A Zebrafish Model of Retinitis Pigmentosa Shows Continuous Degeneration and Regeneration of Rod Photoreceptors
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Husbandry
2.2. Transgene Construction and Development of Transgenic Fish
2.3. BrdU Labeling
2.4. Tissue Preparation, Histology, Immunocytochemistry (ICCH) and Imaging
2.5. TUNEL Staining
2.6. Quantitative Real-Time PCR
2.7. Statistical Analysis
3. Results
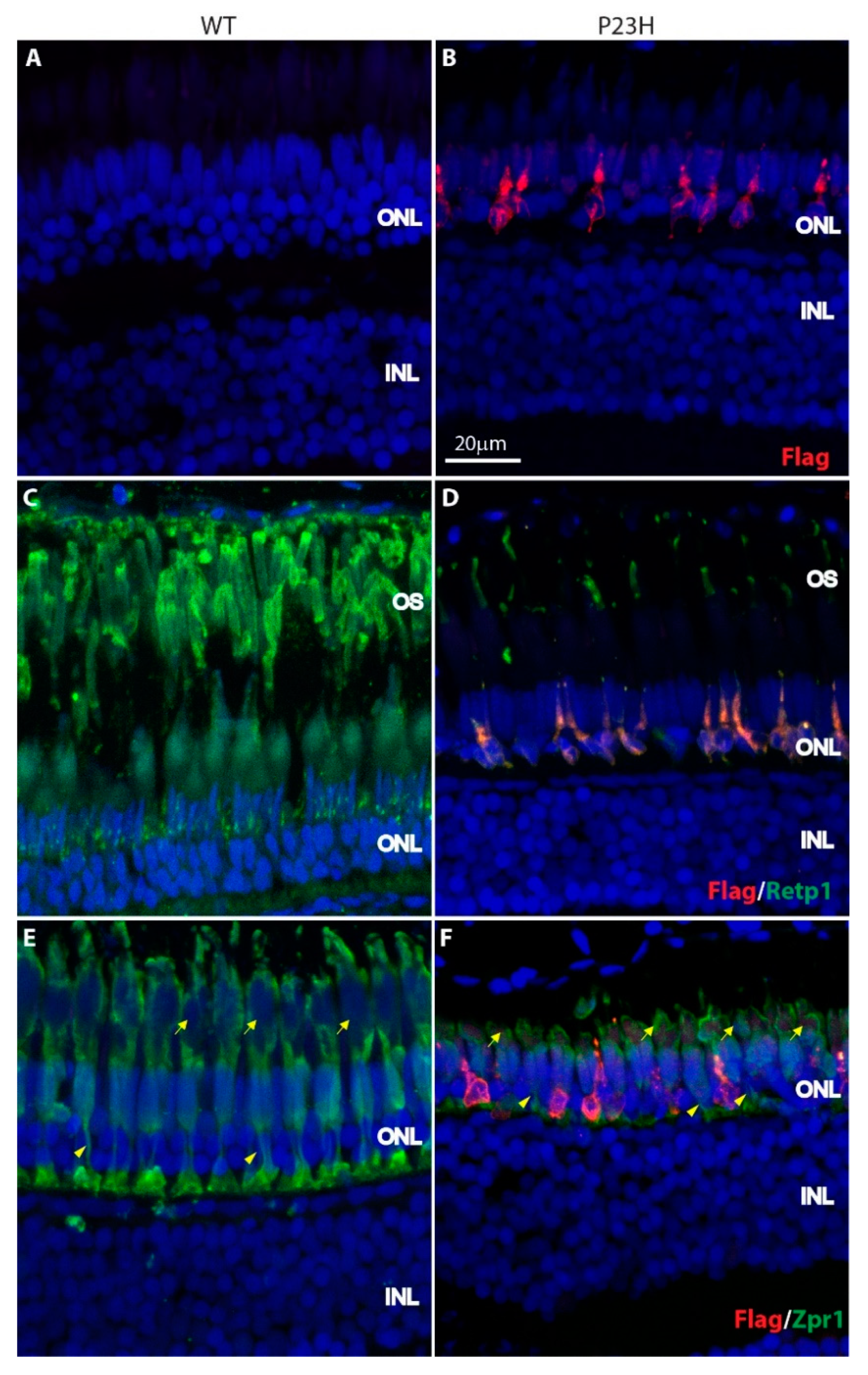
3.1. Mutant Rhodopsin is Expressed in the Rod Photoreceptors
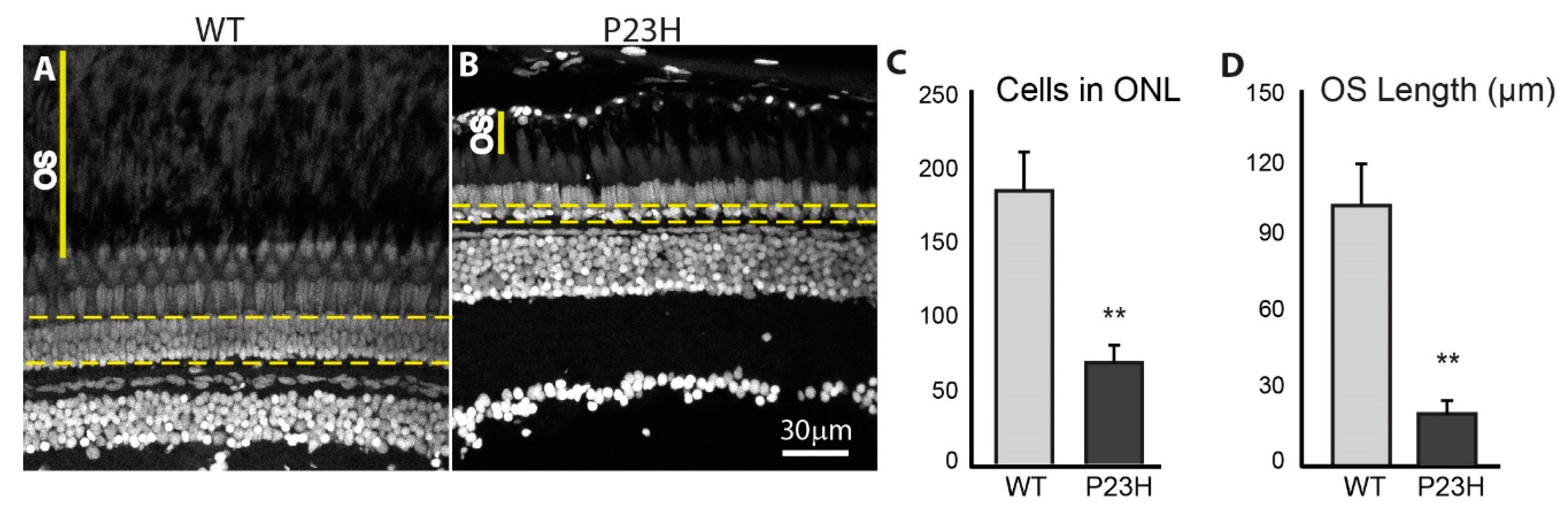
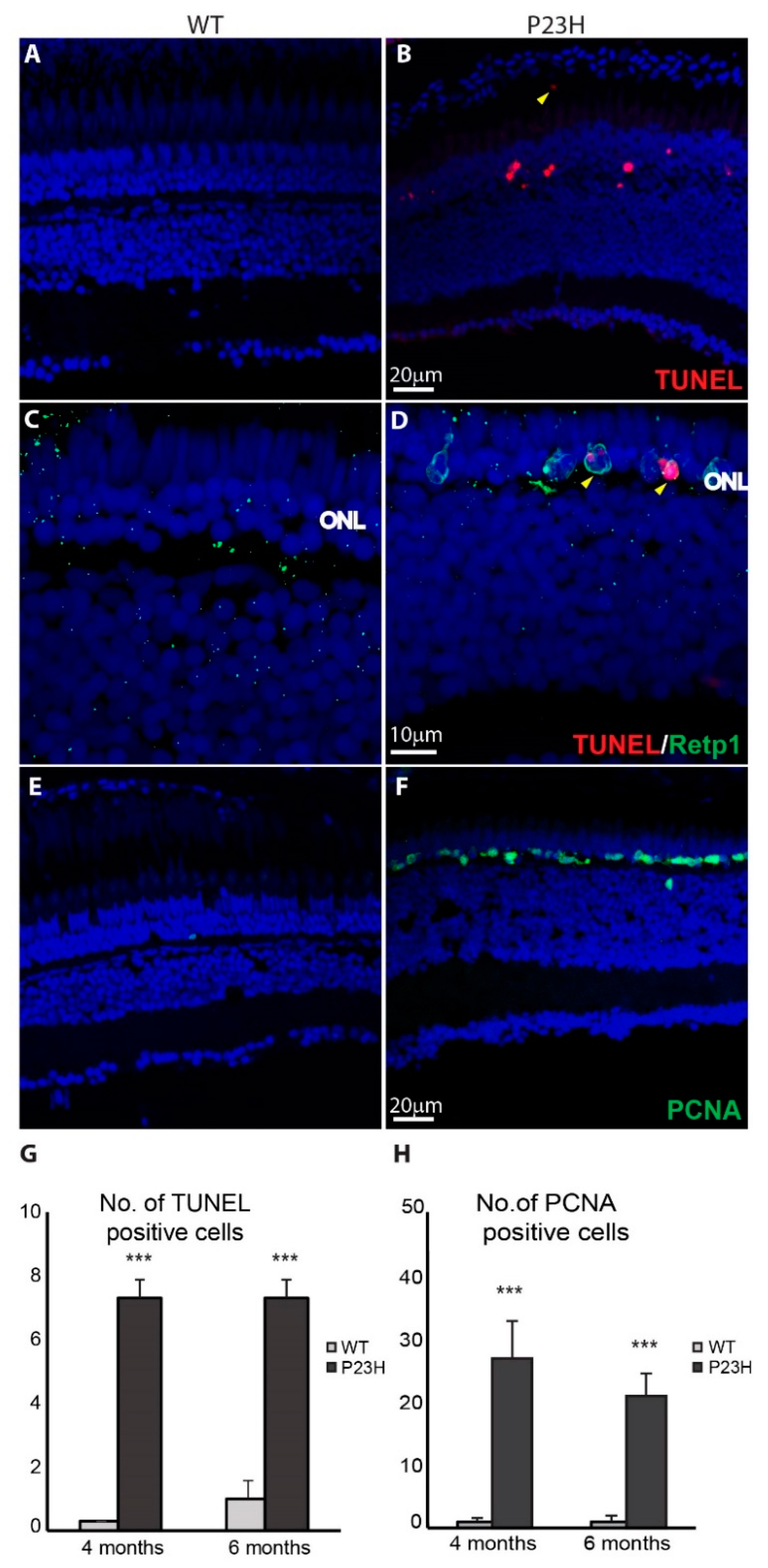
3.2. Degeneration of Rod Photoreceptors in the P23H Transgenic Zebrafish
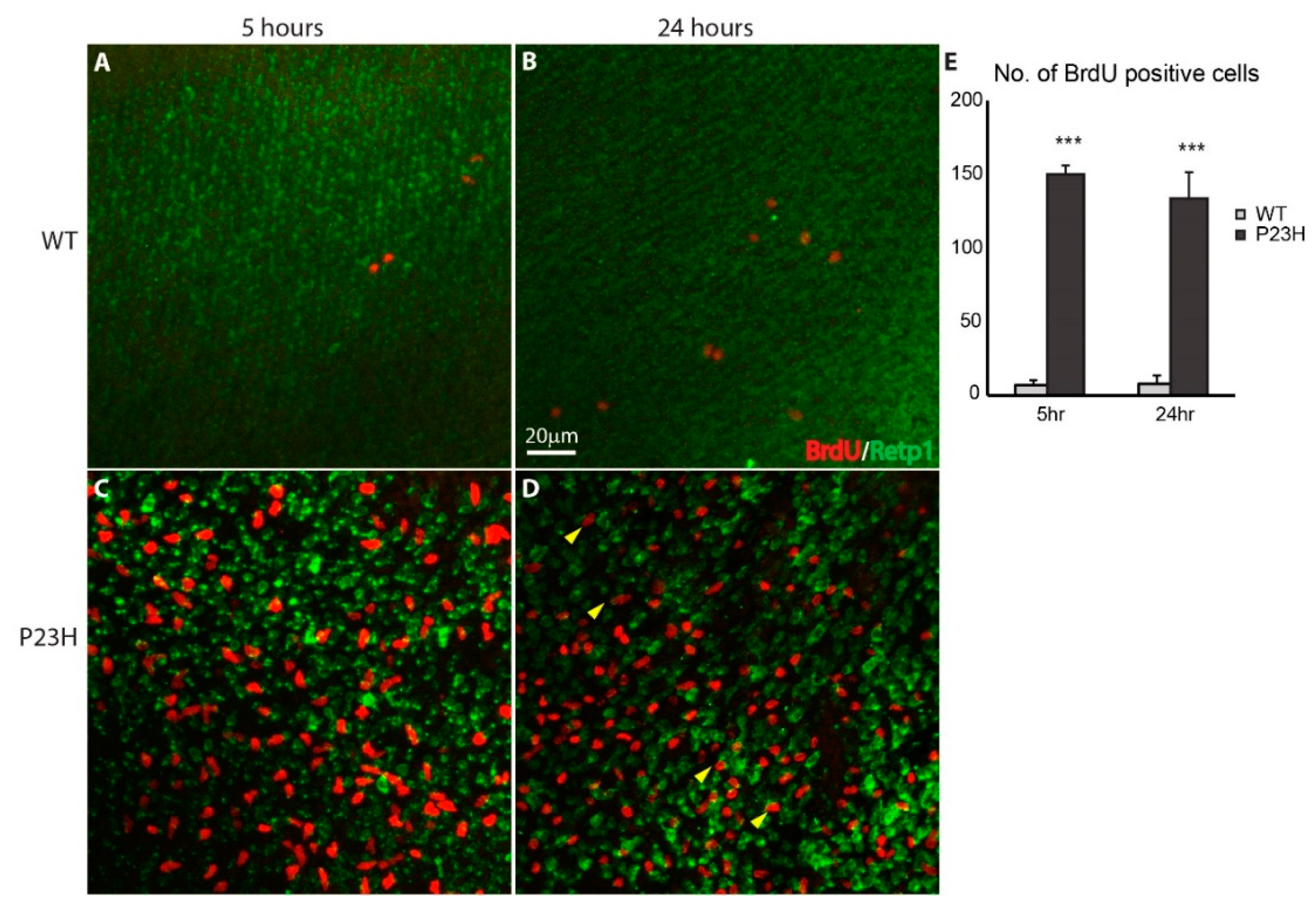
3.3. Regeneration in the P23H Transgenic Zebrafish
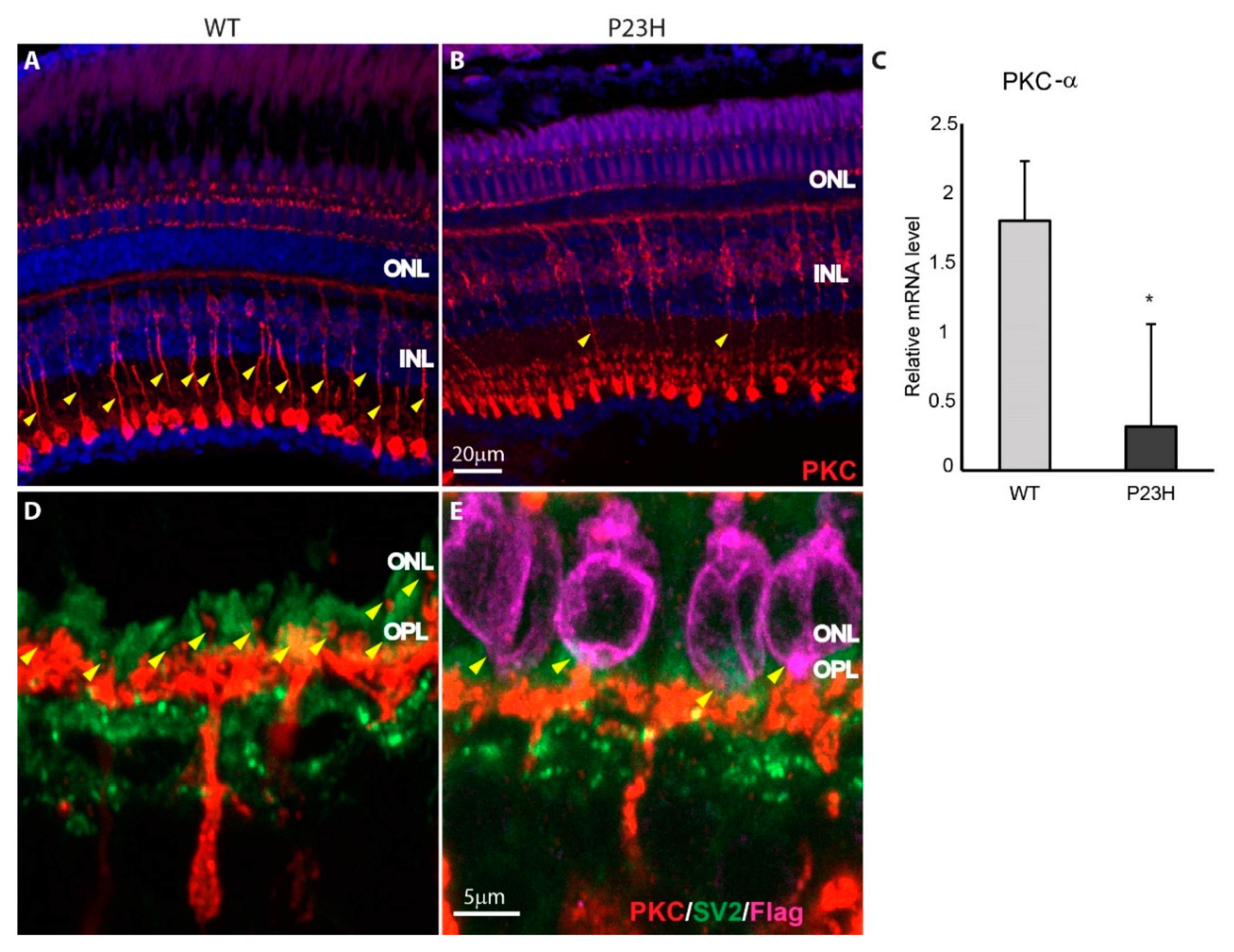
3.4. Retina Remodeling in the P23H Transgenic Zebrafish
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Berson, E.L. Retinitis pigmentosa: Unfolding its mystery. Proc. Natl. Acad. Sci. USA 1996, 93, 4526–4528. [Google Scholar] [CrossRef] [PubMed]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- Frick, K.D.; Roebuck, M.C.; Feldstein, J.I.; McCarty, C.A.; Grover, L.L. Health services utilization and cost of retinitis pigmentosa. Arch. Ophthalmol. 2012, 130, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Daiger, S.P.; Bowne, S.J.; Sullivan, L.S. Perspective on genes and mutations causing retinitis pigmentosa. Arch. Ophthalmol. 2007, 125, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Rivolta, C.; Sharon, D.; DeAngelis, M.M.; Dryja, T.P. Retinitis pigmentosa and allied diseases: Numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 2002, 11, 1219–1227. [Google Scholar] [CrossRef]
- Daiger, S.P.; Bowne, S.J.; Sullivan, L.S. Genes and mutations causing autosomal dominant retinitis pigmentosa. Cold Spring Harb. Perspect. Med. 2015, 5. [Google Scholar] [CrossRef]
- Ziviello, C.; Simonelli, F.; Testa, F.; Anastasi, M.; Marzoli, S.B.; Falsini, B.; Ghiglione, D.; Macaluso, C.; Manitto, M.P.; Garrè, C.; et al. Molecular genetics of autosomal dominant retinitis pigmentosa (ADRP): A comprehensive study of 43 Italian families. J. Med. Genet. 2005, 42, e47. [Google Scholar] [CrossRef]
- Sullivan, L.S.; Bowne, S.J.; Birch, D.G.; Hughbanks-Wheaton, D.; Heckenlively, J.R.; Lewis, R.A.; Garcia, C.A.; Ruiz, R.S.; Blanton, S.H.; Northrup, H.; et al. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3052–3064. [Google Scholar] [CrossRef]
- Chen, Y.; Jastrzebska, B.; Cao, P.; Zhang, J.; Wang, B.; Sun, W.; Yuan, Y.; Feng, Z.; Palczewski, K. Inherent instability of the retinitis pigmentosa P23H mutant opsin. J. Biol. Chem. 2014, 289, 9288–9303. [Google Scholar] [CrossRef]
- Kaushal, S.; Khorana, H.G. Structure and function in rhodopsin. 7. Point mutations associated with autosomal dominant retinitis pigmentosa. Biochemistry 1994, 33, 6121–6128. [Google Scholar] [CrossRef]
- Noorwez, S.M.; Kuksa, V.; Imanishi, Y.; Zhu, L.; Filipek, S.; Palczewski, K.; Kaushal, S. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J. Biol. Chem. 2003, 278, 14442–14450. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Hawes, N.L.; Hurd, R.E.; Davisson, M.T.; Nusinowitz, S.; Heckenlively, J.R. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef]
- Beltran, W.A.; Hammond, P.; Acland, G.M.; Aguirre, G.D. A frameshift mutation in RPGR exon ORF15 causes photoreceptor degeneration and inner retina remodeling in a model of X-linked retinitis pigmentosa. Investig. Ophthalmol. Vis. Sci. 2006, 47, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Falasconi, A.; Biagioni, M.; Novelli, E.; Piano, I.; Gargini, C.; Strettoi, E. Retinal phenotype in the rd9 mutant mouse, a model of X-linked RP. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed]
- Olsson, J.E.; Gordon, J.W.; Pawlyk, B.S.; Roof, D.; Hayes, A.; Molday, R.S.; Mukai, S.; Cowley, G.S.; Berson, E.L.; Dryja, T.P. Transgenic mice with a rhodopsin mutation (Pro23His): A mouse model of autosomal dominant retinitis pigmentosa. Neuron 1992, 9, 815–830. [Google Scholar] [CrossRef]
- Naash, M.I.; Hollyfield, J.G.; al-Ubaidi, M.R.; Baehr, W. Simulation of human autosomal dominant retinitis pigmentosa in transgenic mice expressing a mutated murine opsin gene. Proc. Natl. Acad. Sci. USA 1993, 90, 5499–5503. [Google Scholar] [CrossRef] [PubMed]
- Lewin, A.S.; Drenser, K.A.; Hauswirth, W.W.; Nishikawa, S.; Yasumura, D.; Flannery, J.G.; LaVail, M.M. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat. Med. 1998, 4, 967–971. [Google Scholar] [CrossRef]
- Sakami, S.; Maeda, T.; Bereta, G.; Okano, K.; Golczak, M.; Sumaroka, A.; Roman, A.J.; Cideciyan, A.V.; Jacobson, S.G.; Palczewski, K. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J. Biol. Chem. 2011, 286, 10551–10567. [Google Scholar] [CrossRef]
- Wilken, M.S.; Reh, T.A. Retinal regeneration in birds and mice. Curr. Opin. Genet. Dev. 2016, 40, 57–64. [Google Scholar] [CrossRef]
- Wan, J.; Goldman, D. Retina regeneration in zebrafish. Curr. Opin. Genet. Dev. 2016, 40, 41–47. [Google Scholar] [CrossRef]
- Rueda, E.M.; Hall, B.M.; Hill, M.C.; Swinton, P.G.; Tong, X.; Martin, J.F.; Poché, R.A. The hippo pathway blocks mammalian retinal müller glial cell reprogramming. Cell Rep. 2019, 27, 1637–1649. [Google Scholar] [CrossRef] [PubMed]
- Hamon, A.; Roger, J.E.; Yang, X.-J.; Perron, M. Müller glial cell-dependent regeneration of the neural retina: An overview across vertebrate model systems. Dev. Dyn. 2016, 245, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Goldman, D. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014, 15, 431–442. [Google Scholar] [CrossRef] [PubMed]
- Dooley, K.; Zon, L.I. Zebrafish: A model system for the study of human disease. Curr. Opin. Genet. Dev. 2000, 10, 252–256. [Google Scholar] [CrossRef]
- Wikler, K.C.; Rakic, P. Distribution of photoreceptor subtypes in the retina of diurnal and nocturnal primates. J. Neurosci. 1990, 10, 3390–3401. [Google Scholar] [CrossRef] [PubMed]
- Bilotta, J.; Saszik, S. The zebrafish as a model visual system. Int. J. Dev. Neurosci. 2001, 19, 621–629. [Google Scholar] [CrossRef]
- Yoshimatsu, T.; Schröder, C.; Nevala, N.E.; Berens, P.; Baden, T. Fovea-like photoreceptor specializations underlie single UV cone driven prey-capture behavior in Zebrafish. Neuron 2020, 107, 320–337. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000; Available online: https://zfin.org/zf_info/zfbook/cont.html (accessed on 19 April 2020).
- Urasaki, A.; Morvan, G.; Kawakami, K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics 2006, 174, 639–649. [Google Scholar] [CrossRef]
- Kawakami, K.; Takeda, H.; Kawakami, N.; Kobayashi, M.; Matsuda, N.; Mishina, M. A transposon-mediated gene trap approach identifies developmentally regulated genes in Zebrafish. Dev. Cell 2004, 7, 133–144. [Google Scholar] [CrossRef]
- Meeker, N.D.; Hutchinson, S.A.; Ho, L.; Trede, N.S. Method for isolation of PCR-ready genomic DNA from zebrafish tissues. BioTechniques 2007, 43, 610–612. [Google Scholar] [CrossRef]
- Arocho, A.; Chen, B.; Ladanyi, M.; Pan, Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn. Mol. Pathol. 2006, 15, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Larison, K.D.; Bremiller, R. Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 1990, 109, 567–576. [Google Scholar] [PubMed]
- Nakao, T.; Tsujikawa, M.; Notomi, S.; Ikeda, Y.; Nishida, K. The Role of mislocalized phototransduction in photoreceptor cell death of retinitis pigmentosa. PLoS ONE 2012, 7, e0032472. [Google Scholar] [CrossRef] [PubMed]
- Hargrave, P.A.; Adamus, G.; Arendt, A.; McDowell, J.H.; Wang, J.; Szary, A.; Curtis, D.; Jackson, R.W. Rhodopsin’s amino terminus is a principal antigenic site. Exp. Eye Res. 1986, 42, 363–373. [Google Scholar] [CrossRef]
- Morris, A.C.; Schroeter, E.H.; Bilotta, J.; Wong, R.O.L.; Fadool, J.M. Cone Survival Despite Rod Degeneration in XOPS-mCFP Transgenic Zebrafish. Investig. Ophthalmol. Vis. Sci. 2005, 46, 4762–4771. [Google Scholar] [CrossRef]
- Fadool, J.M. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev. Biol. 2003, 258, 277–290. [Google Scholar] [CrossRef]
- Rashid, K.; Akhtar-Schaefer, I.; Langmann, T. Microglia in retinal degeneration. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef]
- Noailles, A.; Maneu, V.; Campello, L.; Gómez-Vicente, V.; Lax, P.; Cuenca, N. Persistent inflammatory state after photoreceptor loss in an animal model of retinal degeneration. Sci. Rep. 2016, 6, 33356. [Google Scholar] [CrossRef]
- Wolf, H.K.; Dittrich, K.L. Detection of proliferating cell nuclear antigen in diagnostic histopathology. J. Histochem. Cytochem. 1992, 40, 1269–1273. [Google Scholar] [CrossRef]
- Rao, M.B.; Didiano, D.; Patton, J.G. Neurotransmitter-regulated regeneration in the Zebrafish retina. Stem Cell Rep. 2017, 8, 831–842. [Google Scholar] [CrossRef]
- Fimbel, S.M.; Montgomery, J.E.; Burket, C.T.; Hyde, D.R. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J. Neurosci. 2007, 27, 1712–1724. [Google Scholar] [CrossRef] [PubMed]
- Meyn, R.E.; Hewitt, R.R.; Humphrey, R.M. Evaluation of S phase synchronization by analysis of DNA replication in 5-bromodeoxyuridine. Exp. Cell Res. 1973, 82, 137–142. [Google Scholar] [CrossRef]
- Haug, M.F.; Berger, M.; Gesemann, M.; Neuhauss, S.C.F. Differential expression of PKCα and -β in the zebrafish retina. Histochem. Cell Biol. 2019, 151, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Iandiev, I.; Pannicke, T.; Wurm, A.; Hollborn, M.; Wiedemann, P.; Osborne, N.N.; Reichenbach, A. Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Progress Retin. Eye Res. 2009, 28, 423–451. [Google Scholar] [CrossRef] [PubMed]
- Strettoi, E.; Pignatelli, V. Modifications of retinal neurons in a mouse model of retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2000, 97, 11020–11025. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Sánchez, L.; Lax, P.; Isiegas, C.; Ayuso, E.; Ruiz, J.M.; de la Villa, P.; Bosch, F.; de la Rosa, E.J.; Cuenca, N. Proinsulin slows retinal degeneration and vision loss in the P23H rat model of retinitis pigmentosa. Hum. Gene Ther. 2012, 23, 1290–1300. [Google Scholar] [CrossRef] [PubMed]
- Bringmann, A.; Pannicke, T.; Biedermann, B.; Francke, M.; Iandiev, I.; Grosche, J.; Wiedemann, P.; Albrecht, J.; Reichenbach, A. Role of retinal glial cells in neurotransmitter uptake and metabolism. Neurochem. Int. 2009, 54, 143–160. [Google Scholar] [CrossRef]
- Roesch, K.; Stadler, M.B.; Cepko, C.L. Gene expression changes within Müller glial cells in retinitis pigmentosa. Mol. Vis. 2012, 18, 1197–1214. [Google Scholar]
- Brockerhoff, S.E.; Fadool, J.M. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell. Mol. Life Sci. 2011, 68, 651–659. [Google Scholar] [CrossRef]
- Brockerhoff, S.E.; Hurley, J.B.; Janssen-Bienhold, U.; Neuhauss, S.C.; Driever, W.; Dowling, J.E. A behavioral screen for isolating zebrafish mutants with visual system defects. Proc. Natl. Acad. Sci. USA 1995, 92, 10545–10549. [Google Scholar] [CrossRef]
- Stearns, G.; Evangelista, M.; Fadool, J.M.; Brockerhoff, S.E. A Mutation in the cone-specific pde6 gene causes rapid cone photoreceptor degeneration in Zebrafish. J. Neurosci. 2007, 27, 13866–13874. [Google Scholar] [CrossRef] [PubMed]
- Chang, B.; Grau, T.; Dangel, S.; Hurd, R.; Jurklies, B.; Sener, E.C.; Andreasson, S.; Dollfus, H.; Baumann, B.; Bolz, S.; et al. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc. Natl. Acad. Sci. USA 2009, 106, 19581–19586. [Google Scholar] [CrossRef] [PubMed]
- Tsujikawa, M.; Malicki, J. Intraflagellar transport genes are essential for differentiation and survival of vertebrate sensory neurons. Neuron 2004, 42, 703–716. [Google Scholar] [CrossRef]
- Becker, T.S.; Burgess, S.M.; Amsterdam, A.H.; Allende, M.L.; Hopkins, N. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor-1 and avian initiation binding repressor. Development 1998, 125, 4369–4378. [Google Scholar] [PubMed]
- Omori, Y.; Zhao, C.; Saras, A.; Mukhopadhyay, S.; Kim, W.; Furukawa, T.; Sengupta, P.; Veraksa, A.; Malicki, J. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat. Cell Biol. 2008, 10, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Gross, J.M.; Perkins, B.D.; Amsterdam, A.; Egaña, A.; Darland, T.; Matsui, J.I.; Sciascia, S.; Hopkins, N.; Dowling, J.E. Identification of zebrafish insertional mutants with defects in visual system development and function. Genetics 2005, 170, 245–261. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dowling, J.E. Effects of Dopamine Depletion on Visual Sensitivity of Zebrafish. J. Neurosci. 2000, 20, 1893–1903. [Google Scholar] [CrossRef]
- Maaswinkel, H.; Riesbeck, L.E.; Riley, M.E.; Carr, A.L.; Mullin, J.P.; Nakamoto, A.T.; Li, L. Behavioral screening for nightblindness mutants in zebrafish reveals three new loci that cause dominant photoreceptor cell degeneration. Mech. Ageing Dev. 2005, 126, 1079–1089. [Google Scholar] [CrossRef]
- Gross, J.M.; Perkins, B.D. Zebrafish mutants as models for congenital ocular disorders in humans. Mol. Reprod. Dev. 2008, 75, 547–555. [Google Scholar] [CrossRef]
- Liu, F.; Qin, Y.; Yu, S.; Soares, D.C.; Yang, L.; Weng, J.; Li, C.; Gao, M.; Lu, Z.; Hu, X.; et al. Pathogenic mutations in retinitis pigmentosa 2 predominantly result in loss of RP2 protein stability in humans and zebrafish. J. Biol. Chem. 2017, 292, 6225–6239. [Google Scholar] [CrossRef]
- Shu, X.; Zeng, Z.; Gautier, P.; Lennon, A.; Gakovic, M.; Cheetham, M.E.; Patton, E.E.; Wright, A.F. Knockdown of the Zebrafish ortholog of the retinitis pigmentosa 2 (RP2) gene results in retinal degeneration. Investig. Ophthalmol. Vis. Sci. 2011, 52, 2960–2966. [Google Scholar] [CrossRef] [PubMed]
- Kassen, S.C.; Ramanan, V.; Montgomery, J.E.; T Burket, C.; Liu, C.-G.; Vihtelic, T.S.; Hyde, D.R. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev. Neurobiol. 2007, 67, 1009–1031. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.A.; Gentile, K.L.; Middleton, F.A.; Yurco, P. Gene expression profiles of intact and regenerating zebrafish retina. Mol. Vis. 2005, 11, 775–791. [Google Scholar] [PubMed]
- Bernardos, R.L.; Barthel, L.K.; Meyers, J.R.; Raymond, P.A. Late-Stage neuronal progenitors in the retina are radial müller glia that function as retinal stem cells. J. Neurosci. 2007, 27, 7028–7040. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.C.; Scholz, T.L.; Brockerhoff, S.E.; Fadool, J.M. Genetic dissection reveals two separate pathways for rod and cone regeneration in the teleost retina. Dev. Neurobiol. 2008, 68, 605–619. [Google Scholar] [CrossRef]



| Antibody | Host | Antigen | Source | Catalog Number | Dilution |
|---|---|---|---|---|---|
| Retp1 | Ms | Rat Rhodopsin | Novus Biologicals | NB120-3267-0 | 1:200 |
| Centennial, CO, USA | |||||
| Flag-DDK | Ms | DYKDDDDK | Origene | TA50011 | 1:250 |
| Rockville, MD, USA | |||||
| Zpr1/Fret43 | Ms | Fixed Zebrafish retinal cells | ZIRC | AB_10013803 | 1:10 |
| Eugene, OR, USA | |||||
| PCNA | Ms | Protein A-Proliferating Cell Nuclear Antigen fusion protein | Abcam Cambridge, MA, USA | ab29 | 1:100 |
| PCNA | Rb | Synthetic peptide corresponding to Human PCNA aa 200 to the C-terminus | Abcam | Ab18197 | 1:100 |
| SV2 | Ms | Synaptic Vesicle Protein 2a | Developmental Studies Hybridoma Bank | SV2 | 1:100 |
| Iowa City, IA, USA | |||||
| GS-6 | Ms | Glutamine Synthetase | Millipore-Sigma | MAB302 | 1:1000 |
| Burlington, MA, USA | |||||
| BU-1 | Ms | 5-bromo-2-deoxyuridine (BrdU) | Invitrogen | MA3-071 | 1:100 |
| Carlsbad, CA, USA | |||||
| PKC-α | Rb | Protein Kinase Cα | Millipore, Sigma | P4334 | 1:400 |
| Cy3 | Gt | Goat Anti-Mouse IgG Fcγ subclass 2a specific | Jackson ImmunoResearch | 115-165-206 | 1:500 |
| West Grove, PA, USA | |||||
| Alexa Flour 488 | Dk | Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | 715-545-150 | 1:500 |
| Alexa Fluor 488 | Dk | Donkey Anti-Rabbit IgG | Jackson ImmunoResearch | 711-545-152 | 1:500 |
| Alexa Fluor 488 | Gt | Goat Anti Mouse IgG Fcγ subclass 1 specific | Jackson ImmunoResearch | 115-545-205 | 1:500 |
| DAPI | Nuclear Counterstaining | Vector Laboratories | H-1000 | ||
| Burlingame, CA, USA |
| Gene | Primer Sequence (5′-3′) | Size (bp) | GenBank Accession |
|---|---|---|---|
| prkca-FP | TCCCCAGTATGTGGCTGGTA | 119 | NM_001256241.1 |
| prkca-RP | TTGGCTATCTCAAATTTCTGTCG | ||
| glula-FP | CGCATTACAGAGCCTGCCTA | 212 | NM_181559.2 |
| glula-RP | ATTCCAGTTGCCTGGGATCG | ||
| GAPDH-FP | ATGACCCCTCCAGCATGA | 134 | NM_213094.2 |
| GAPDH-RP | GGCGGTGTAGGCATGAAC |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santhanam, A.; Shihabeddin, E.; Atkinson, J.A.; Nguyen, D.; Lin, Y.-P.; O’Brien, J. A Zebrafish Model of Retinitis Pigmentosa Shows Continuous Degeneration and Regeneration of Rod Photoreceptors. Cells 2020, 9, 2242. https://doi.org/10.3390/cells9102242
Santhanam A, Shihabeddin E, Atkinson JA, Nguyen D, Lin Y-P, O’Brien J. A Zebrafish Model of Retinitis Pigmentosa Shows Continuous Degeneration and Regeneration of Rod Photoreceptors. Cells. 2020; 9(10):2242. https://doi.org/10.3390/cells9102242
Chicago/Turabian StyleSanthanam, Abirami, Eyad Shihabeddin, Joshua A. Atkinson, Duc Nguyen, Ya-Ping Lin, and John O’Brien. 2020. "A Zebrafish Model of Retinitis Pigmentosa Shows Continuous Degeneration and Regeneration of Rod Photoreceptors" Cells 9, no. 10: 2242. https://doi.org/10.3390/cells9102242
APA StyleSanthanam, A., Shihabeddin, E., Atkinson, J. A., Nguyen, D., Lin, Y.-P., & O’Brien, J. (2020). A Zebrafish Model of Retinitis Pigmentosa Shows Continuous Degeneration and Regeneration of Rod Photoreceptors. Cells, 9(10), 2242. https://doi.org/10.3390/cells9102242





