The Intrinsically Disordered C-Terminal Domain Triggers Nucleolar Localization and Function Switch of PARN in Response to DNA Damage
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmid Construction, Protein Expression and Purification
2.3. Size-Exclusion Chromatography Analysis
2.4. Spectroscopy
2.5. Enzyme Assay
2.6. Cell Culture
2.7. Immunofluorescence Microscopy
2.8. Co-Immunoprecipitation (Co-IP) Assay
2.9. Cell Apoptosis Assay
2.10. Real-Time PCR
2.11. Statistical Analysis
3. Results
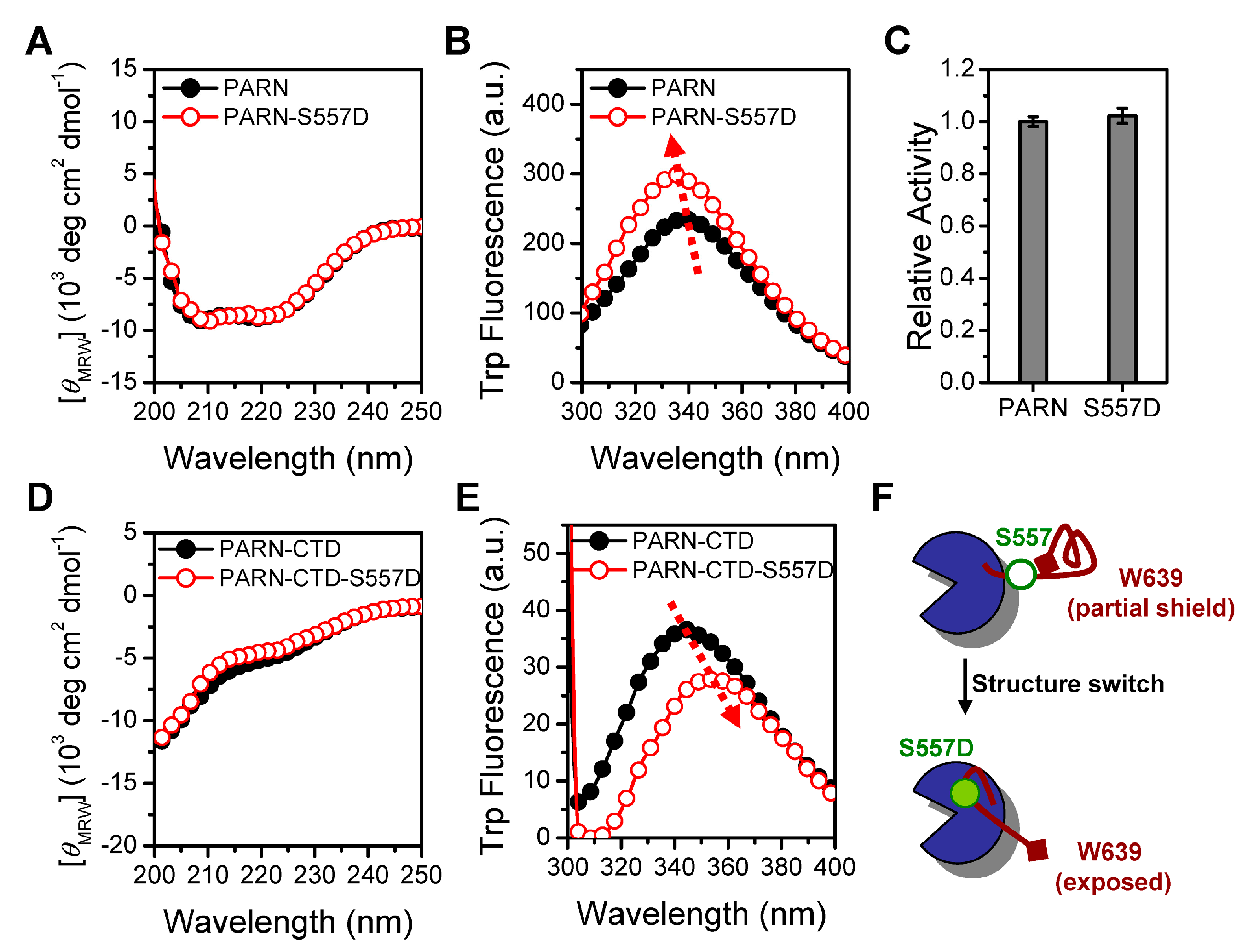
3.1. PARN-CTD Is Intrinsically Disordered with Loosely Packed Local Structures
3.2. Identification of NLS and NoLS in the CTD of PARN
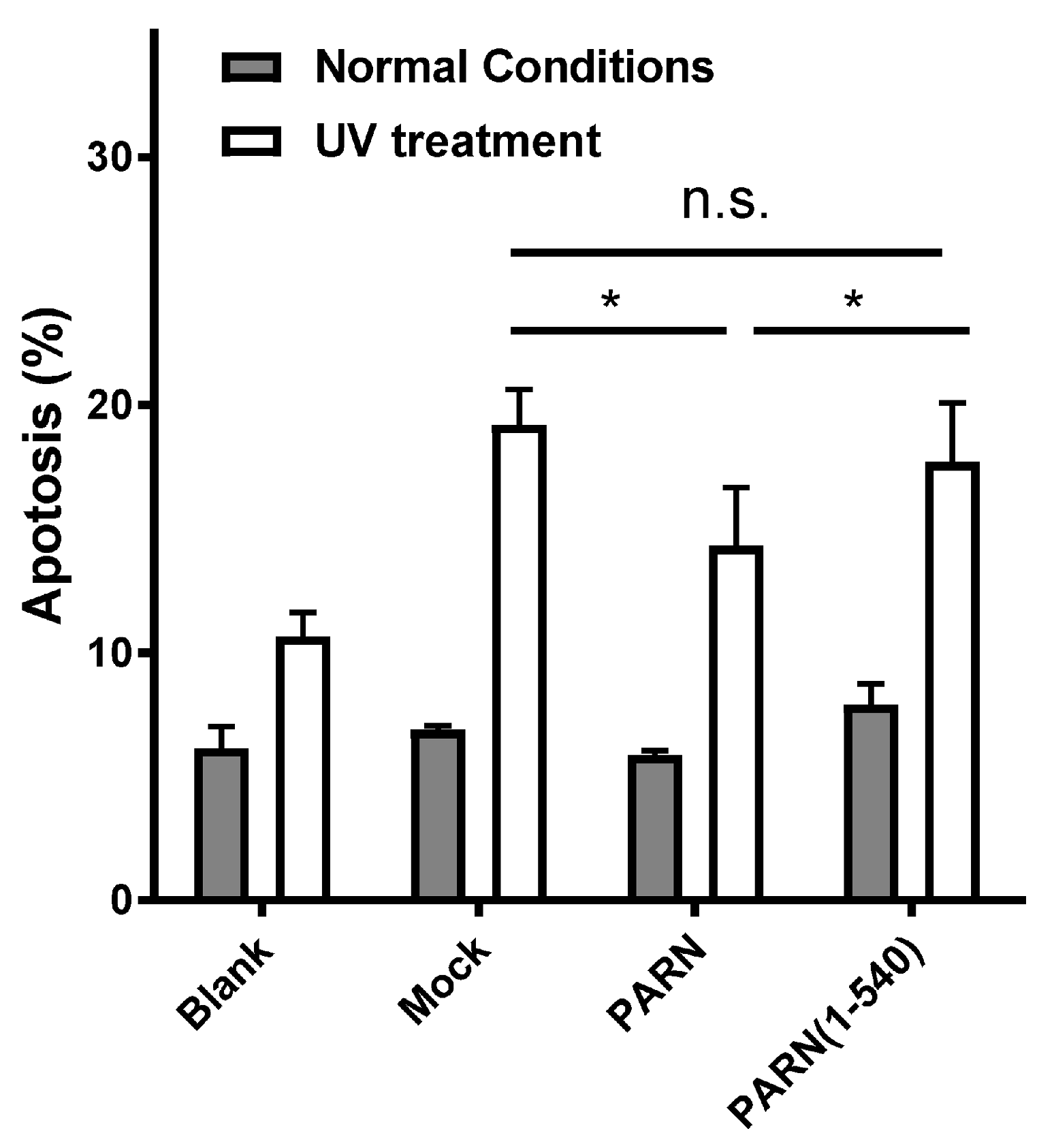
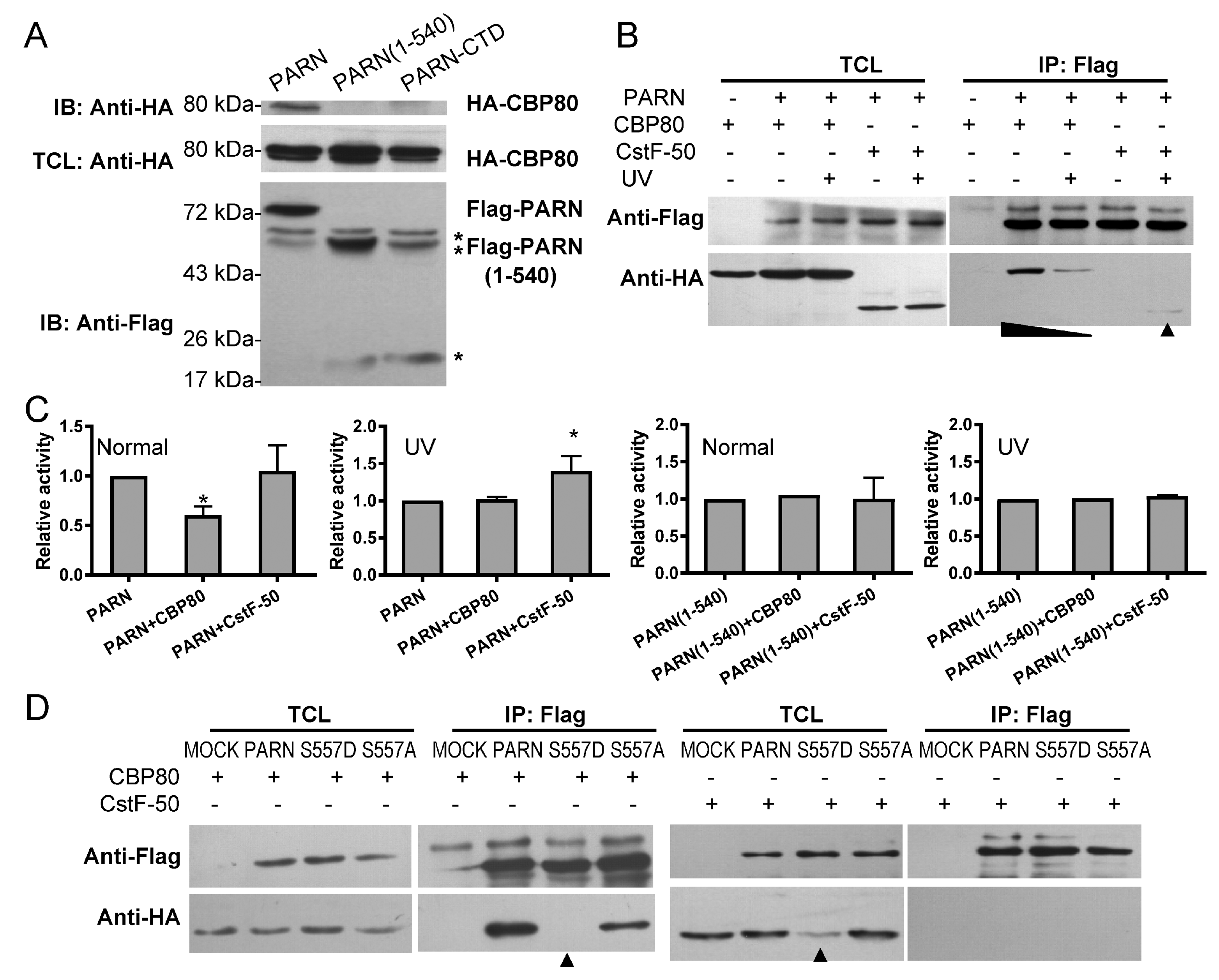
3.3. The CTD Contributes to PARN Function in DDR by Modulating the Protein–Protein Interaction Network
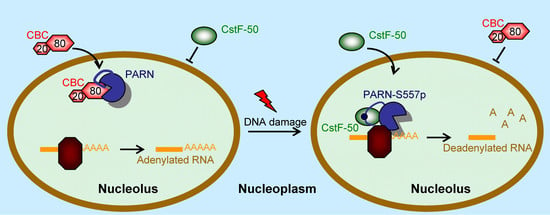
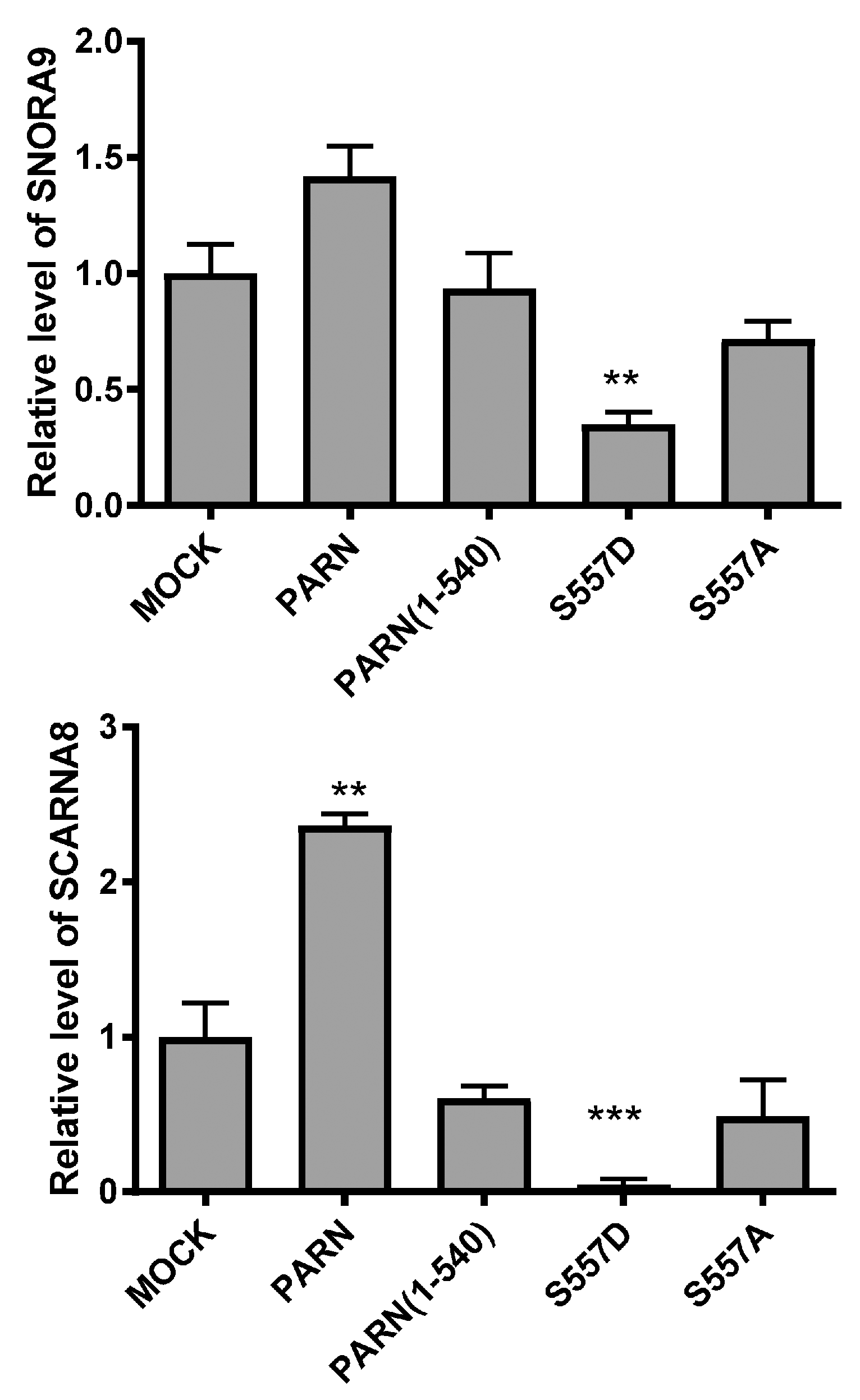
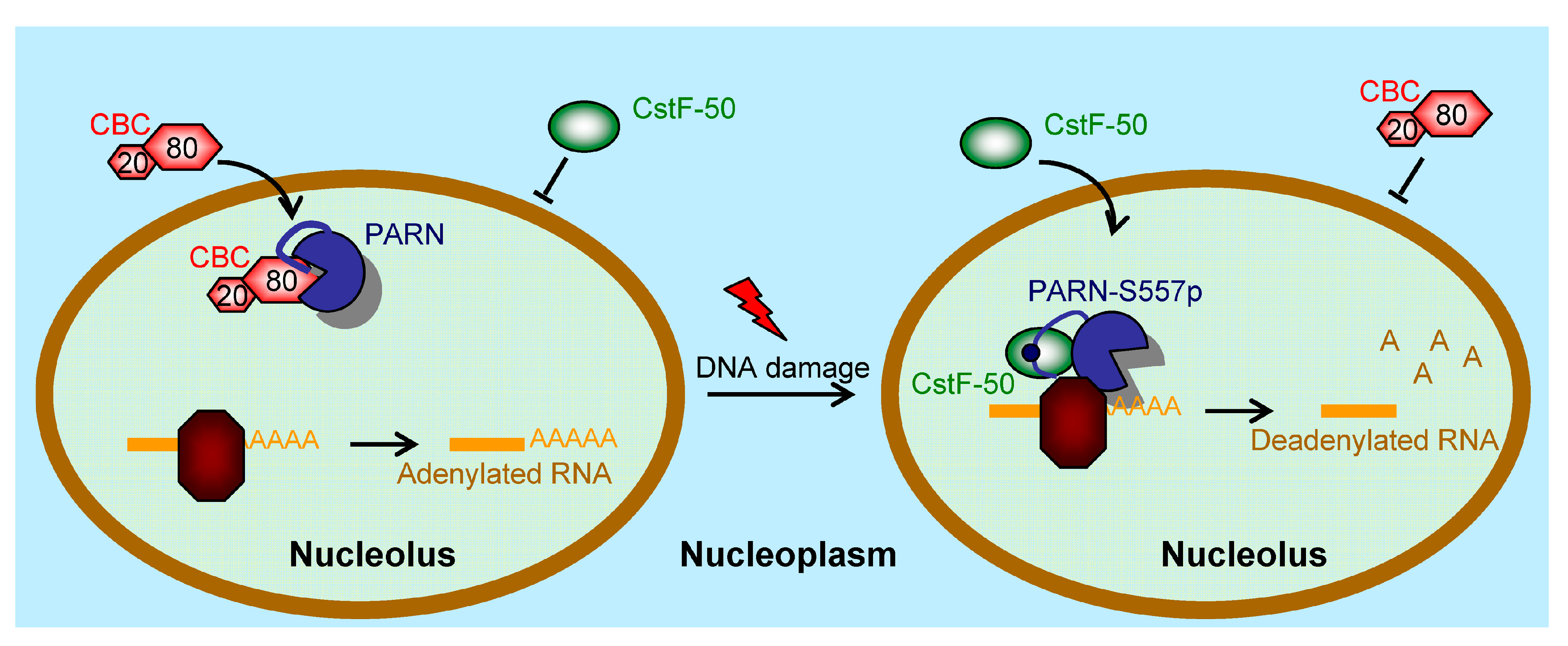
3.4. DNA Damage-Induced Phosphorylation Redefines PARN-Binding Partners in the Nucleoli to Reshape the Profile of Small Nuclear RNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, J.H.; Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 2006, 24, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.B. Deadenylation: Enzymes, regulation, and functional implications. Wiley Interdiscip. Rev. RNA 2014, 5, 421–443. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Virtanen, A.; Kleiman, F.E. To polyadenylate or to deadenylate: That is the question. Cell Cycle 2010, 9, 4437–4449. [Google Scholar] [CrossRef] [PubMed]
- Goldstrohm, A.C.; Wickens, M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008, 9, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M. A history of poly A sequences: From formation to factors to function. Prog Nucleic Acid Res. Mol. Biol. 2002, 71, 285–389. [Google Scholar] [PubMed]
- Garneau, N.L.; Wilusz, J.; Wilusz, C.J. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 2007, 8, 113–126. [Google Scholar] [CrossRef]
- Subtelny, A.O.; Eichhorn, S.W.; Chen, G.R.; Sive, H.; Bartel, D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014, 508, 66–71. [Google Scholar] [CrossRef]
- Jackson, R.J.; Standart, N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell 1990, 62, 15–24. [Google Scholar] [CrossRef]
- Lima, S.A.; Chipman, L.B.; Nicholson, A.L.; Chen, Y.H.; Yee, B.A.; Yeo, G.W.; Coller, J.; Pasquinelli, A.E. Short poly(A) tails are a conserved feature of highly expressed genes. Nat. Struct Mol. Biol. 2017, 24, 1057–1063. [Google Scholar] [CrossRef]
- Chorghade, S.; Seimetz, J.; Emmons, R.; Yang, J.; Bresson, S.M.; Lisio, M.; Parise, G.; Conrad, N.K.; Kalsotra, A. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. eLife 2017. [Google Scholar] [CrossRef]
- Park, J.E.; Yi, H.; Kim, Y.; Chang, H.; Kim, V.N. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol. Cell 2016, 62, 462–471. [Google Scholar] [CrossRef]
- Berndt, H.; Harnisch, C.; Rammelt, C.; Stohr, N.; Zirkel, A.; Dohm, J.C.; Himmelbauer, H.; Tavanez, J.P.; Huttelmaier, S.; Wahle, E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA 2012, 18, 958–972. [Google Scholar] [CrossRef]
- Ding, D.; Liu, J.; Dong, K.; Midic, U.; Hess, R.A.; Xie, H.; Demireva, E.Y.; Chen, C. PNLDC1 is essential for piRNA 3′ end trimming and transposon silencing during spermatogenesis in mice. Nat. Commun. 2017. [Google Scholar] [CrossRef]
- Izumi, N.; Shoji, K.; Sakaguchi, Y.; Honda, S.; Kirino, Y.; Suzuki, T.; Katsuma, S.; Tomari, Y. Identification and functional analysis of the pre-piRNA 3′ trimmer in silkworms. Cell 2016, 164, 962–973. [Google Scholar] [CrossRef]
- Yoda, M.; Cifuentes, D.; Izumi, N.; Sakaguchi, Y.; Suzuki, T.; Giraldez, A.J.; Tomari, Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013, 5, 715–726. [Google Scholar] [CrossRef]
- Nishimura, T.; Nagamori, I.; Nakatani, T.; Izumi, N.; Tomari, Y.; Kuramochi-Miyagawa, S.; Nakano, T. PNLDC1, mouse pre-piRNA Trimmer, is required for meiotic and post-meiotic male germ cell development. EMBO Rep. 2018. [Google Scholar] [CrossRef]
- Zhang, L.-N.; Yan, Y.-B. Depletion of poly(A)-specific ribonuclease (PARN) inhibits proliferation of human gastric cancer cells by blocking cell cycle progression. Biochim. Biophys. Acta 2015, 1853, 522–534. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, R.; Cui, Y.; Zhu, Z.; Zhang, Y.; Wu, H.; Zheng, B.; Yue, Q.; Bai, S.; Zeng, W.; et al. An essential role for PNLDC1 in piRNA 3′ end trimming and male fertility in mice. Cell Res. 2017, 27, 1392–1396. [Google Scholar] [CrossRef]
- Balatsos, N.A.; Maragozidis, P.; Anastasakis, D.; Stathopoulos, C. Modulation of poly(A)-specific ribonuclease (PARN): Current knowledge and perspectives. Curr. Med. Chem. 2012, 19, 4838–4849. [Google Scholar] [CrossRef]
- Virtanen, A.; Henriksson, N.; Nilsson, P.; Nissbeck, M. Poly(A)-specific ribonuclease (PARN): An allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol 2013, 48, 192–209. [Google Scholar] [CrossRef]
- Devany, E.; Zhang, X.; Park, J.Y.; Tian, B.; Kleiman, F.E. Positive and negative feedback loops in the p53 and mRNA 3′ processing pathways. Proc. Natl. Acad. Sci. USA 2013, 110, 3351–3356. [Google Scholar] [CrossRef]
- Reinhardt, H.C.; Hasskamp, P.; Schmedding, I.; Morandell, S.; van Vugt, M.A.; Wang, X.; Linding, R.; Ong, S.E.; Weaver, D.; Carr, S.A.; et al. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Mol. Cell 2010, 40, 34–49. [Google Scholar] [CrossRef]
- Cevher, M.A.; Zhang, X.; Fernandez, S.; Kim, S.; Baquero, J.; Nilsson, P.; Lee, S.; Virtanen, A.; Kleiman, F.E. Nuclear deadenylation/polyadenylation factors regulate 3′ processing in response to DNA damage. EMBO J. 2010, 29, 1674–1687. [Google Scholar] [CrossRef]
- Balatsos, N.A.; Nilsson, P.; Mazza, C.; Cusack, S.; Virtanen, A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC). J. Biol. Chem. 2006, 281, 4517–4522. [Google Scholar] [CrossRef]
- Tummala, H.; Walne, A.; Collopy, L.; Cardoso, S.; de la Fuente, J.; Lawson, S.; Powell, J.; Cooper, N.; Foster, A.; Mohammed, S.; et al. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. J. Clin. Invest. 2015, 125, 2151–2160. [Google Scholar] [CrossRef]
- Roake, C.M.; Chen, L.; Chakravarthy, A.L.; Ferrell, J.E., Jr.; Raffa, G.D.; Artandi, S.E. Disruption of telomerase RNA maturation kinetics precipitates disease. Mol. Cell 2019. [Google Scholar] [CrossRef]
- Shukla, S.; Parker, R. PARN modulates Y RNA stability and its 3′-end formation. Mol. Cell Biol. 2017. [Google Scholar] [CrossRef]
- Shukla, S.; Bjerke, G.A.; Muhlrad, D.; Yi, R.; Parker, R. The RNase PARN controls the levels of specific miRNAs that contribute to p53 regulation. Mol. Cell 2019, 73, 1204–1216. [Google Scholar] [CrossRef]
- He, G.J.; Zhang, A.; Liu, W.F.; Cheng, Y.; Yan, Y.B. Conformational stability and multistate unfolding of poly(A)-specific ribonuclease. FEBS J. 2009, 276, 2849–2860. [Google Scholar] [CrossRef]
- Niedzwiecka, A.; Lekka, M.; Nilsson, P.; Virtanen, A. Global architecture of human poly(A)-specific ribonuclease by atomic force microscopy in liquid and dynamic light scattering. Biophys. Chem. 2011, 158, 141–149. [Google Scholar] [CrossRef]
- Liu, W.F.; Zhang, A.; Cheng, Y.; Zhou, H.M.; Yan, Y.B. Allosteric regulation of human poly(A)-specific ribonuclease by cap and potassium ions. Biochem. Biophys. Res. Commun. 2009, 379, 341–345. [Google Scholar] [CrossRef]
- He, G.J.; Zhang, A.; Liu, W.F.; Yan, Y.B. Distinct roles of the R3H and RRM domains in poly(A)-specific ribonuclease structural integrity and catalysis. Biochim. Biophys. Acta 2013, 1834, 1089–1098. [Google Scholar] [CrossRef]
- He, G.J.; Yan, Y.B. Contributions of the C-terminal domain to poly(A)-specific ribonuclease (PARN) stability and self-association. Biochem. Biophys. Rep. 2019. [Google Scholar] [CrossRef]
- Copeland, P.R.; Wormington, M. The mechanism and regulation of deadenylation: Identification and characterization of Xenopus PARN. RNA 2001, 7, 875–886. [Google Scholar] [CrossRef]
- Martìnez, J.; Ren, Y.G.; Thuresson, A.C.; Hellma, U.; Åström, J.; Virtanen, A. A 54-kDa fragment of the poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting poly(A)-specific 3′ exonuclease. J. Biol. Chem. 2000, 275, 24222–24230. [Google Scholar] [CrossRef]
- Liu, W.F.; Zhang, A.; Cheng, Y.; Zhou, H.M.; Yan, Y.B. Effect of magnesium ions on the thermal stability of human poly(A)-specific ribonuclease. FEBS Lett. 2007, 581, 1047–1052. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–252. [Google Scholar] [CrossRef]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Reshetnyak, Y.K.; Burstein, E.A. Decomposition of protein tryptophan fluorescence spectra into log-normal components. II. The statistical proof of discreteness of tryptophan classes in proteins. Biophys. J. 2001, 81, 1710–1734. [Google Scholar] [CrossRef]
- Burstein, E.A.; Abornev, S.M.; Reshetnyak, Y.K. Decomposition of protein tryptophan fluorescence spectra into Log-Normal components. I. Decomposition algorithms. Biophys. J. 2001, 81, 1699–1709. [Google Scholar] [CrossRef]
- He, H.W.; Zhang, J.; Zhou, H.M.; Yan, Y.B. Conformational change in the C-terminal domain is responsible for the initiation of creatine kinase thermal aggregation. Biophys J. 2005, 89, 2650–2658. [Google Scholar] [CrossRef]
- He, G.J.; Yan, Y.B. A deadenylase assay by size-exclusion chromatography. PLoS ONE 2012, 7, e33700. [Google Scholar] [CrossRef][Green Version]
- Wu, M.; Nilsson, P.; Henriksson, N.; Niedzwiecka, A.; Lim, M.K.; Cheng, Z.; Kokkoris, K.; Virtanen, A.; Song, H. Structural basis of m7GpppG binding to poly(A)-specific ribonuclease. Structure 2009, 17, 276–286. [Google Scholar] [CrossRef]
- Horiuchi, M.; Takeuchi, K.; Noda, N.; Muroya, N.; Suzuki, T.; Nakamura, T.; Kawamura-Tsuzuku, J.; Takahasi, K.; Yamamoto, T.; Inagaki, F. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J. Biol. Chem. 2009, 284, 13244–13255. [Google Scholar] [CrossRef]
- Wu, M.S.; Reuter, M.; Lilie, H.; Liu, Y.Y.; Wahle, E.; Song, H.W. Structural insight into poly(A) binding and catalytic mechanism of human PARN. EMBO J. 2005, 24, 4082–4093. [Google Scholar] [CrossRef]
- Monecke, T.; Schell, S.; Dickmanns, A.; Ficner, R. Crystal structure of the RRM domain of poly(A)-specific ribonuclease reveals a novel m7G-cap-binding mode. J. Mol. Biol. 2008, 382, 827–834. [Google Scholar] [CrossRef]
- Turoverov, K.K.; Kuznetsova, I.M.; Uversky, V.N. The protein kingdom extended: Ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog. Biophys. Mol. Biol. 2010, 102, 73–84. [Google Scholar] [CrossRef]
- He, G.J.; Yan, Y.B. Self-association of poly(A)-specific ribonuclease (PARN) triggered by the R3H domain. Biochim. Biophys Acta 2014, 1844, 2077–2085. [Google Scholar] [CrossRef]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–36. [Google Scholar] [CrossRef]
- Scott, M.S.; Troshin, P.V.; Barton, G.J. NoD: A Nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinf. 2011, 12, 317. [Google Scholar] [CrossRef]
- Bah, A.; Vernon, R.M.; Siddiqui, Z.; Krzeminski, M.; Muhandiram, R.; Zhao, C.; Sonenberg, N.; Kay, L.E.; Forman-Kay, J.D. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 2015, 519, 106–109. [Google Scholar] [CrossRef]
- Zinchuk, V.; Zinchuk, O.; Okada, T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: Pushing pixels to explore biological phenomena. Acta Histochem. Cytochem. 2007, 40, 101–111. [Google Scholar] [CrossRef]
- Zhang, X.; Devany, E.; Murphy, M.R.; Glazman, G.; Persaud, M.; Kleiman, F.E. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic Acids Res. 2015, 43, 10925–10938. [Google Scholar] [CrossRef]
- Cammas, A.; Sanchez, B.J.; Lian, X.J.; Dormoy-Raclet, V.; van der Giessen, K.; de Silanes, I.L.; Ma, J.; Wilusz, C.; Richardson, J.; Gorospe, M.; et al. Destabilization of nucleophosmin mRNA by the HuR/KSRP complex is required for muscle fibre formation. Nat. Commun. 2014. [Google Scholar] [CrossRef]
- Moraes, K.C.; Wilusz, C.J.; Wilusz, J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA 2006, 12, 1084–1091. [Google Scholar] [CrossRef]
- Henriksson, N.; Nilsson, P.; Wu, M.; Song, H.; Virtanen, A. Recognition of adenosine residues by the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2010, 285, 163–170. [Google Scholar] [CrossRef]
- Lee, D.; Park, D.; Park, J.H.; Kim, J.H.; Shin, C. Poly(A)-specific ribonuclease sculpts the 3′ ends of microRNAs. RNA 2019, 25, 388–405. [Google Scholar] [CrossRef]
- Katoh, T.; Hojo, H.; Suzuki, T. Destabilization of microRNAs in human cells by 3′ deadenylation mediated by PARN and CUGBP1. Nucleic Acids Res. 2015, 43, 7521–7534. [Google Scholar] [CrossRef]
- Son, A.; Park, J.E.; Kim, V.N. PARN and TOE1 constitute a 3′ end maturation module for nuclear non-coding RNAs. Cell Rep. 2018, 23, 888–898. [Google Scholar] [CrossRef]
- Montellese, C.; Montel-Lehry, N.; Henras, A.K.; Kutay, U.; Gleizes, P.E.; O’Donohue, M.F. Poly(A)-specific ribonuclease is a nuclear ribosome biogenesis factor involved in human 18S rRNA maturation. Nucleic Acids Res. 2017, 45, 6822–6836. [Google Scholar] [CrossRef]
- Moon, D.H.; Segal, M.; Boyraz, B.; Guinan, E.; Hofmann, I.; Cahan, P.; Tai, A.K.; Agarwal, S. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat. Genet. 2015, 47, 1482–1488. [Google Scholar] [CrossRef]
- Shukla, S.; Schmidt, J.C.; Goldfarb, K.C.; Cech, T.R.; Parker, R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol. 2016, 23, 286–292. [Google Scholar] [CrossRef]
- Tseng, C.K.; Wang, H.F.; Burns, A.M.; Schroeder, M.R.; Gaspari, M.; Baumann, P. Human Telomerase RNA Processing and Quality Control. Cell Rep. 2015, 13, 2232–2243. [Google Scholar] [CrossRef]
- Nguyen, D.; Grenier St-Sauveur, V.; Bergeron, D.; Dupuis-Sandoval, F.; Scott, M.S.; Bachand, F. A Polyadenylation-Dependent 3′ End Maturation Pathway Is Required for the Synthesis of the Human Telomerase RNA. Cell Rep. 2015, 13, 2244–2257. [Google Scholar] [CrossRef]
- Seal, R.; Temperley, R.; Wilusz, J.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M. Serum-deprivation stimulates cap-binding by PARN at the expense of eIF4E, consistent with the observed decrease in mRNA stability. Nucleic Acids Res. 2005, 33, 376–387. [Google Scholar] [CrossRef]
- Maragozidis, P.; Karangeli, M.; Labrou, M.; Dimoulou, G.; Papaspyrou, K.; Salataj, E.; Pournaras, S.; Matsouka, P.; Gourgoulianis, K.I.; Balatsos, N.A. Alterations of deadenylase expression in acute leukemias: Evidence for poly(a)-specific ribonuclease as a potential biomarker. Acta Haematol. 2012, 128, 39–46. [Google Scholar] [CrossRef]
- Schneider, R.; Blackledge, M.; Jensen, M.R. Elucidating binding mechanisms and dynamics of intrinsically disordered protein complexes using NMR spectroscopy. Curr. Opin. Struct. Biol. 2018, 54, 10–18. [Google Scholar] [CrossRef]
- Gianni, S.; Dogan, J.; Jemth, P. Coupled binding and folding of intrinsically disordered proteins: What can we learn from kinetics? Curr. Opin. Struct. Biol. 2016, 36, 18–24. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, T.-L.; He, G.-J.; Hu, L.-D.; Yan, Y.-B. The Intrinsically Disordered C-Terminal Domain Triggers Nucleolar Localization and Function Switch of PARN in Response to DNA Damage. Cells 2019, 8, 836. https://doi.org/10.3390/cells8080836
Duan T-L, He G-J, Hu L-D, Yan Y-B. The Intrinsically Disordered C-Terminal Domain Triggers Nucleolar Localization and Function Switch of PARN in Response to DNA Damage. Cells. 2019; 8(8):836. https://doi.org/10.3390/cells8080836
Chicago/Turabian StyleDuan, Tian-Li, Guang-Jun He, Li-Dan Hu, and Yong-Bin Yan. 2019. "The Intrinsically Disordered C-Terminal Domain Triggers Nucleolar Localization and Function Switch of PARN in Response to DNA Damage" Cells 8, no. 8: 836. https://doi.org/10.3390/cells8080836
APA StyleDuan, T.-L., He, G.-J., Hu, L.-D., & Yan, Y.-B. (2019). The Intrinsically Disordered C-Terminal Domain Triggers Nucleolar Localization and Function Switch of PARN in Response to DNA Damage. Cells, 8(8), 836. https://doi.org/10.3390/cells8080836