Interleukin-22 Mediates the Chemotactic Migration of Breast Cancer Cells and Macrophage Infiltration of the Bone Microenvironment by Potentiating S1P/SIPR Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Cell Cultures
2.2. RNA Isolation, RT-PCR, and qPCR
2.3. Immunoblotting
2.4. ELISA
2.5. Transwell Assay
2.6. Matrigel Invasion Assay
2.7. Immunohistochemical Staining Analysis
2.8. Immunofluorescent Staining for S1PR1
2.9. Statistics
3. Results
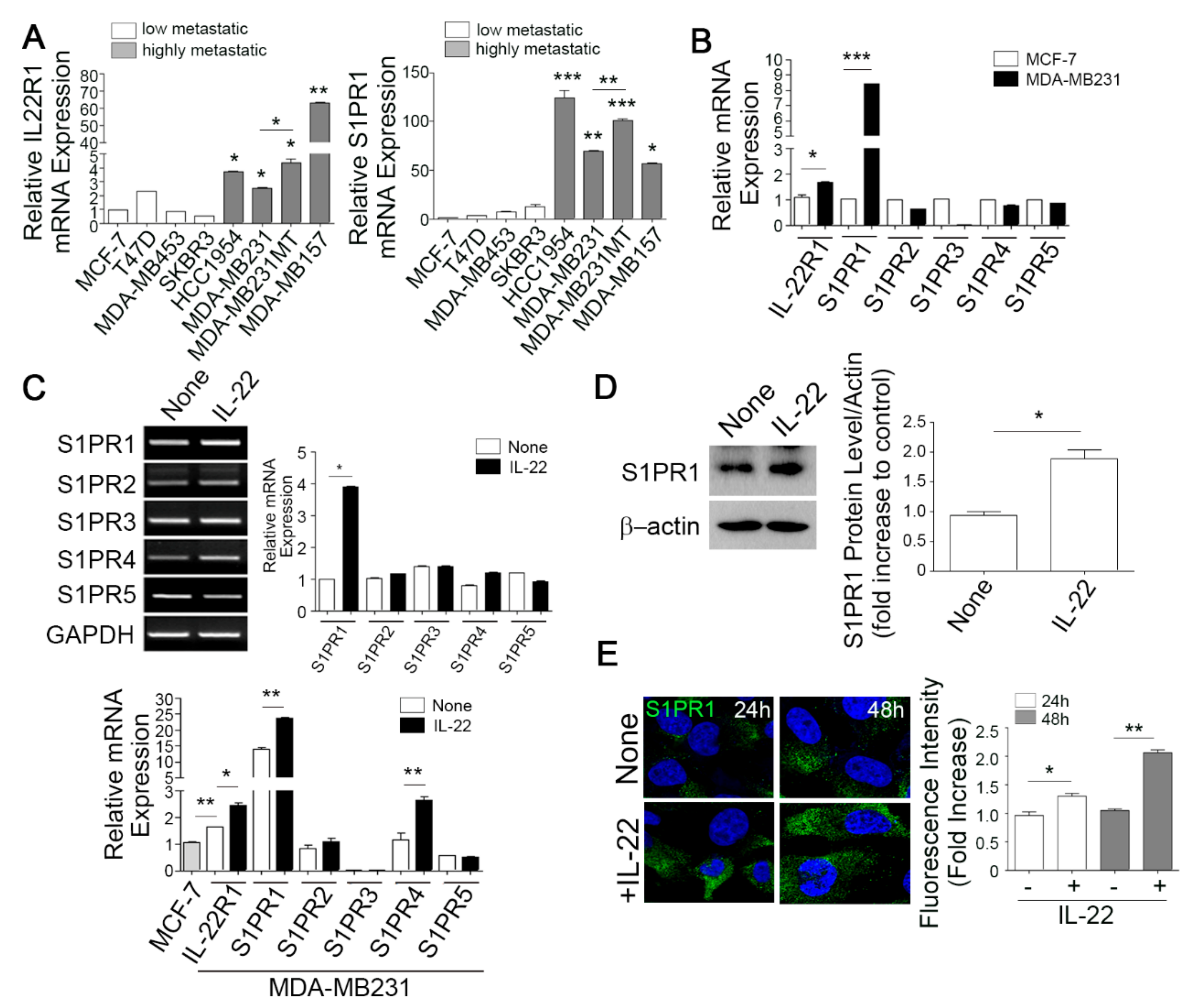
3.1. The Elevated Co-Expression of IL-22R1 and S1PR1 Is Associated with Advanced Human Breast Cancers with Bone Metastatic Potential
3.2. IL-22 Enhances IL-22R1 and S1PR1 Expression in Invasive Metastatic Breast Cancer Cells
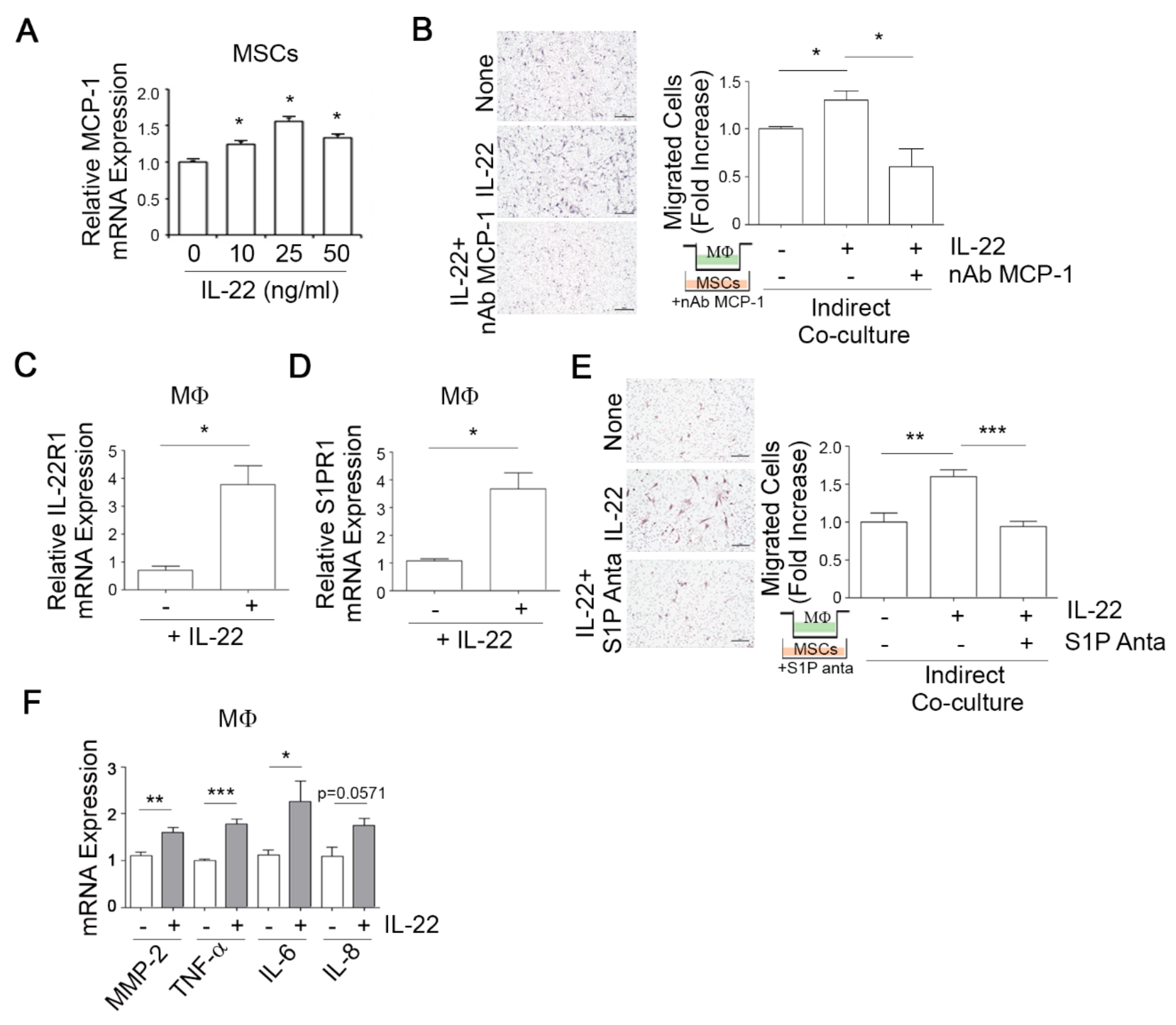
3.3. IL-22-Induced S1P in Mesenchymal Stem Cells (MSCs) Promotes the Migratory and Invasive Potential of Breast Cancer Cells
3.4. IL-22-Induced MCP-1 in MSCs Promotes Macrophage Chemotaxis
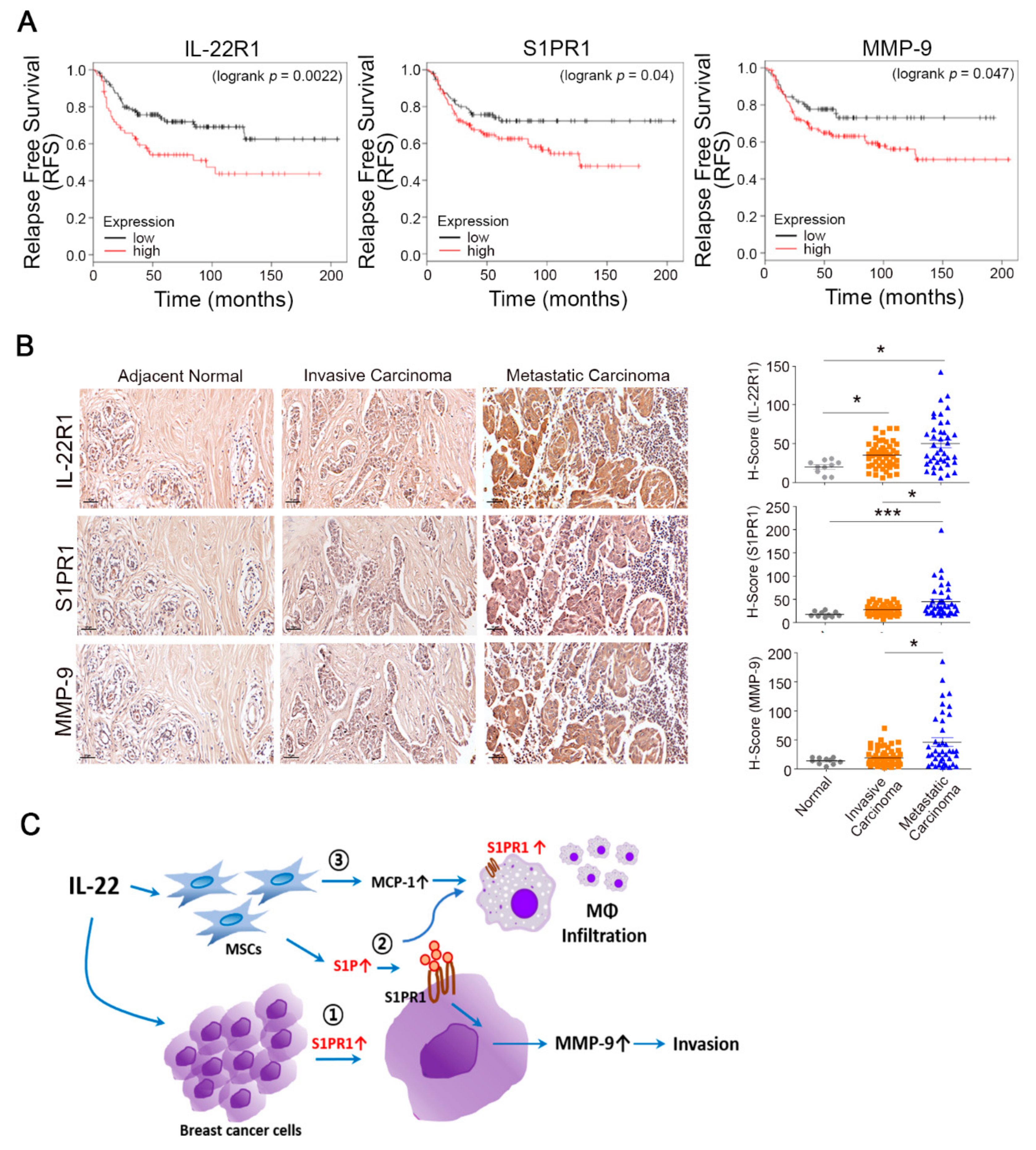
3.5. Prognostic Value of the IL-22R1, S1PR1, and MMP-9 Expression Levels in Breast Tumors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BMMs | Bone marrow-derived macrophages |
| CM | conditioned media |
| FBS | fetal bovine serum |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| HCC | hepatocellular carcinoma |
| IL-22 | interleukin 22 |
| IL-22R1 | interleukin-22 receptor 1 |
| MMP-2 | matrix metalloproteinase-2 |
| MMP-9 | matrix metalloproteinase-9 |
| MSCs | mesenchymal stem cells |
| MCP-1 | monocyte chemoattractant protein 1 |
| PBS | phosphate-buffered saline |
| S1P | sphingosine-1-phosphate |
| S1PR1 | sphingosine-1-phosphate receptor 1 |
References
- Dias, A.S.; Almeida, C.R.; Helguero, L.A.; Duarte, I.F. Metabolic crosstalk in the breast cancer microenvironment. Euro. J. Cancer (Oxford, England: 1990) 2019, 121, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Graham, N.; Qian, B.Z. Mesenchymal Stromal Cells: Emerging Roles in Bone Metastasis. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Linde, N.; Casanova-Acebes, M.; Sosa, M.S.; Mortha, A.; Rahman, A.; Farias, E.; Harper, K.; Tardio, E.; Reyes Torres, I.; Jones, J.; et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- De Miguel, M.P.; Fuentes-Julian, S.; Blazquez-Martinez, A.; Pascual, C.Y.; Aller, M.A.; Arias, J.; Arnalich-Montiel, F. Immunosuppressive properties of mesenchymal stem cells: Advances and applications. Curr. Mol. Med. 2012, 12, 574–591. [Google Scholar] [CrossRef]
- Shi, Y.; Du, L.; Lin, L.; Wang, Y. Tumour-associated mesenchymal stem/stromal cells: Emerging therapeutic targets. Nat. Rev. Drug Discov. 2017, 16, 35–52. [Google Scholar] [CrossRef]
- Ridge, S.M.; Sullivan, F.J.; Glynn, S.A. Mesenchymal stem cells: Key players in cancer progression. Mol. Cancer 2017, 16, 31. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Tower, H.; Ruppert, M.; Britt, K. The Immune Microenvironment of Breast Cancer Progression. Cancers 2019, 11. [Google Scholar] [CrossRef]
- Sabat, R.; Ouyang, W.; Wolk, K. Therapeutic opportunities of the IL-22-IL-22R1 system. Nat. Rev. Drug Discov. 2014, 13, 21–38. [Google Scholar] [CrossRef]
- Li, P.; Shi, X.; Xu, Y.; Zhong, B.; Lu, Y.; Sun, Y. Interleukin-22 Promotes Osteosarcoma Cell Proliferation and Invasion via STAT3 Activation. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2018, 24, 7802–7808. [Google Scholar] [CrossRef]
- Ye, Z.J.; Zhou, Q.; Yin, W.; Yuan, M.L.; Yang, W.B.; Xiang, F.; Zhang, J.C.; Xin, J.B.; Xiong, X.Z.; Shi, H.Z. Interleukin 22-producing CD4+ T cells in malignant pleural effusion. Cancer Lett. 2012, 326, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Chen, Y.; Wei, H.; Zheng, C.; Sun, R.; Zhang, J.; Tian, Z. Antiapoptotic activity of autocrine interleukin-22 and therapeutic effects of interleukin-22-small interfering RNA on human lung cancer xenografts. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 6432–6439. [Google Scholar] [CrossRef] [PubMed]
- Bard, J.D.; Gelebart, P.; Anand, M.; Amin, H.M.; Lai, R. Aberrant expression of IL-22 receptor 1 and autocrine IL-22 stimulation contribute to tumorigenicity in ALK+ anaplastic large cell lymphoma. Leukemia 2008, 22, 1595–1603. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wu, J.; Shi, J.; Huo, Y.M.; Dai, W.; Geng, J.; Lu, P.; Yang, M.W.; Fang, Y.; Wang, W.; et al. IL22RA1/STAT3 Signaling Promotes Stemness and Tumorigenicity in Pancreatic Cancer. Cancer Res. 2018, 78, 3293–3305. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, N.; Caetano, M.S.; Cumpian, A.M.; Unver, N.; De la Garza Ramos, C.; Noble, O.; Daliri, S.; Hernandez, B.J.; Gutierrez, B.A.; Evans, S.E.; et al. IL22 Promotes Kras-Mutant Lung Cancer by Induction of a Protumor Immune Response and Protection of Stemness Properties. Cancer Immunol. Res. 2018, 6, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Khare, V.; Paul, G.; Movadat, O.; Frick, A.; Jambrich, M.; Krnjic, A.; Marian, B.; Wrba, F.; Gasche, C. IL10R2 Overexpression Promotes IL22/STAT3 Signaling in Colorectal Carcinogenesis. Cancer Immunol. Res. 2015, 3, 1227–1235. [Google Scholar] [CrossRef]
- Huber, S.; Gagliani, N.; Zenewicz, L.A.; Huber, F.J.; Bosurgi, L.; Hu, B.; Hedl, M.; Zhang, W.; O’Connor, W., Jr.; Murphy, A.J.; et al. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 2012, 491, 259–263. [Google Scholar] [CrossRef]
- Kirchberger, S.; Royston, D.J.; Boulard, O.; Thornton, E.; Franchini, F.; Szabady, R.L.; Harrison, O.; Powrie, F. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J. Exp. Med. 2013, 210, 917–931. [Google Scholar] [CrossRef]
- Jiang, R.; Tan, Z.; Deng, L.; Chen, Y.; Xia, Y.; Gao, Y.; Wang, X.; Sun, B. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology 2011, 54, 900–909. [Google Scholar] [CrossRef]
- Zhuang, Y.; Peng, L.S.; Zhao, Y.L.; Shi, Y.; Mao, X.H.; Guo, G.; Chen, W.; Liu, X.F.; Zhang, J.Y.; Liu, T.; et al. Increased intratumoral IL-22-producing CD4(+) T cells and Th22 cells correlate with gastric cancer progression and predict poor patient survival. Cancer Immunol. Immunother. Cii 2012, 61, 1965–1975. [Google Scholar] [CrossRef]
- Sukocheva, O.A. Expansion of Sphingosine Kinase and Sphingosine-1-Phosphate Receptor Function in Normal and Cancer Cells: From Membrane Restructuring to Mediation of Estrogen Signaling and Stem Cell Programming. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, J.; Nagahashi, M.; Takabe, K.; Wakai, T. Clinical Impact of Sphingosine-1-Phosphate in Breast Cancer. Mediat. Inflamm. 2017, 2017, 2076239. [Google Scholar] [CrossRef] [PubMed]
- Pyne, N.J.; Pyne, S. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer 2010, 10, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Xia, P.; Di, W. Sphingosine 1-phosphate: A potential molecular target for ovarian cancer therapy? Cancer Investig. 2014, 32, 71–80. [Google Scholar] [CrossRef]
- Takabe, K.; Paugh, S.W.; Milstien, S.; Spiegel, S. “Inside-out” signaling of sphingosine-1-phosphate: Therapeutic targets. Pharmacol. Rev. 2008, 60, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Zondag, G.C.; Postma, F.R.; Etten, I.V.; Verlaan, I.; Moolenaar, W.H. Sphingosine 1-phosphate signalling through the G-protein-coupled receptor Edg-1. Biochem. J. 1998, 330, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V. Sphingosine 1-phosphate receptors in health and disease: Mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol. Ther. 2007, 115, 84–105. [Google Scholar] [CrossRef]
- Watson, C.; Long, J.S.; Orange, C.; Tannahill, C.L.; Mallon, E.; McGlynn, L.M.; Pyne, S.; Pyne, N.J.; Edwards, J. High expression of sphingosine 1-phosphate receptors, S1P1 and S1P3, sphingosine kinase 1, and extracellular signal-regulated kinase-1/2 is associated with development of tamoxifen resistance in estrogen receptor-positive breast cancer patients. Am. J. Pathol. 2010, 177, 2205–2215. [Google Scholar] [CrossRef]
- Choi, B.; Lee, E.J.; Park, Y.S.; Kim, S.M.; Kim, E.Y.; Song, Y.; Kang, S.W.; Rhu, M.H.; Chang, E.J. Pentraxin-3 Silencing Suppresses Gastric Cancer-related Inflammation by Inhibiting Chemotactic Migration of Macrophages. Anticancer Res. 2015, 35, 2663–2668. [Google Scholar]
- Li, H.; Qiu, Z.; Li, F.; Wang, C. The relationship between MMP-2 and MMP-9 expression levels with breast cancer incidence and prognosis. Oncol. Lett. 2017, 14, 5865–5870. [Google Scholar] [CrossRef]
- Jacob, A.; Jing, J.; Lee, J.; Schedin, P.; Gilbert, S.M.; Peden, A.A.; Junutula, J.R.; Prekeris, R. Rab40b regulates trafficking of MMP2 and MMP9 during invadopodia formation and invasion of breast cancer cells. J. Cell Sci. 2013, 126, 4647–4658. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Pollard, J.W. Macrophage diversity enhances tumor progression and metastasis. Cell 2010, 141, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Nounou, M.I.; ElAmrawy, F.; Ahmed, N.; Abdelraouf, K.; Goda, S.; Syed-Sha-Qhattal, H. Breast Cancer: Conventional Diagnosis and Treatment Modalities and Recent Patents and Technologies. Breast Cancer Basic Clin. Res. 2015, 9, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Aoyagi, T.; Nagahashi, M.; Yamada, A.; Takabe, K. The role of sphingosine-1-phosphate in breast cancer tumor-induced lymphangiogenesis. Lymphat. Res. Biol. 2012, 10, 97–106. [Google Scholar] [CrossRef]
- Nagahashi, M.; Ramachandran, S.; Kim, E.Y.; Allegood, J.C.; Rashid, O.M.; Yamada, A.; Zhao, R.; Milstien, S.; Zhou, H.; Spiegel, S.; et al. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012, 72, 726–735. [Google Scholar] [CrossRef]
- Nagahashi, M.; Yamada, A.; Katsuta, E.; Aoyagi, T.; Huang, W.C.; Terracina, K.P.; Hait, N.C.; Allegood, J.C.; Tsuchida, J.; Yuza, K.; et al. Targeting the SphK1/S1P/S1PR1 Axis That Links Obesity, Chronic Inflammation, and Breast Cancer Metastasis. Cancer Res. 2018, 78, 1713–1725. [Google Scholar] [CrossRef]
- Sutphen, R.; Xu, Y.; Wilbanks, G.D.; Fiorica, J.; Grendys, E.C., Jr.; LaPolla, J.P.; Arango, H.; Hoffman, M.S.; Martino, M.; Wakeley, K.; et al. Lysophospholipids are potential biomarkers of ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2004, 13, 1185–1191. [Google Scholar]
- Alberg, A.J.; Armeson, K.; Pierce, J.S.; Bielawski, J.; Bielawska, A.; Visvanathan, K.; Hill, E.G.; Ogretmen, B. Plasma sphingolipids and lung cancer: A population-based, nested case-control study. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1374–1382. [Google Scholar] [CrossRef]
- Jiang, R.; Wang, H.; Deng, L.; Hou, J.; Shi, R.; Yao, M.; Gao, Y.; Yao, A.; Wang, X.; Yu, L.; et al. IL-22 is related to development of human colon cancer by activation of STAT3. BMC Cancer 2013, 13, 59. [Google Scholar] [CrossRef]
- Kobold, S.; Volk, S.; Clauditz, T.; Kupper, N.J.; Minner, S.; Tufman, A.; Duwell, P.; Lindner, M.; Koch, I.; Heidegger, S.; et al. Interleukin-22 is frequently expressed in small- and large-cell lung cancer and promotes growth in chemotherapy-resistant cancer cells. J. Thorac. Oncol. 2013, 8, 1032–1042. [Google Scholar] [CrossRef]
- Wolk, K.; Witte, E.; Hoffmann, U.; Doecke, W.D.; Endesfelder, S.; Asadullah, K.; Sterry, W.; Volk, H.D.; Wittig, B.M.; Sabat, R. IL-22 induces lipopolysaccharide-binding protein in hepatocytes: A potential systemic role of IL-22 in Crohn’s disease. J. Immunol. 2007, 178, 5973–5981. [Google Scholar] [CrossRef] [PubMed]
- Souza, J.M.; Matias, B.F.; Rodrigues, C.M.; Murta, E.F.; Michelin, M.A. IL-17 and IL-22 serum cytokine levels in patients with squamous intraepithelial lesion and invasive cervical carcinoma. Eur. J. Gynaecol. Oncol. 2013, 34, 466–468. [Google Scholar] [PubMed]
- Ikeuchi, H.; Kuroiwa, T.; Hiramatsu, N.; Kaneko, Y.; Hiromura, K.; Ueki, K.; Nojima, Y. Expression of interleukin-22 in rheumatoid arthritis: Potential role as a proinflammatory cytokine. Arthritis Rheum. 2005, 52, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, L.; Martin, C.; Jeannot, P.; Ader, I.; Gstalder, C.; Andrieu, G.; Bocquet, M.; Laffosse, J.M.; Gomez-Brouchet, A.; Malavaud, B.; et al. Osteoblast-derived sphingosine 1-phosphate to induce proliferation and confer resistance to therapeutics to bone metastasis-derived prostate cancer cells. Mol. Oncol. 2014, 8, 1181–1195. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, E.-Y.; Choi, B.; Kim, J.-E.; Park, S.-O.; Kim, S.-M.; Chang, E.-J. Interleukin-22 Mediates the Chemotactic Migration of Breast Cancer Cells and Macrophage Infiltration of the Bone Microenvironment by Potentiating S1P/SIPR Signaling. Cells 2020, 9, 131. https://doi.org/10.3390/cells9010131
Kim E-Y, Choi B, Kim J-E, Park S-O, Kim S-M, Chang E-J. Interleukin-22 Mediates the Chemotactic Migration of Breast Cancer Cells and Macrophage Infiltration of the Bone Microenvironment by Potentiating S1P/SIPR Signaling. Cells. 2020; 9(1):131. https://doi.org/10.3390/cells9010131
Chicago/Turabian StyleKim, Eun-Young, Bongkun Choi, Ji-Eun Kim, Si-On Park, Sang-Min Kim, and Eun-Ju Chang. 2020. "Interleukin-22 Mediates the Chemotactic Migration of Breast Cancer Cells and Macrophage Infiltration of the Bone Microenvironment by Potentiating S1P/SIPR Signaling" Cells 9, no. 1: 131. https://doi.org/10.3390/cells9010131
APA StyleKim, E.-Y., Choi, B., Kim, J.-E., Park, S.-O., Kim, S.-M., & Chang, E.-J. (2020). Interleukin-22 Mediates the Chemotactic Migration of Breast Cancer Cells and Macrophage Infiltration of the Bone Microenvironment by Potentiating S1P/SIPR Signaling. Cells, 9(1), 131. https://doi.org/10.3390/cells9010131



