Modeling Non-Alcoholic Fatty Liver Disease (NAFLD) Using “Good-Fit” Genome-Editing Tools
Abstract
1. Introduction
1.1. Pathophysiology of Non-Alcoholic Fatty Liver Disease
1.2. Genetic Variants Associated with NAFLD
1.2.1. PNPLA3
1.2.2. TM6SF2
1.2.3. GCKR
1.2.4. Other Genetic Variants that Influence NAFLD Susceptibility
2. NAFLD Models Generated Using Genome Editing
2.1. CRISPR/Cas9 Genome Editing
2.2. CRISPR/Cas9-Mediated NAFLD Models
2.2.1. PNPLA3-Targeted Models
2.2.2. TM6SF2-Targeted Models
2.2.3. Other NAFLD Models
3. Selection of a “Good-Fit” Genome-Editing Tool for Replicating NAFLD Generic Variants
3.1. Advanced Cas9 Variants
3.1.1. Base Editor
3.1.2. Prime Editor
3.2. Various Gene-Delivery Methods for Optimizing Genome-Editing Efficiency
4. Discussion
5. Future Directions
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Brunt, E.M.; Wong, V.W.; Nobili, V.; Day, C.P.; Sookoian, S.; Maher, J.J.; Bugianesi, E.; Sirlin, C.B.; Neuschwander-Tetri, B.A.; Rinella, M.E. Nonalcoholic fatty liver disease. Nat. Rev. Dis. Primers 2015, 1, 15080. [Google Scholar] [CrossRef] [PubMed]
- Anstee, Q.M.; Day, C.P. The genetics of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Bettermann, K.; Hohensee, T.; Haybaeck, J. Steatosis and steatohepatitis: Complex disorders. Int. J. Mol. Sci. 2014, 15, 9924–9944. [Google Scholar] [CrossRef] [PubMed]
- Haybaeck, J.; Stumptner, C.; Thueringer, A.; Kolbe, T.; Magin, T.M.; Hesse, M.; Fickert, P.; Tsybrovskyy, O.; Muller, H.; Trauner, M.; et al. Genetic background effects of keratin 8 and 18 in a DDC-induced hepatotoxicity and Mallory-Denk body formation mouse model. Lab. Investig. 2012, 92, 857–867. [Google Scholar] [CrossRef]
- Zatloukal, B.; Kufferath, I.; Thueringer, A.; Landegren, U.; Zatloukal, K.; Haybaeck, J. Sensitivity and specificity of in situ proximity ligation for protein interaction analysis in a model of steatohepatitis with Mallory-Denk bodies. PLoS ONE 2014, 9, e96690. [Google Scholar] [CrossRef]
- Younossi, Z.M.; Koenig, A.B.; Abdelatif, D.; Fazel, Y.; Henry, L.; Wymer, M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, 73–84. [Google Scholar] [CrossRef]
- Bettermann, K.; Mehta, A.K.; Hofer, E.M.; Wohlrab, C.; Golob-Schwarzl, N.; Svendova, V.; Schimek, M.G.; Stumptner, C.; Thuringer, A.; Speicher, M.R.; et al. Keratin 18-deficiency results in steatohepatitis and liver tumors in old mice: A model of steatohepatitis-associated liver carcinogenesis. Oncotarget 2016, 7, 73309–73322. [Google Scholar] [CrossRef]
- Golob-Schwarzl, N.; Bettermann, K.; Mehta, A.K.; Kessler, S.M.; Unterluggauer, J.; Krassnig, S.; Kojima, K.; Chen, X.; Hoshida, Y.; Bardeesy, N.M.; et al. High Keratin 8/18 Ratio Predicts Aggressive Hepatocellular Cancer Phenotype. Transl. Oncol. 2019, 12, 256–268. [Google Scholar] [CrossRef]
- Severson, T.J.; Besur, S.; Bonkovsky, H.L. Genetic factors that affect nonalcoholic fatty liver disease: A systematic clinical review. World J. Gastroenterol. 2016, 22, 6742–6756. [Google Scholar] [CrossRef]
- Machado, M.V.; Cortez-Pinto, H. Non-alcoholic fatty liver disease: What the clinician needs to know. World J. Gastroenterol. 2014, 20, 12956–12980. [Google Scholar] [CrossRef]
- Romeo, S.; Kozlitina, J.; Xing, C.; Pertsemlidis, A.; Cox, D.; Pennacchio, L.A.; Boerwinkle, E.; Cohen, J.C.; Hobbs, H.H. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008, 40, 1461–1465. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Castano, G.O.; Burgueno, A.L.; Gianotti, T.F.; Rosselli, M.S.; Pirola, C.J. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 2009, 50, 2111–2116. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Patman, G.L.; Leathart, J.B.; Piguet, A.C.; Burt, A.D.; Dufour, J.F.; Day, C.P.; Daly, A.K.; Reeves, H.L.; Anstee, Q.M. Carriage of the PNPLA3 rs738409 C > G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J. Hepatol. 2014, 61, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Kozlitina, J.; Smagris, E.; Stender, S.; Nordestgaard, B.G.; Zhou, H.H.; Tybjaerg-Hansen, A.; Vogt, T.F.; Hobbs, H.H.; Cohen, J.C. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014, 46, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Reeves, H.L.; Burt, A.D.; Tiniakos, D.; McPherson, S.; Leathart, J.B.; Allison, M.E.; Alexander, G.J.; Piguet, A.C.; Anty, R.; et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014, 5, 4309. [Google Scholar] [CrossRef] [PubMed]
- Mahdessian, H.; Taxiarchis, A.; Popov, S.; Silveira, A.; Franco-Cereceda, A.; Hamsten, A.; Eriksson, P.; van’t Hooft, F. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. USA 2014, 111, 8913–8918. [Google Scholar] [CrossRef]
- Beer, N.L.; Tribble, N.D.; McCulloch, L.J.; Roos, C.; Johnson, P.R.; Orho-Melander, M.; Gloyn, A.L. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum. Mol. Genet. 2009, 18, 4081–4088. [Google Scholar] [CrossRef]
- Speliotes, E.K.; Yerges-Armstrong, L.M.; Wu, J.; Hernaez, R.; Kim, L.J.; Palmer, C.D.; Gudnason, V.; Eiriksdottir, G.; Garcia, M.E.; Launer, L.J.; et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011, 7, e1001324. [Google Scholar] [CrossRef]
- Petta, S.; Miele, L.; Bugianesi, E.; Camma, C.; Rosso, C.; Boccia, S.; Cabibi, D.; Di Marco, V.; Grimaudo, S.; Grieco, A.; et al. Glucokinase regulatory protein gene polymorphism affects liver fibrosis in non-alcoholic fatty liver disease. PLoS ONE 2014, 9, e87523. [Google Scholar] [CrossRef]
- Buch, S.; Stickel, F.; Trepo, E.; Way, M.; Herrmann, A.; Nischalke, H.D.; Brosch, M.; Rosendahl, J.; Berg, T.; Ridinger, M.; et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. Nat. Genet. 2015, 47, 1443–1448. [Google Scholar] [CrossRef]
- Mancina, R.M.; Dongiovanni, P.; Petta, S.; Pingitore, P.; Meroni, M.; Rametta, R.; Boren, J.; Montalcini, T.; Pujia, A.; Wiklund, O.; et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology 2016, 150, 1219–1230.e6. [Google Scholar] [CrossRef] [PubMed]
- Viitasalo, A.; Eloranta, A.M.; Atalay, M.; Romeo, S.; Pihlajamaki, J.; Lakka, T.A. Association of MBOAT7 gene variant with plasma ALT levels in children: The PANIC study. Pediatr. Res. 2016, 80, 651–655. [Google Scholar] [CrossRef] [PubMed]
- Donati, B.; Dongiovanni, P.; Romeo, S.; Meroni, M.; McCain, M.; Miele, L.; Petta, S.; Maier, S.; Rosso, C.; De Luca, L.; et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017, 7, 4492. [Google Scholar] [CrossRef] [PubMed]
- Salley, T.N.; Mishra, M.; Tiwari, S.; Jadhav, A.; Ndisang, J.F. The heme oxygenase system rescues hepatic deterioration in the condition of obesity co-morbid with type-2 diabetes. PLoS ONE 2013, 8, e79270. [Google Scholar] [CrossRef]
- Hinds, T.D., Jr.; Sodhi, K.; Meadows, C.; Fedorova, L.; Puri, N.; Kim, D.H.; Peterson, S.J.; Shapiro, J.; Abraham, N.G.; Kappas, A. Increased HO-1 levels ameliorate fatty liver development through a reduction of heme and recruitment of FGF21. Obesity 2014, 22, 705–712. [Google Scholar] [CrossRef]
- Chang, P.F.; Lin, Y.C.; Liu, K.; Yeh, S.J.; Ni, Y.H. Heme oxygenase-1 gene promoter polymorphism and the risk of pediatric nonalcoholic fatty liver disease. Int. J. Obes. 2015, 39, 1236–1240. [Google Scholar] [CrossRef]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F., 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Furey, N.; Johnson, C.G.; Bissig, K.D. Using CRISPR/Cas9 to model human liver disease. JHEP Rep. 2019, 1, 392–402. [Google Scholar] [CrossRef]
- Barrangou, R. The roles of CRISPR-Cas systems in adaptive immunity and beyond. Curr. Opin. Immunol. 2015, 32, 36–41. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Wang, C.; Lee, H.K.; Yoo, K.H.; Zeng, X.; Kuhns, T.; Yang, C.M.; Mohr, T.; Liu, C.; Hennighausen, L. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017, 8, 15464. [Google Scholar] [CrossRef]
- Luukkonen, P.K.; Nick, A.; Holtta-Vuori, M.; Thiele, C.; Isokuortti, E.; Lallukka-Bruck, S.; Zhou, Y.; Hakkarainen, A.; Lundbom, N.; Peltonen, M.; et al. Human PNPLA3-I148M variant increases hepatic retention of polyunsaturated fatty acids. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Fan, Y.; Lu, H.; Guo, Y.; Zhu, T.; Garcia-Barrio, M.T.; Jiang, Z.; Willer, C.J.; Zhang, J.; Chen, Y.E. Hepatic Transmembrane 6 Superfamily Member 2 Regulates Cholesterol Metabolism in Mice. Gastroenterology 2016, 150, 1208–1218. [Google Scholar] [CrossRef]
- O’Hare, E.A.; Yang, R.; Yerges-Armstrong, L.M.; Sreenivasan, U.; McFarland, R.; Leitch, C.C.; Wilson, M.H.; Narina, S.; Gorden, A.; Ryan, K.A.; et al. TM6SF2 rs58542926 impacts lipid processing in liver and small intestine. Hepatology 2017, 65, 1526–1542. [Google Scholar] [CrossRef]
- Codner, G.F.; Mianne, J.; Caulder, A.; Loeffler, J.; Fell, R.; King, R.; Allan, A.J.; Mackenzie, M.; Pike, F.J.; McCabe, C.V.; et al. Application of long single-stranded DNA donors in genome editing: Generation and validation of mouse mutants. BMC Biol. 2018, 16, 70. [Google Scholar] [CrossRef]
- Meroni, M.; Dongiovanni, P.; Longo, M.; Carli, F.; Baselli, G.; Rametta, R.; Pelusi, S.; Badiali, S.; Maggioni, M.; Gaggini, M.; et al. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine 2020, 52, 102658. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for Improving HDR Efficiency. Front. Genet. 2018, 9, 691. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Paquet, D.; Kwart, D.; Chen, A.; Sproul, A.; Jacob, S.; Teo, S.; Olsen, K.M.; Gregg, A.; Noggle, S.; Tessier-Lavigne, M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 2016, 533, 125–129. [Google Scholar] [CrossRef]
- Chen, S.; Lee, B.; Lee, A.Y.; Modzelewski, A.J.; He, L. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J. Biol. Chem. 2016, 291, 14457–14467. [Google Scholar] [CrossRef]
- Shin, H.Y.; Willi, M.; HyunYoo, K.; Zeng, X.; Wang, C.; Metser, G.; Hennighausen, L. Hierarchy within the mammary STAT5-driven Wap super-enhancer. Nat. Genet. 2016, 48, 904–911. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef]
- Liang, X.; Potter, J.; Kumar, S.; Zou, Y.; Quintanilla, R.; Sridharan, M.; Carte, J.; Chen, W.; Roark, N.; Ranganathan, S.; et al. Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J. Biotechnol. 2015, 208, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Yesilbag Tonga, G.; Lee, Y.W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Arai, Y.; Yamazaki, M.; Morita, S.; Kimura, M.; Itoh, M.; Abe, Y.; Hatada, I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci. Rep. 2014, 4, 4513. [Google Scholar] [CrossRef]
- Chuang, C.K.; Chen, C.H.; Huang, C.L.; Su, Y.H.; Peng, S.H.; Lin, T.Y.; Tai, H.C.; Yang, T.S.; Tu, C.F. Generation of GGTA1 Mutant Pigs by Direct Pronuclear Microinjection of CRISPR/Cas9 Plasmid Vectors. Anim. Biotechnol. 2017, 28, 174–181. [Google Scholar] [CrossRef]
- Crispo, M.; Mulet, A.P.; Tesson, L.; Barrera, N.; Cuadro, F.; dos Santos-Neto, P.C.; Nguyen, T.H.; Creneguy, A.; Brusselle, L.; Anegon, I.; et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS ONE 2015, 10, e0136690. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
- Biagioni, A.; Laurenzana, A.; Margheri, F.; Chilla, A.; Fibbi, G.; Del Rosso, M. Delivery systems of CRISPR/Cas9-based cancer gene therapy. J. Biol. Eng. 2018, 12, 33. [Google Scholar] [CrossRef]
- Raveux, A.; Vandormael-Pournin, S.; Cohen-Tannoudji, M. Optimization of the production of knock-in alleles by CRISPR/Cas9 microinjection into the mouse zygote. Sci. Rep. 2017, 7, 42661. [Google Scholar] [CrossRef]
- Metser, G.; Shin, H.Y.; Wang, C.; Yoo, K.H.; Oh, S.; Villarino, A.V.; O’Shea, J.J.; Kang, K.; Hennighausen, L. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 2016, 44, 1052–1063. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef]
- Hung, S.S.; Chrysostomou, V.; Li, F.; Lim, J.K.; Wang, J.H.; Powell, J.E.; Tu, L.; Daniszewski, M.; Lo, C.; Wong, R.C.; et al. AAV-Mediated CRISPR/Cas Gene Editing of Retinal Cells In Vivo. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3470–3476. [Google Scholar] [CrossRef]
- Lau, C.H.; Suh, Y. In vivo genome editing in animals using AAV-CRISPR system: Applications to translational research of human disease. F1000Res 2017, 6, 2153. [Google Scholar] [CrossRef]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Swiech, L.; Heidenreich, M.; Banerjee, A.; Habib, N.; Li, Y.; Trombetta, J.; Sur, M.; Zhang, F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 2015, 33, 102–106. [Google Scholar] [CrossRef]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef]
- Carroll, K.J.; Makarewich, C.A.; McAnally, J.; Anderson, D.M.; Zentilin, L.; Liu, N.; Giacca, M.; Bassel-Duby, R.; Olson, E.N. A mouse model for adult cardiac-specific gene deletion with CRISPR/Cas9. Proc. Natl. Acad. Sci. USA 2016, 113, 338–343. [Google Scholar] [CrossRef]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef]
- Voets, O.; Tielen, F.; Elstak, E.; Benschop, J.; Grimbergen, M.; Stallen, J.; Janssen, R.; van Marle, A.; Essrich, C. Highly efficient gene inactivation by adenoviral CRISPR/Cas9 in human primary cells. PLoS ONE 2017, 12, e0182974. [Google Scholar] [CrossRef]
- Maddalo, D.; Manchado, E.; Concepcion, C.P.; Bonetti, C.; Vidigal, J.A.; Han, Y.C.; Ogrodowski, P.; Crippa, A.; Rekhtman, N.; de Stanchina, E.; et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature 2014, 516, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Mou, H.; Li, S.; Li, Y.; Hough, S.; Tran, K.; Li, J.; Yin, H.; Anderson, D.G.; Sontheimer, E.J.; et al. Adenovirus-Mediated Somatic Genome Editing of Pten by CRISPR/Cas9 in Mouse Liver in Spite of Cas9-Specific Immune Responses. Hum. Gene. Ther. 2015, 26, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Strong, A.; Patel, K.M.; Ng, S.L.; Gosis, B.S.; Regan, S.N.; Cowan, C.A.; Rader, D.J.; Musunuru, K. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ. Res. 2014, 115, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Lindel, F.; Dodt, C.R.; Weidner, N.; Noll, M.; Bergemann, F.; Behrendt, R.; Fischer, S.; Dietrich, J.; Cartellieri, M.; Hamann, M.V.; et al. TraFo-CRISPR: Enhanced Genome Engineering by Transient Foamy Virus Vector-Mediated Delivery of CRISPR/Cas9 Components. Mol. Ther. Nucleic Acids 2019, 18, 708–726. [Google Scholar] [CrossRef]
- Stenger, D.; Stief, T.A.; Kaeuferle, T.; Willier, S.; Rataj, F.; Schober, K.; Vick, B.; Lotfi, R.; Wagner, B.; Grunewald, T.G.P.; et al. Endogenous TCR promotes in vivo persistence of CD19-CAR-T cells compared to a CRISPR/Cas9-mediated TCR knockout CAR. Blood 2020, 136, 1407–1418. [Google Scholar] [CrossRef]
- Vila-Navarro, E.; Fernandez-Castaner, E.; Rovira-Rigau, M.; Raimondi, G.; Vila-Casadesus, M.; Lozano, J.J.; Soubeyran, P.; Iovanna, J.; Castells, A.; Fillat, C.; et al. MiR-93 is related to poor prognosis in pancreatic cancer and promotes tumor progression by targeting microtubule dynamics. Oncogenesis 2020, 9, 43. [Google Scholar] [CrossRef]
- Huo, W.; Zhao, G.; Yin, J.; Ouyang, X.; Wang, Y.; Yang, C.; Wang, B.; Dong, P.; Wang, Z.; Watari, H.; et al. Lentiviral CRISPR/Cas9 vector mediated miR-21 gene editing inhibits the epithelial to mesenchymal transition in ovarian cancer cells. J. Cancer 2017, 8, 57–64. [Google Scholar] [CrossRef]
- Kabadi, A.M.; Ousterout, D.G.; Hilton, I.B.; Gersbach, C.A. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014, 42, e147. [Google Scholar] [CrossRef]
- Annunziato, S.; Kas, S.M.; Nethe, M.; Yucel, H.; Del Bravo, J.; Pritchard, C.; Bin Ali, R.; van Gerwen, B.; Siteur, B.; Drenth, A.P.; et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016, 30, 1470–1480. [Google Scholar] [CrossRef]
- Hindriksen, S.; Bramer, A.J.; Truong, M.A.; Vromans, M.J.M.; Post, J.B.; Verlaan-Klink, I.; Snippert, H.J.; Lens, S.M.A.; Hadders, M.A. Baculoviral delivery of CRISPR/Cas9 facilitates efficient genome editing in human cells. PLoS ONE 2017, 12, e0179514. [Google Scholar] [CrossRef]
- Mansouri, M.; Ehsaei, Z.; Taylor, V.; Berger, P. Baculovirus-based genome editing in primary cells. Plasmid 2017, 90, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.H.; Hennighausen, L.; Shin, H.Y. Dissecting Tissue-Specific Super-Enhancers by Integrating Genome-Wide Analyses and CRISPR/Cas9 Genome Editing. J. Mammary Gland Biol. Neoplasia 2019, 24, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Song, M.; Lee, J.; Menon, A.V.; Jung, S.; Kang, Y.M.; Choi, J.W.; Woo, E.; Koh, H.C.; Nam, J.W.; et al. In vivo high-throughput profiling of CRISPR-Cpf1 activity. Nat. Methods 2017, 14, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Ono, C.; Okamoto, T.; Abe, T.; Matsuura, Y. Baculovirus as a Tool for Gene Delivery and Gene Therapy. Viruses 2018, 10, 510. [Google Scholar] [CrossRef] [PubMed]
- Volkman, L.E.; Goldsmith, P.A. In Vitro Survey of Autographa californica Nuclear Polyhedrosis Virus Interaction with Nontarget Vertebrate Host Cells. Appl. Environ. Microbiol. 1983, 45, 1085–1093. [Google Scholar] [CrossRef] [PubMed]
- Smagris, E.; BasuRay, S.; Li, J.; Huang, Y.; Lai, K.M.; Gromada, J.; Cohen, J.C.; Hobbs, H.H. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology 2015, 61, 108–118. [Google Scholar] [CrossRef]
- O’Hare, E.A.; Yerges-Armstrong, L.M.; Perry, J.A.; Shuldiner, A.R.; Zaghloul, N.A. Assignment of Functional Relevance to Genes at Type 2 Diabetes-Associated Loci Through Investigation of beta-Cell Mass Deficits. Mol. Endocrinol. 2016, 30, 429–445. [Google Scholar] [CrossRef]
- Tanaka, Y.; Shimanaka, Y.; Caddeo, A.; Kubo, T.; Mao, Y.; Kubota, T.; Kubota, N.; Yamauchi, T.; Mancina, R.M.; Baselli, G.; et al. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut 2020. [Google Scholar] [CrossRef]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, M.; Zheng, T.; Hou, Y.; Zhang, P.; Tang, T.; Wei, J.; Du, Q. Specificity profiling of CRISPR system reveals greatly enhanced off-target gene editing. Sci. Rep. 2020, 10, 2269. [Google Scholar] [CrossRef]
- Lee, H.K.; Willi, M.; Miller, S.M.; Kim, S.; Liu, C.; Liu, D.R.; Hennighausen, L. Targeting fidelity of adenine and cytosine base editors in mouse embryos. Nat. Commun. 2018, 9, 4804. [Google Scholar] [CrossRef] [PubMed]
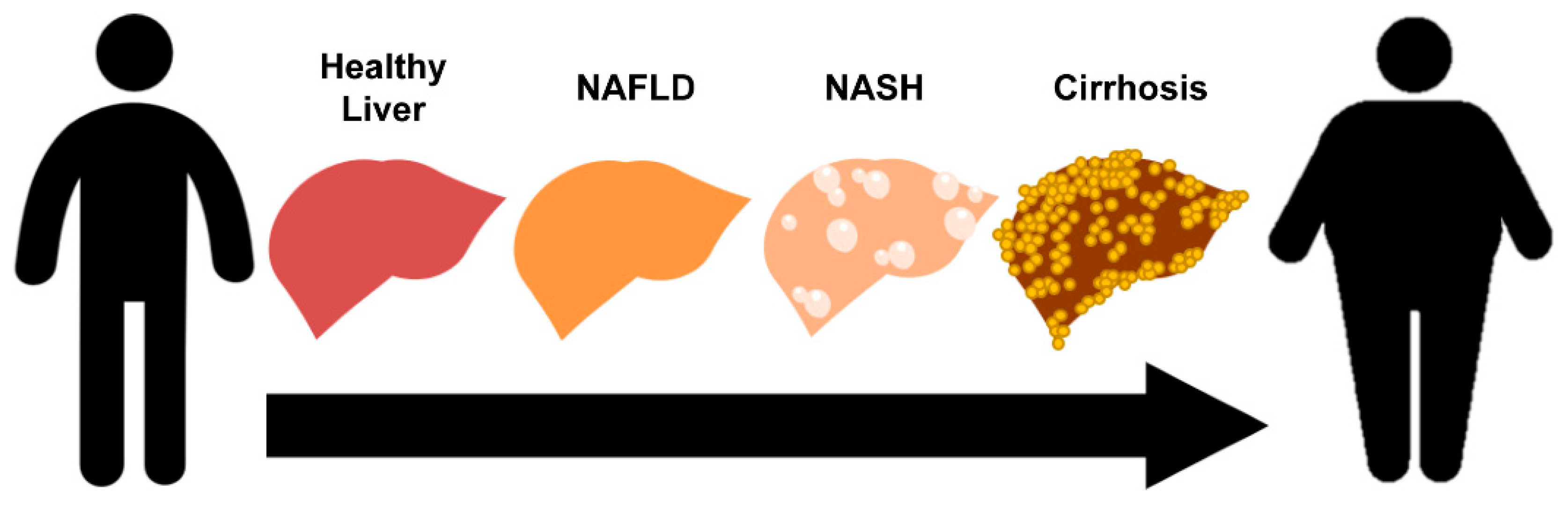
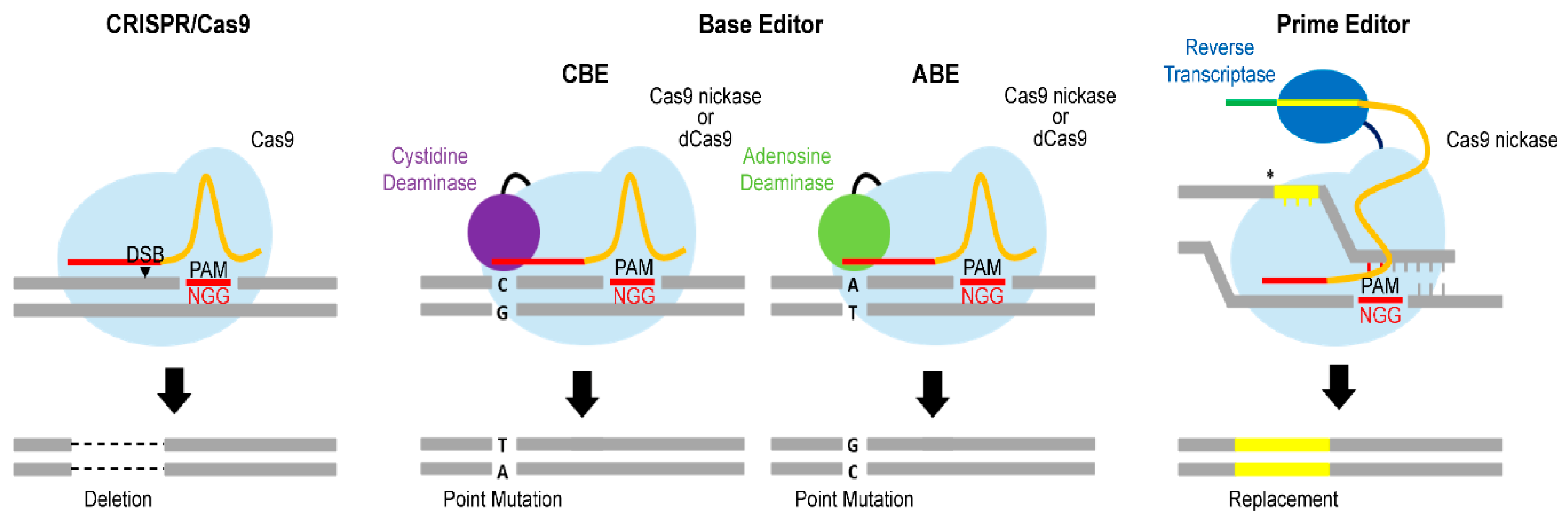
| Gene | Function | Variant | Phenotype of Genetic Variant | Reference |
|---|---|---|---|---|
| PNPLA3 | TG lipase | rs738409 C > G (I148M) | Increased hepatic fat content, high risk of hepatic steatosis, fibrosis, and HCC | [11,12,13] |
| TM6SF2 | Involved in very low-density lipoprotein (VLDL) secretion and hepatic TG metabolism | rs58542926 C > T (E167K) | Elevated hepatic TG, serum ALT, and AST levels | [14,15] |
| rs10401969 C > T (intron) | Low hepatic TM6SF2 mRNA levels correlated with larger hepatocellular lipid droplets | [16] | ||
| GCKR | Regulator of glucokinase | rs1260326 T>C/T>G (P446L) | Increased hepatic TG and LDL-cholesterol levels, correlated with the severity of NASH | [17] |
| rs780094 C > T | Increased hepatic TG levels, high risk of liver fibrosis | [18,19] | ||
| MBOAT7 | Reacylation of phospholipid | rs641738 C > T | Increased hepatic TG levels and severity of NAFLD, high risk of HCC | [20,21,22,23] |
| HMOX1 | Protects the liver against oxidative stress | rs2071746 A > T (promoter) | Increased serum ALT levels in pediatric NAFLD patients | [24,25,26] |
| Target Gene | Model Organism | Genotypic Outcomes | Delivery of Cas9 | Phenotypic Effects | Ref. |
|---|---|---|---|---|---|
| PNPLA3 (rs738409) | Human epidermal carcinoma cell line (A431) | 2-bp deletion after 146C | Transfection | Increased lipid droplet accumulation | [35] |
| I148M | Transfection | Increased levels of neutral lipids | [35] | ||
| TM6SF2 (rs58542926) | Mouse | C/G insertion after start codon | Microinjection | Increased serum TG levels with a high-fat diet | [36] |
| Zebrafish | Partial deletion of exon 3 or exon 4 | Microinjection | Increased hepatic lipid accumulation | [37] | |
| GCKR (rs1260326) | Mouse | P446L | Microinjection | N/A | [38] |
| MBOAT7 (rs641738) | Human HCC cell line (HepG2) | 31-bp, 91-bp, or 101-bp deletion | N/A | Increased fat accumulation | [39] |
| Type | Delivery Methods | Advantages | Limitations | Reference |
|---|---|---|---|---|
| Non-viral | Electroporation/nucleofection | Delivery to cell population, well-characterized | In vitro only; some cells are not suitable | [32,51] |
| Microinjection | Efficient germ line delivery | Technically challenging, laborious | [48,49,59,60] | |
| Liposomes | Simple, easy to manipulate, low cost | In vitro only, endosomal degradation of cargo, specific cell tropism | [31,42,44,45] | |
| Gold nanoparticles (AuNPs) | Inert, low immune responses | Not well characterized | [52,61] | |
| Viral | Adeno-associated virus (AAV) | Efficient delivery to both dividing and non-dividing cells | Very low genome capacity (<5 kb nucleic acid) | [62,63,64,65,66,67,68,69] |
| Adenovirus | High delivery efficiency, high genome capacity (up to 30 kb nucleic acid) | Adaptive immune responses | [70,71,72,73] | |
| Retrovirus | High delivery efficiency, persistent gene expression | Only dividing cells can be infected, unwanted viral genome integration | [74,75,76] | |
| Lentivirus | Long-term gene expression, relatively high genome capacity, (~10 kb) | Unwanted viral genome integration, strong immune responses | [77,78,79] | |
| Baculovirus | Very high genome capacity (>100 kb), minimal immunogenicity | Not well characterized | [80,81] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, U.; Kim, N.; Shin, H.Y. Modeling Non-Alcoholic Fatty Liver Disease (NAFLD) Using “Good-Fit” Genome-Editing Tools. Cells 2020, 9, 2572. https://doi.org/10.3390/cells9122572
Kim U, Kim N, Shin HY. Modeling Non-Alcoholic Fatty Liver Disease (NAFLD) Using “Good-Fit” Genome-Editing Tools. Cells. 2020; 9(12):2572. https://doi.org/10.3390/cells9122572
Chicago/Turabian StyleKim, Uijin, Nahyun Kim, and Ha Youn Shin. 2020. "Modeling Non-Alcoholic Fatty Liver Disease (NAFLD) Using “Good-Fit” Genome-Editing Tools" Cells 9, no. 12: 2572. https://doi.org/10.3390/cells9122572
APA StyleKim, U., Kim, N., & Shin, H. Y. (2020). Modeling Non-Alcoholic Fatty Liver Disease (NAFLD) Using “Good-Fit” Genome-Editing Tools. Cells, 9(12), 2572. https://doi.org/10.3390/cells9122572





