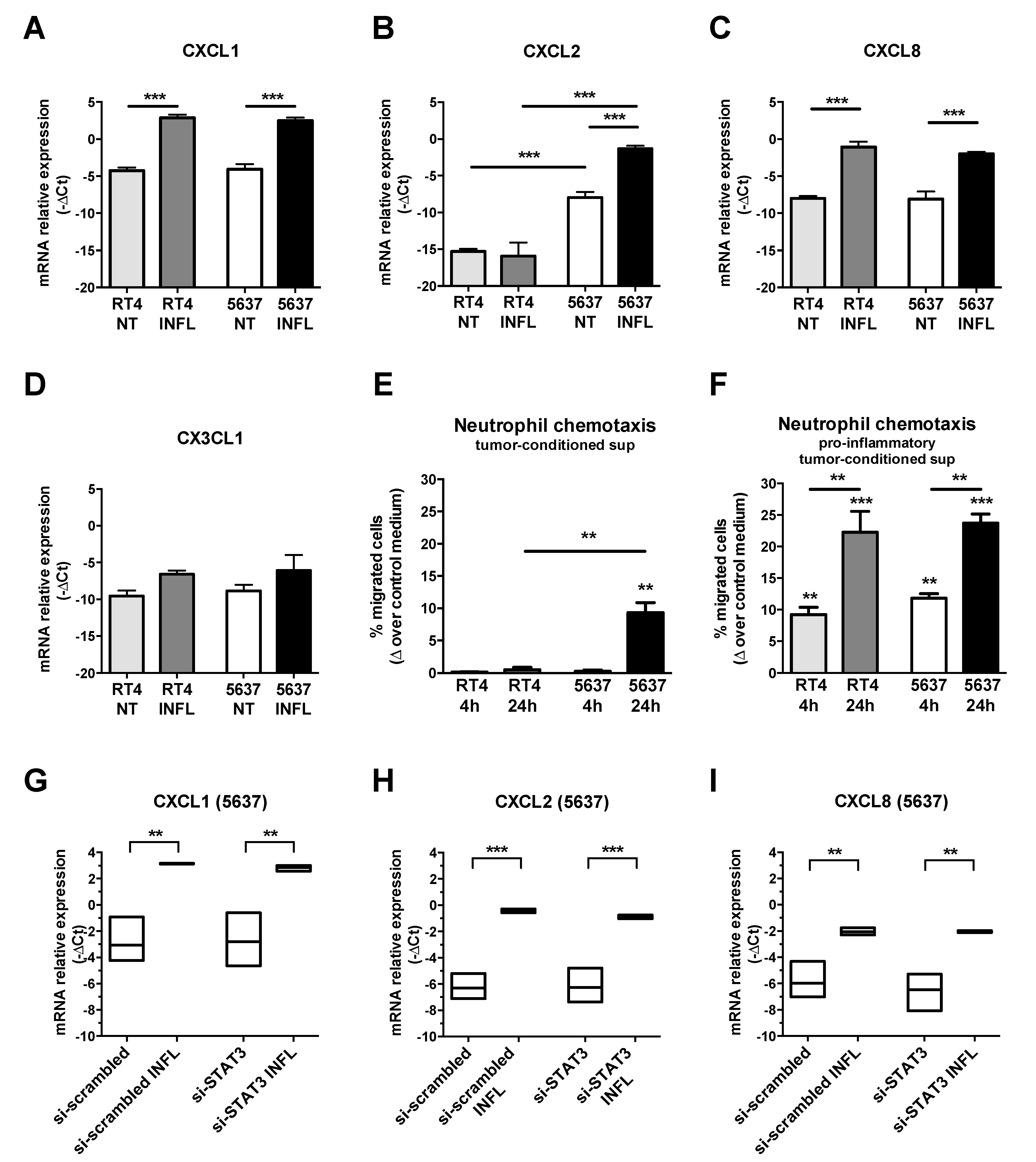
3.5. Expression of the TAN-Attracting Chemokines in Basal-Type UBC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Antoni, S.; Ferlay, J.; Soerjomataram, I.; Znaor, A.; Jemal, A.; Bray, F. Bladder Cancer Incidence and Mortality: A Global Overview and Recent Trends. Eur. Urol. 2017, 71, 96–108. [Google Scholar] [CrossRef]
- Babjuk, M.; Oosterlinck, W.; Sylvester, R.; Kaasinen, E.; Böhle, A.; Palou-Redorta, J.; Rouprêt, M. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur. Urol. 2011, 59, 997–1008. [Google Scholar] [CrossRef]
- Bellmunt, J.; Petrylak, D.P. New Therapeutic Challenges in Advanced Bladder Cancer. YSONC 2012, 39, 598–607. [Google Scholar] [CrossRef]
- Sridhar, S.S. Evolving Treatment of Advanced Urothelial Cancer. J. Oncol. Pract. 2018, 13, 309–316. [Google Scholar] [CrossRef]
- Alifrangis, C.; McGovern, U.; Freeman, A.; Powles, T.; Linch, M. Molecular and histopathology directed therapy for advanced bladder cancer. Nat. Rev. Urol. 2019, 16, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Inokuchi, J.; Eto, M. Profile of pembrolizumab in the treatment of patients with unresectable or metastatic urothelial carcinoma. Cancer Manag. Res. 2019, 11, 4519–4528. [Google Scholar] [CrossRef] [PubMed]
- Balar, A.V.; Galsky, M.D.; Rosenberg, J.E.; Powles, T.; Petrylak, D.P.; Bellmunt, J.; Loriot, Y.; Necchi, A.; Hoffman-Censits, J.; Perez-Gracia, J.L.; et al. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: A single-arm, multicentre, phase 2 trial. Lancet 2017, 389, 67–76. [Google Scholar] [CrossRef]
- McConkey, D.J.; Choi, W. Molecular Subtypes of Bladder Cancer. Curr. Oncol. Rep. 2018, 20, 1–7. [Google Scholar] [CrossRef]
- Czerniak, B.; Dinney, C.; McConkey, D. Origins of Bladder Cancer. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 149–174. [Google Scholar] [CrossRef]
- McConkey, D.J.; Choi, W.; Dinney, C.P.N. New insights into subtypes of invasive bladder cancer: Considerations of the clinician. Eur. Urol. 2014, 609–610. [Google Scholar] [CrossRef]
- Loriot, Y.; Necchi, A.; Park, S.H.; Garcia-Donas, J.; Huddart, R.; Burgess, E.; Fleming, M.; Rezazadeh, A.; Mellado, B.; Varlamov, S.; et al. Erdafitinib in Locally Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2019, 381, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of Distinct Basal and Luminal Subtypes of Muscle-Invasive Bladder Cancer with Different Sensitivities to Frontline Chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Zitvogel, L.; Sautès-Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Sjödahl, G.; Lövgren, K.; Lauss, M.; Chebil, G.; Patschan, O.; Gudjonsson, S.; Månsson, W.; Fernö, M.; Leandersson, K.; Lindgren, D.; et al. Infiltration of CD3+and CD68+cells in bladder cancer is subtype specific and affects the outcome of patients with muscle-invasive tumors. Urol. Oncol. Semin. Orig. Investig. 2014, 32, 791–797. [Google Scholar] [CrossRef]
- Pfannstiel, C.; Strissel, P.L.; Chiappinelli, K.B.; Sikic, D.; Wach, S.; Wirtz, R.M.; Wullweber, A.; Taubert, H.; Breyer, J.; Otto, W.; et al. The tumor immune microenvironment drives a prognostic relevance that correlates with bladder cancer subtypes. Cancer Immunol. Res. 2019, 7, 923–938. [Google Scholar] [CrossRef]
- Hodgson, A.; Liu, S.K.; Vesprini, D.; Xu, B.; Downes, M.R. Basal-subtype bladder tumours show a ‘hot’ immunophenotype. Histopathology 2018, 73, 748–757. [Google Scholar] [CrossRef]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2018, 174, 1033. [Google Scholar] [CrossRef]
- Sjödahl, G.; Lauss, M.; Lövgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Fernö, M.; Ringnér, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef]
- Dowell, A.C.; Cobby, E.; Wen, K.; Devall, A.J.; During, V.; Anderson, J.; James, N.D.; Cheng, K.K.; Zeegers, M.P.; Bryan, R.T.; et al. Interleukin-17-positive mast cells influence outcomes from BCG for patients with CIS: Data from a comprehensive characterisation of the immune microenvironment of urothelial bladder cancer. PLoS ONE 2017, 12, e0184841. [Google Scholar] [CrossRef]
- Nunez-Nateras, R.; Castle, E.P.; Protheroe, C.A.; Stanton, M.L.; Ocal, T.I.; Ferrigni, E.N.; Ochkur, S.I.; Jacobsen, E.A.; Hou, Y.X.; Andrews, P.E.; et al. Predicting response to bacillus Calmette-Guérin (BCG) in patients with carcinoma in situ of the bladder. Urol. Oncol. Semin. Orig. Investig. 2014, 32, e23–e45. [Google Scholar] [CrossRef]
- Yu, A.; Mansure, J.J.; Solanki, S.; Siemens, D.R.; Koti, M.; Dias, A.B.T.; Burnier, M.M.; Brimo, F.; Kassouf, W. Presence of lymphocytic infiltrate cytotoxic T lymphocyte CD3 +, CD8 +, and immunoscore as prognostic marker in patients after radical cystectomy. PLoS ONE 2018, 13, 1–13. [Google Scholar] [CrossRef]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.G.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of Molecular Subtypes in Muscle-invasive Bladder Cancer on Predicting Response and Survival after Neoadjuvant Chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.E.; Hoffman-Censits, J.; Powles, T.; Van Der Heijden, M.S.; Balar, A.V.; Necchi, A.; Dawson, N.; O’Donnell, P.H.; Balmanoukian, A.; Loriot, Y.; et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: A single-arm, multicentre, phase 2 trial. Lancet 2016, 387, 1909–1920. [Google Scholar] [CrossRef]
- Liakou, C.I.; Narayanan, S.; Ng Tang, D.; Logothetis, C.J.; Sharma, P. Focus on TILs: Prognostic significance of tumor infiltrating lymphocytes in human bladder cancer. Cancer Immun. J. Acad. Cancer Immunol. 2007, 7, 10. [Google Scholar]
- Bellmunt, J.; De Wit, R.; Vaughn, D.J.; Fradet, Y.; Lee, J.L.; Fong, L.; Vogelzang, N.J.; Climent, M.A.; Petrylak, D.P.; Choueiri, T.K.; et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N. Engl. J. Med. 2017, 376, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Retz, M.; Siefker-Radtke, A.; Baron, A.; Necchi, A.; Bedke, J.; Plimack, E.R.; Vaena, D.; Grimm, M.O.; Bracarda, S.; et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): A multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017, 18, 312–322. [Google Scholar] [CrossRef]
- Rosaria, M.; Bonavita, E.; Barajon, I.; Garlanda, C.; Mantovani, A.; Jaillon, S. Tumor associated macrophages and neutrophils in cancer. Immunobiology 2013, 218, 1402–1410. [Google Scholar]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of Tumor-Associated Neutrophil Phenotype by TGF-β: “N1” versus “N2” TAN. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef]
- Andzinski, L.; Kasnitz, N.; Stahnke, S.; Wu, C.F.; Gereke, M.; Von Köckritz-Blickwede, M.; Schilling, B.; Brandau, S.; Weiss, S.; Jablonska, J. Type i IFNs induce anti-tumor polarization of tumor associated neutrophils in mice and human. Int. J. Cancer 2016, 138, 1982–1993. [Google Scholar] [CrossRef]
- Giese, M.A.; Hind, L.E.; Huttenlocher, A. Neutrophil plasticity in the tumor microenvironment. Blood 2019, 133, 2159–2167. [Google Scholar] [CrossRef]
- Gentles, A.J.; Newman, A.M.; Liu, C.L.; Bratman, S.V.; Feng, W.; Kim, D.; Nair, V.S.; Xu, Y.; Khuong, A.; Hoang, C.D.; et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat. Med. 2015, 21, 938–945. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Hu, P.; Donskov, F.; Wang, G.; Liu, Q.; Du, J. Tumor-associated neutrophils as a new prognostic factor in cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, 2–10. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.; Bhojnagarwala, P.S.; O’Brien, S.; Moon, E.K.; Garfall, A.L.; Rao, A.S.; Quatromoni, J.G.; Stephen, T.L.; Litzky, L.; Deshpande, C.; et al. Origin and Role of a Subset of Tumor-Associated Neutrophils with Antigen-Presenting Cell Features in Early-Stage Human Lung Cancer. Cancer Cell 2016, 30, 120–135. [Google Scholar] [CrossRef]
- Kawahara, T.; Furuya, K.; Nakamura, M.; Sakamaki, K.; Osaka, K.; Ito, H.; Ito, Y.; Izumi, K.; Ohtake, S.; Miyoshi, Y.; et al. Neutrophil-to-lymphocyte ratio is a prognostic marker in bladder cancer patients after radical cystectomy. BMC Cancer 2016, 16, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Morizawa, Y.; Miyake, M.; Shimada, K.; Hori, S.; Tatsumi, Y.; Nakai, Y.; Anai, S.; Tanaka, N.; Konishi, N.; Fujimoto, K. Neutrophil-to-lymphocyte ratio as a detection marker of tumor recurrence in patients with muscle-invasive bladder cancer after radical cystectomy. Urol. Oncol. Semin. Orig. Investig. 2016, 34, e11–e257. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhao, K.; Wang, L.; Sun, E. The prognostic values of tumor-infiltrating neutrophils, lymphocytes and neutrophil/lymphocyte rates in bladder urothelial cancer. Pathol. Res. Pract. 2018, 214, 1074–1080. [Google Scholar] [CrossRef]
- Zhou, L.; Xu, L.; Chen, L.; Fu, Q.; Liu, Z.; Chang, Y.; Lin, Z.; Xu, J. Tumor-infiltrating neutrophils predict benefit from adjuvant chemotherapy in patients with muscle invasive bladder cancer. Oncoimmunology 2017, 6, e1293211. [Google Scholar] [CrossRef]
- Pagès, F.; Mlecnik, B.; Marliot, F.; Bindea, G.; Ou, F.S.; Bifulco, C.; Lugli, A.; Zlobec, I.; Rau, T.T.; Berger, M.D.; et al. International validation of the consensus Immunoscore for the classification of colon cancer: A prognostic and accuracy study. Lancet 2018, 391, 2128–2139. [Google Scholar] [CrossRef]
- Sobin, L.H.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumors, 7th ed.; Wiley-Blackwell: Chichester, West Sussex, UK, 2010. [Google Scholar]
- Gatta, L.B.; Melocchi, L.; Bugatti, M.; Missale, F.; Lonardi, S.; Zanetti, B.; Cristinelli, L.; Belotti, S.; Simeone, C.; Ronca, R.; et al. Hyper-Activation of STAT3 Sustains Progression of Non-Papillary Basal-Type Bladder Cancer via FOSL1 Regulome. Cancers (Basel) 2019, 11, 1219. [Google Scholar] [CrossRef]
- Hurvich, C.M.; Simonoff, J.S.; Tsai, C.L. Smoothing parameter selection in nonparametric regression using an improved Akaike information criterion. J. R. Stat. Soc. Ser. B Stat. Methodol. 1998, 60, 271–293. [Google Scholar] [CrossRef]
- Calzetti, F.; Tamassia, N.; Arruda-Silva, F.; Gasperini, S.; Cassatella, M.A. The importance of being “pure” neutrophils. J. Allergy Clin. Immunol. 2017, 139, 352–355.e6. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, M.; Maggi, L.; Micheletti, A.; Lazzeri, E.; Tamassia, N.; Costantini, C.; Cosmi, L.; Lunardi, C.; Annunziato, F.; Romagnani, S.; et al. Evidence for a cross-talk between human neutrophils and Th17 cells. Blood 2010, 115, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Seiler, R.; Gibb, E.A.; Wang, N.Q.; Oo, H.Z.; Lam, H.-M.; van Kessel, K.E.; Voskuilen, C.S.; Winters, B.; Erho, N.; Takhar, M.M.; et al. Divergent Biological Response to Neoadjuvant Chemotherapy in Muscle-invasive Bladder Cancer. Clin. Cancer Res. 2018, 25, 5082–5094. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-Seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Zlotnik, A.; Yoshie, O. The Chemokine Superfamily Revisited. Immunity 2012, 36, 705–716. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Smyth, G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58. [Google Scholar] [CrossRef]
- Dadhania, V.; Zhang, M.; Zhang, L.; Bondaruk, J.; Majewski, T.; Siefker-Radtke, A.; Guo, C.C.; Dinney, C.; Cogdell, D.E.; Zhang, S.; et al. Meta-Analysis of the Luminal and Basal Subtypes of Bladder Cancer and the Identification of Signature Immunohistochemical Markers for Clinical Use. EBioMedicine 2016, 12, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.L.; Lay, E.J.; Jian, W.; Parra, D.; Chan, K.S. Stat3 activation in urothelial stem cells leads to direct progression to invasive bladder cancer. Cancer Res. 2012, 72, 3135–3142. [Google Scholar] [CrossRef] [PubMed]
- Annis, M.G.; Ouellet, V.; Rennhack, J.P.; L’Esperance, S.; Rancourt, C.; Mes-Masson, A.M.; Andrechek, E.R.; Siegel, P.M. Integrin-uPAR signaling leads to FRA-1 phosphorylation and enhanced breast cancer invasion. Breast Cancer Res. 2018, 20, 9. [Google Scholar] [CrossRef]
- Sokol, C.L.; Luster, A.D. The chemokine system in innate immunity. Cold Spring Harb. Perspect. Biol. 2015, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Bonavita, O.; Massara, M.; Bonecchi, R. Chemokine regulation of neutrophil function in tumors. Cytokine Growth Factor Rev. 2016, 30, 81–86. [Google Scholar] [CrossRef]
- Vidotto, T.; Nersesian, S.; Graham, C.; Siemens, D.R.; Koti, M. DNA damage repair gene mutations and their association with tumor immune regulatory gene expression in muscle invasive bladder cancer subtypes. J. Immunother. Cancer 2019, 7, 1–12. [Google Scholar] [CrossRef]
- Kortylewski, M.; Kujawski, M.; Wang, T.; Wei, S.; Zhang, S.; Pilon-Thomas, S.; Niu, G.; Kay, H.; Mulé, J.; Kerr, W.G.; et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat. Med. 2005, 11, 1314–1321. [Google Scholar] [CrossRef]
- Taniguchi, K.; Karin, M. IL-6 and related cytokines as the critical lynchpins between inflammation and cancer. Semin. Immunol. 2014, 26, 54–74. [Google Scholar] [CrossRef]
- Liu, Q.; Yu, S.; Li, A.; Xu, H.; Han, X.; Wu, K. Targeting interlukin-6 to relieve immunosuppression in tumor microenvironment. Tumor Biol. 2017, 39. [Google Scholar] [CrossRef]
- Wang, J.; Jia, Y.; Wang, N.; Zhang, X.; Tan, B.; Zhang, G.; Cheng, Y. The clinical significance of tumor-infiltrating neutrophils and neutrophil-to-CD8+ lymphocyte ratio in patients with resectable esophageal squamous cell carcinoma. J. Transl. Med. 2014, 12, 7. [Google Scholar] [CrossRef]
- Kargl, J.; Bolouri, H.; Houghton, A.M.; Kargl, J.; Zhu, X.; Zhang, H.; Yang, G.H.Y.; Friesen, T.J.; Shipley, M.; Maeda, D.Y.; et al. Neutrophil content predicts lymphocyte depletion and anti-PD1 treatment failure in NSCLC Find the latest version: Neutrophil content predicts lymphocyte depletion and anti-PD1 treatment failure in NSCLC. JCI Insight 2019, 4, e130850. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, M.R.; Bianchi, P.; Grizzi, F.; Di Caro, G.; Basso, G.; Ponzetta, A.; Bonavita, E.; Barbagallo, M.; Tartari, S.; Polentarutti, N.; et al. Occurrence and significance of tumor-Associated neutrophils in patients with colorectal cancer. Int. J. Cancer 2016, 139, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Eruslanov, E.B.; Bhojnagarwala, P.S.; Quatromoni, J.G.; Stephen, T.L.; Ranganathan, A.; Deshpande, C.; Akimova, T.; Vachani, A.; Litzky, L.; Hancock, W.W.; et al. Tumor-associated neutrophils stimulate T cell responses in early-stage human lung cancer. J. Clin. Invest. 2014, 124, 5466–5480. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.P.; O’Donnell, M.A.; Griffith, T.S. Role of neutrophils in BCG immunotherapy for bladder cancer. Urol. Oncol. Semin. Orig. Investig. 2008, 26, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, D.; Feng, Y.; Zhai, J.; Lu, C. Improved antitumor efficacy of neutrophils stimulated by bacillus Calmette-Guérin. Mol. Med. Rep. 2019, 20, 2909–2915. [Google Scholar] [PubMed]
- Koga, Y.; Matsuzaki, A.; Suminoe, A.; Hattori, H.; Hara, T. Neutrophil-Derived TNF-Related Apoptosis-Inducing Ligand (TRAIL): A Novel Mechanism of Antitumor Effect by Neutrophils. Cancer Res. 2004, 64, 1037–1043. [Google Scholar] [CrossRef]
- Ludwig, A.T.; Moore, J.M.; Luo, Y.; Chen, X.; Saltsgaver, N.A.; O’Donnell, M.A.; Griffith, T.S. Tumor necrosis factor-related apoptosis-inducing ligand: A novel mechanism for Bacillus Calmette-Guérin-induced antitumor activity. Cancer Res. 2004, 64, 3386–3390. [Google Scholar] [CrossRef]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; Amaral, F.A. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef]
- Thalmann, G.N.; Sermier, A.; Rentsch, C.; Möhrle, K.; Cecchini, M.G.; Studer, U.E. Urinary interleukin-8 and 18 predict the response of superficial bladder cancer to intravesical therapy with bacillus Calmette-Guerin. J. Urol. 2000, 164, 2129–2133. [Google Scholar] [CrossRef]
- Zilionis, R.; Engblom, C.; Pfirschke, C.; Savova, V.; Zemmour, D.; Saatcioglu, H.D.; Krishnan, I.; Maroni, G.; Meyerovitz, C.V.; Kerwin, C.M.; et al. Single-Cell Transcriptomics of Human and Mouse Lung Cancers Reveals Conserved Myeloid Populations across Individuals and Species. Immunity 2019, 50, 1317–1334.e10. [Google Scholar] [CrossRef]


| Total | N (%) | CIS Concomitant | N (%) |
|---|---|---|---|
| 84 (100) | |||
| Yes | 50 (60) | ||
| Gender | No | 34 (40) | |
| Female | 16 (19) | Global Stage | |
| Male | 68 (81) | II | 22 (26) |
| Age (years) | III | 33 (39) | |
| Mean (SD) | 70.9 (10.44) | IV | 29 (35) |
| Median (IQR) | 72 (14) | Subtype | |
| Stage pT | Luminal | 41 (49) | |
| pT2 | 25 (30) | Basal | 20 (24) |
| pT3 | 41 (49) | Non-type | 23 (27) |
| pT4 | 18 (21) | pSTAT3 | |
| Stage pN | Score 0–1 | 43 (51) | |
| pN0 | 57 (68) | Score 2–3 | 41 (49) |
| pN1 | 16 (19) | FOSL1 | |
| pN2 | 10 (12) | Score 0–1 | 19 (23) |
| pN3 | 1 (1) | Score 2–3 NA | 22 (26) 43 (51) |
| Clinicopathological Variables | Overall Survival | Progression Free Survival | ||
|---|---|---|---|---|
| H.R. (CI 95%) | p | H.R. (CI 95%) | p | |
| Age (years) | 1.04 (1.01–1.08) | 0.024 | 0.99 (0.96–1.02) | 0.65 |
| Gender (F vs. M) | 2.93 (1.45–5.94) | 0.003 | 1.94 (0.94–4.00) | 0.072 |
| pT (pT3-4 vs. pT2) | 4.21 (1.63–10.85) | 0.003 | 3.54 (1.47–8.55) | 0.005 |
| pN (pN+ vs. pN0) | 2.22 (1.16–4.23) | 0.016 | 2.25 (1.17–4.33) | 0.015 |
| CIS (yes vs. no) | 0.63 (0.33–1.19) | 0.155 | 0.86 (0.45–1.65) | 0.657 |
| Overall Stage (SIII-IV vs. SII) | 5.07 (1.78–14.41) | 0.002 | 3.94 (1.52–10.21) | 0.005 |
| Subtype (basal vs. luminal) | 1.3 (0.59–2.87) | 0.520 | 0.70 (0.32–1.55) | 0.383 |
| Subtype (Non-type vs. luminal) | 1.66 (0.78–3.53) | 0.191 | 0.73 (0.33–1.60) | 0.431 |
| pSTAT3 (High vs. Low) | 1.19 (0.63–2.26) | 0.595 | 0.69 (0.36–1.32) | 0.265 |
| CD66b (Low vs. High) | 2.53 (1.30–4.92) | 0.006 | 2.68 (1.37–5.24) | 0.004 |
| CD3 (Low vs. High) | 2.62 (1.35–5.08) | 0.004 | 1.69 (0.88–3.22) | 0.11 |
| CD3CD66b Immunoscore (LoLo vs. HiHi) | 5.27 (2.12–13.11) | <0.001 | 3.95 (1.53–10.16) | 0.004 |
| CD3CD66b Immunoscore (LoHi vs. HiHi) | 2.26 (0.79–6.49) | 0.13 | 2.15 (0.75–6.14) | 0.155 |
| CD3CD66b Immunoscore (HiLo vs. HiHi) | 2.18 (0.79–6.04) | 0.133 | 3.43 (1.32–8.89) | 0.011 |
| tNLR (CD66b+/CD3+) | 1.04 (0.75–1.44) | 0.828 | 1.07 (0.75–1.53) | 0.699 |
| Clinicopathological Variables | CD66b+ TAN Density | CD3+ T Cells Density | tNLR (CD66b+/CD3+) | ||||
|---|---|---|---|---|---|---|---|
| N° (%) | Median | p | Median | p | Median | p | |
| Total | 84 (100) | 78 | 396 | ||||
| Gender | 0.76 * | 0.47 * | 0.90 * | ||||
| Female | 16 (19) | 51 | 328 | 0.186 | |||
| Male | 68 (81) | 86 | 399 | 0.216 | |||
| Age | 0.80 * | 0.26 * | 0.55 * | ||||
| < median | 39 (46) | 83 | 327 | 0.186 | |||
| ≥ median | 45 (54) | 73 | 415 | 0.224 | |||
| pT category | 0.11 * | 0.003 * | 0.35 * | ||||
| pT2 | 25 (30) | 106 | 544 | 0.136 | |||
| pT3-pT4 | 59 (70) | 60 | 304 | 0.292 | |||
| pN category | 0.10 * | 0.19 * | 0.50 * | ||||
| pN+ | 27 (32) | 106 | 403 | 0.154 | |||
| pN0 | 57 (68) | 48 | 357 | 0.234 | |||
| Overall Stage | 0.14 * | 0.001 * | 0.32 * | ||||
| II | 22 (26) | 123 | 587 | 0.149 | |||
| III-IV | 62 (74) | 56 | 312 | 0.263 | |||
| CIS | 0.46 * | 0.65 * | 0.90 * | ||||
| No | 34 (40) | 107 | 381 | 0.183 | |||
| Yes | 50 (60) | 68 | 402 | 0.239 | |||
| pSTAT3 | 0.048 * | 0.43 * | 0.28 * | ||||
| Low | 43 (51) | 51 | 357 | 0.176 | |||
| High | 41 (49) | 126 | 400 | 0.243 | |||
| Subtype | 0.007# | 0.75 # | 0.06 # | ||||
| Luminal | 41 (49) | 51 | 401 | 0.136 | |||
| Basal | 20 (24) | 178 | 398 | 0.359 | |||
| Non-Type | 23 (27) | 60 | 366 | 0.304 | |||
| FOSL1 | 0.054 * | 0.227 * | 0.55 * | ||||
| Low | 19 (22) | 58 | 418 | 0.291 | |||
| High | 22 (26) | 155 | 398 | 0.281 | |||
| Overall Survival | ||
|---|---|---|
| H.R. (CI 95%) | p | |
| Age (year) | 1.041 (1.006–1.077) | 0.019 |
| Gender (F) | 2.83 (1.33–5.99) | 0.01 |
| pT (pT3-4) | 3.67 (1.28–10.51) | 0.016 |
| CD3CD66b Immunoscore (LoLo) | 6.67 (2.43–18.25) | <0.001 |
| CD3CD66b Immunoscore (LoHi) | 4.20 (1.35–13.11) | 0.013 |
| CD3CD66b Immunoscore (HiLo) | 1.59 (0.55–4.61) | 0.40 |
| Progression Free Survival | ||
| pT (pT3-4) | 3.58 (1.44–8.93) | 0.006 |
| CD3CD66b Immunoscore (LoLo) | 3.55 (1.35–9.34) | 0.01 |
| CD3CD66b Immunoscore (LoHi) | 2.66 (0.91–7.84) | 0.08 |
| CD3CD66b Immunoscore (HiLo) | 3.69 (1.41–9.64) | 0.008 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandelli, G.E.; Missale, F.; Bresciani, D.; Benerini Gatta, L.; Scapini, P.; Caveggion, E.; Roca, E.; Bugatti, M.; Monti, M.; Cristinelli, L.; et al. Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer. Cells 2020, 9, 291. https://doi.org/10.3390/cells9020291
Mandelli GE, Missale F, Bresciani D, Benerini Gatta L, Scapini P, Caveggion E, Roca E, Bugatti M, Monti M, Cristinelli L, et al. Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer. Cells. 2020; 9(2):291. https://doi.org/10.3390/cells9020291
Chicago/Turabian StyleMandelli, Giulio Eugenio, Francesco Missale, Debora Bresciani, Luisa Benerini Gatta, Patrizia Scapini, Elena Caveggion, Elisa Roca, Mattia Bugatti, Matilde Monti, Luca Cristinelli, and et al. 2020. "Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer" Cells 9, no. 2: 291. https://doi.org/10.3390/cells9020291
APA StyleMandelli, G. E., Missale, F., Bresciani, D., Benerini Gatta, L., Scapini, P., Caveggion, E., Roca, E., Bugatti, M., Monti, M., Cristinelli, L., Belotti, S., Simeone, C., Calza, S., Melocchi, L., & Vermi, W. (2020). Tumor Infiltrating Neutrophils Are Enriched in Basal-Type Urothelial Bladder Cancer. Cells, 9(2), 291. https://doi.org/10.3390/cells9020291






