Long Non-Coding RNAs Target Pathogenetically Relevant Genes and Pathways in Rheumatoid Arthritis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Microarray Analysis
2.3. Protein‒Protein Interaction (PPI) Network Construction and Network Clustering
2.4. Gene Functional Classification and Enrichment Analysis
2.5. Real-Time PCR of LncRNA
2.6. Real-Time PCR of Genes Modulated in RA Patients
2.7. Real-Time PCR of MicroRNA
2.8. Statistical Analysis
3. Results
3.1. High-Throughput Gene and Long Non-Coding RNA Expression Profiling in Peripheral Blood Mononuclear Cells of RA Patients
3.2. Selected Long Non-Coding RNAs Modulated in RA Patients Have the Potential to Regulate Genes Differentially Expressed in the Disease
3.3. LncRNA RP11-498C9.15 Targets RA-Associated Meaningful Signalling Pathways
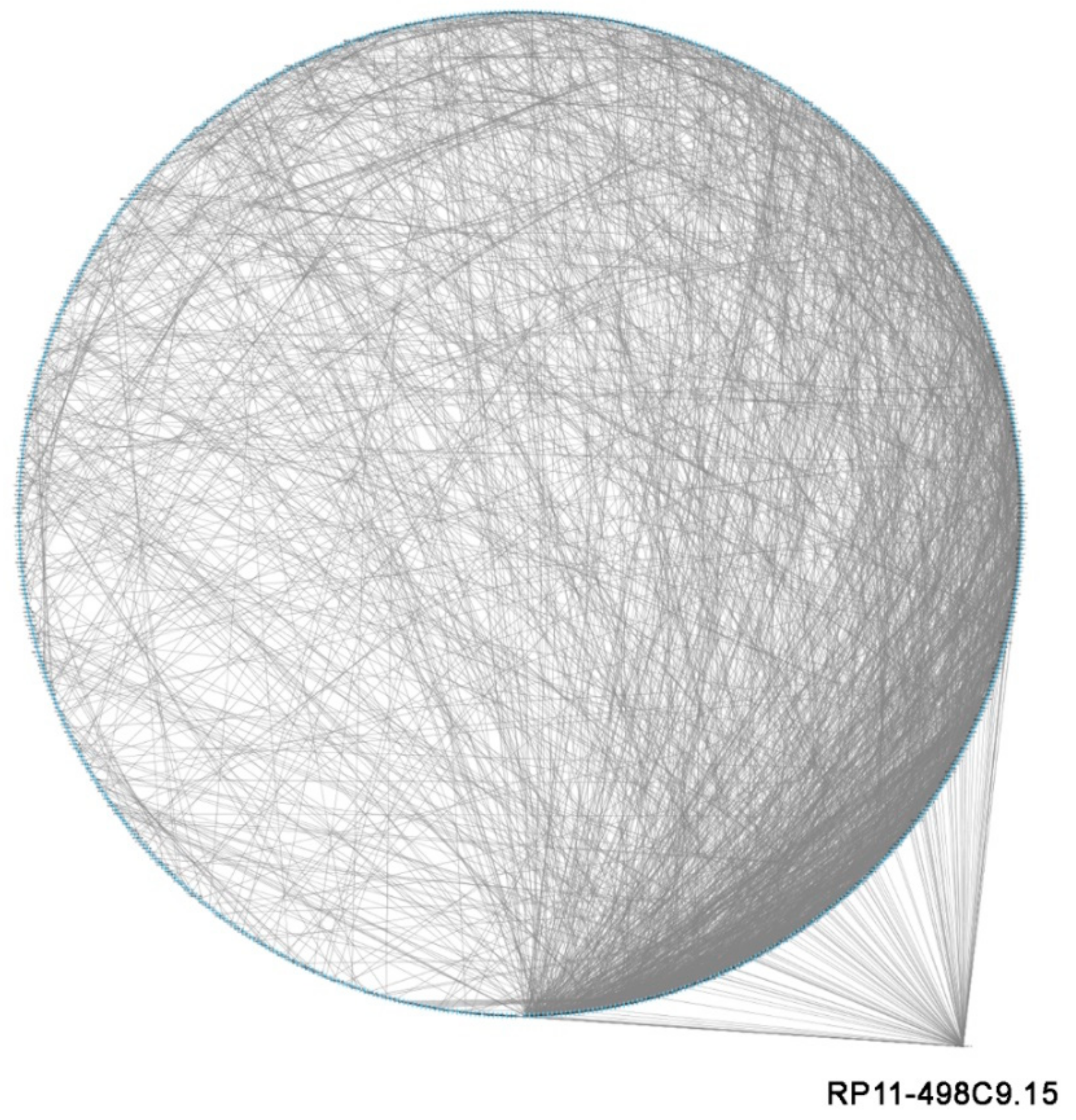
3.4. LncRNA RP11-498C9.15 Targets Highly Connected Genes in the RA Transcriptome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Croia, C.; Bursi, R.; Sutera, D.; Petrelli, F.; Alunno, A.; Puxeddu, I. One year in review 2019: Pathogenesis of rheumatoid arthritis. Clin. Exp. Rheumatol. 2019, 37, 347–357. [Google Scholar] [PubMed]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Pan, H.F.; Leng, R.X.; Wang, D.G.; Li, X.P.; Li, X.M.; Ye, D.Q. Emerging role of long noncoding RNAs in autoimmune diseases. Autoimmun. Rev. 2015, 14, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, M.J.; Jamshidi, A.; Chopra, A.; Aslani, S.; Akhlaghi, M.; Mahmoudi, M. Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J. Cell Physiol. 2018, 234, 335–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Q.; Xu, C.; Li, X.; Zeng, L.; Ye, J.; Guo, Y.; Huang, Z.; Li, J. Comprehensive analysis of long non-coding RNA and mRNA expression profiles in rheumatoid arthritis. Exp. Ther. Med. 2017, 14, 5965–5973. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Wang, S.; Yu, L.; Qu, B.; Xu, L.; Liu, L.; Sun, H.; Li, C.; Shi, Y.; Liu, H. Long noncoding RNA profiling revealed differentially expressed lncRNAs associated with disease activity in PBMCs from patients with rheumatoid arthritis. PLoS ONE 2017, 12, e0186795. [Google Scholar] [CrossRef] [PubMed]
- Aletaha, D.; Neogi, T.; Silman, A.J.; Funovits, J.; Felson, D.T.; Bingham, C.O., 3rd; Birnbaum, N.S.; Burmester, G.R.; Bykerk, V.P.; Cohen, M.D.; et al. 2010 Rheumatoid arthritis classification criteria: An American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum. 2010, 62, 2569–2581. [Google Scholar] [CrossRef]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97, StarBase v2.0. Available online: http://starbase.sysu.edu.cn/starbase2/index.php (accessed on 6 April 2019). [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. Funrich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601, Funrich. Available online: http://www.funrich.org/ (accessed on 12 April 2019). [CrossRef]
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. String 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009, 37, D412–D416, String. Available online: http://string-db.org/ (accessed on 15 April 2019). [CrossRef]
- Cline, M.S.; Smoot, M.; Cerami, E.; Kuchinsky, A.; Landys, N.; Workman, C.; Christmas, R.; Avila-Campilo, I.; Creech, M.; Gross, B.; et al. Integration of biological networks and gene expression data using cytoscape. Nat. Protoc. 2007, 2, 2366–2382, Cytoscape. Available online: http://www.cytoscape.org/ (accessed on 25 April 2019). [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566, Panther. Available online: http://pantherdb.org/ (accessed on 29 June 2019). [CrossRef] [PubMed]
- Yang, D.Q.; Feng, S.; Chen, W.; Zhao, H.; Paulson, C.; Li, Y.P. V-ATPase subunit ATP6AP1 (Ac45) regulates osteoclast differentiation, extracellular acidification, lysosomal trafficking, and protease exocytosis in osteoclast-mediated bone resorption. J. Bone Miner. Res. 2012, 27, 1695–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotake, S.; Nanke, Y.; Kawamoto, M.; Yago, T.; Udagawa, N.; Ichikawa, N.; Kobashigawa, T.; Saito, S.; Momohara, S.; Kamatani, N.; et al. T-cell leukemia translocation-associated gene (TCTA) protein is required for human osteoclastogenesis. Bone 2009, 45, 627–639. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, K.; Uematsu, S.; Kondo, T.; Takeuchi, O.; Martino, M.M.; Kawasaki, T.; Akira, S. Strawberry notch homologue 2 regulates osteoclast fusion by enhancing the expression of DC-STAMP. J. Exp. Med. 2013, 210, 1947–1960. [Google Scholar] [CrossRef]
- Hodge, J.M.; Kirkland, M.A.; Nicholson, G.C. Multiple roles of M-CSF in human osteoclastogenesis. J. Cell Biochem. 2007, 102, 759–768. [Google Scholar] [CrossRef]
- Ramazzotti, G.; Bavelloni, A.; Blalock, W.; Piazzi, M.; Cocco, L.; Faenza, I. BMP-2 Induced Expression of PLCβ1 That is a Positive Regulator of Osteoblast Differentiation. J. Cell Physiol. 2016, 231, 623–629. [Google Scholar] [CrossRef]
- Kutty, R.G.; Xin, G.; Schauder, D.M.; Cossette, S.M.; Bordas, M.; Cui, W.; Ramchandran, R. Dual Specificity Phosphatase 5 Is Essential for T Cell Survival. PLoS ONE 2016, 11, e0167246. [Google Scholar] [CrossRef]
- Alonso, M.A.; Millán, J. The role of lipid rafts in signalling and membrane trafficking in T lymphocytes. J. Cell Sci. 2001, 114, 3957–3965. [Google Scholar]
- Balagopalan, L.; Yi, J.; Nguyen, T.; McIntire, K.M.; Harned, A.S.; Narayan, K.; Samelson, L.E. Plasma membrane LAT activation precedes vesicular recruitment defining two phases of early T-cell activation. Nat. Commun. 2018, 9, 2013. [Google Scholar] [CrossRef] [PubMed]
- Sagi, Y.; Landrigan, A.; Levy, R.; Levy, S. Complementary costimulation of human T-cell subpopulations by cluster of differentiation 28 (CD28) and CD81. Proc. Natl. Acad. Sci. USA 2012, 109, 1613–1618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raychaudhuri, S.; Thomson, B.P.; Remmers, E.F.; Eyre, S.; Hinks, A.; Guiducci, C.; Catanese, J.J.; Xie, G.; Stahl, E.A.; Chen, R.; et al. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009, 41, 1313–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herglotz, J.; Unrau, L.; Hauschildt, F.; Fischer, M.; Kriebitzsch, N.; Alawi, M.; Indenbirken, D.; Spohn, M.; Müller, U.; Ziegler, M.; et al. Essential control of early B-cell development by Mef2 transcription factors. Blood 2016, 127, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Chen, G.; Zhao, Y.; Gao, X.M.; Wang, J. Regulation of the Development and Function of B Cells by ZBTB Transcription Factors. Front. Immunol. 2018, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Arai, Y.; Mori, H.; Matsushita, Y.; Kubo, T.; Nakanishi, T. Small interfering RNA targeting CD81 ameliorated arthritis in rats. Biochem. Biophys. Res. Commun. 2009, 388, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.K.; Kim, J.H.; Kim, J.; Choi, S.; Park, M.; Park, W.; Kim, S.; Lee, K.S.; Kim, T.; Jung, J.; et al. REDD-1 aggravates endotoxin-induced inflammation via atypical NF-κB activation. FASEB J. 2018, 32, 4585–4599. [Google Scholar] [CrossRef]
- Pearson, M.J.; Jones, S.W. Review: Long Noncoding RNAs in the Regulation of Inflammatory Pathways in Rheumatoid Arthritis and Osteoarthritis. Arthritis Rheumatol. 2016, 68, 2575–2583. [Google Scholar] [CrossRef]
- Wagner, S.A.; Satpathy, S.; Beli, P.; Choudhary, C. SPATA2 links CYLD to the TNF-α receptor signaling complex and modulates the receptor signaling outcomes. EMBO J. 2016, 35, 1868–1884. [Google Scholar] [CrossRef]
- Rakonjac, M.; Fischer, L.; Provost, P.; Werz, O.; Steinhilber, D.; Samuelsson, B.; Rådmark, O. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc. Natl. Acad. Sci. USA 2006, 103, 13150–131555. [Google Scholar] [CrossRef]
- Smyth, A.; Gogarty, M.; Crean, D.; Murphy, E.P. Subcellular Localization of NR4A2 Orphan Nuclear Receptor Expression in Human and Mouse Synovial Joint Tissue. Methods Mol. Biol. 2019, 1966, 17–26. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, K.P.; O’Donoghue, G.; Adams, C.; Mulcahy, H.; Molloy, C.; Silke, C.; Molloy, M.; Shanahan, F.; O’Gara, F. High levels of Lymphotoxin-Beta (LT-Beta) gene expression in rheumatoid arthritis synovium: Clinical and cytokine correlations. Rheumatol. Int. 2008, 28, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Caselli, G.; Bonazzi, A.; Lanza, M.; Ferrari, F.; Maggioni, D.; Ferioli, C.; Giambelli, R.; Comi, E.; Zerbi, S.; Perrella, M.; et al. Pharmacological characterisation of CR6086, a potent prostaglandin E(2) receptor 4 antagonist, as a new potential disease-modifying anti-rheumatic drug. Arthritis Res. Ther. 2018, 20, 39. [Google Scholar] [CrossRef]
- Mirsaidi, A.; Tiaden, A.N.; Richards, P.J. Prostaglandin E(2) inhibits matrix mineralization by human bone marrow stromal cell-derived osteoblasts via Epac-dependent cAMP signaling. Sci. Rep. 2017, 7, 2243. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Jeon, W.J.; Kim, E.J.; Jang, W.G. CRTC2 suppresses BMP2-induced osteoblastic differentiation via Smurf1 expression in MC3T3-E1 cells. Life Sci. 2018, 214, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.F.; Matsuo, K. Signalling in osteoclasts and the role of Fos/AP1 proteins. Ann. Rheum. Dis. 2003, 62, 83–85. [Google Scholar] [CrossRef] [PubMed]
- Kinne, R.W.; Boehm, S.; Iftner, T.; Aigner, T.; Vornehm, S.; Weseloh, G.; Bravo, R.; Emmrich, F.; Kroczek, R.A. Synovial fibroblast-like cells strongly express jun-B and C-fos proto-oncogenes in rheumatoid- and osteoarthritis. Scand. J. Rheumatol. 1995, 101, 121–125. [Google Scholar] [CrossRef]
- Carr, T.M.; Wheaton, J.D.; Houtz, G.M.; Ciofani, M. JunB promotes Th17 cell identity and restrains alternative CD4(+) T-cell programs during inflammation. Nat. Commun. 2017, 8, 301. [Google Scholar] [CrossRef]
- Vomero, M.; Barbati, C.; Colasanti, T.; Perricone, C.; Novelli, L.; Ceccarelli, F.; Spinelli, F.R.; Di Franco, M.; Conti, F.; Valesini, G.; et al. Autophagy and Rheumatoid Arthritis: Current Knowledges and Future Perspectives. Front. Immunol. 2018, 9, 1577. [Google Scholar] [CrossRef] [Green Version]
- Ban, J.Y.; Park, H.J.; Kim, S.K.; Kim, J.W.; Lee, Y.A.; Choi, I.A.; Chung, J.H.; Hong, S.J. Association of forkhead box J3 (FOXJ3) polymorphisms with rheumatoid arthritis. Mol. Med. Rep. 2013, 8, 1235–1241. [Google Scholar] [CrossRef]
- Malemud, C.J. Intracellular Signaling Pathways in Rheumatoid Arthritis. J. Clin. Cell Immunol. 2013, 4, 160. [Google Scholar] [CrossRef]
- Jaigirdar, S.A.; Benson, R.A.; Elmesmari, A.; Kurowska-Stolarska, M.S.; McInnes, I.B.; Garside, P.; MacLeod, M.K.L. Sphingosine-1-Phosphate Promotes the Persistence of Activated CD4 T Cells in Inflamed Sites. Front. Immunol. 2017, 8, 1627. [Google Scholar] [CrossRef]
- Lowin, T.; Straub, R.H. Integrins and their ligands in rheumatoid arthritis. Arthritis Res. Ther. 2011, 13, 244. [Google Scholar] [CrossRef]
- Nanki, T.; Shimaoka, T.; Hayashida, K.; Taniguchi, K.; Yonehara, S.; Miyasaka, N. Pathogenic role of the CXCL16-CXCR6 pathway in rheumatoid arthritis. Arthritis Rheum. 2005, 52, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Ohba, T.; Takase, Y.; Ohhara, M.; Kasukawa, R. Thrombin in the synovial fluid of patients with rheumatoid arthritis mediates proliferation of synovial fibroblast-like cells by induction of platelet derived growth factor. J. Rheumatol. 1996, 23, 1505–1511. [Google Scholar]
- Charbonneau, M.; Lavoie, R.R.; Lauzier, A.; Harper, K.; McDonald, P.P.; Dubois, C.M. Platelet-Derived Growth Factor Receptor Activation Promotes the Prodestructive Invadosome-Forming Phenotype of Synoviocytes from Patients with Rheumatoid Arthritis. J. Immunol. 2016, 196, 3264–3275. [Google Scholar] [CrossRef] [Green Version]
- Busso, N.; Hamilton, J.A. Extravascular coagulation and the plasminogen activator/plasmin system in rheumatoid arthritis. Arthritis Rheum. 2002, 46, 2268–2279. [Google Scholar] [CrossRef]
- Pache, M.; Schwarz, H.A.; Kaiser, H.J.; Wüest, P.; Klöti, M.; Dubler, B.; Flammer, J. Elevated plasma endothelin-1 levels and vascular dysregulation in patients with rheumatoid arthritis. Med. Sci. Monit. 2002, 8, CR616–CR619. [Google Scholar]
- Mythreye, K.; Blobe, G.C. Proteoglycan signaling co-receptors: Roles in cell adhesion, migration and invasion. Cell Signal. 2009, 21, 1548–1558. [Google Scholar] [CrossRef] [Green Version]
- Lamoureux, F.; Baud’huin, M.; Duplomb, L.; Heymann, D.; Rédini, F. Proteoglycans: Key partners in bone cell biology. Bioessays 2007, 29, 758–771. [Google Scholar] [CrossRef]
- Szekanecz, Z.; Besenyei, T.; Szentpétery, A.; Koch, A.E. Angiogenesis and vasculogenesis in rheumatoid arthritis. Curr. Opin. Rheumatol. 2010, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Shelef, M.A.; Bennin, D.A.; Yasmin, N.; Warner, T.F.; Ludwig, T.; Beggs, H.E.; Huttenlocher, A. Focal adhesion kinase is required for synovial fibroblast invasion, but not murine inflammatory arthritis. Arthritis Res. Ther. 2014, 16, 464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, F.L.; Li, X.; Lu, W.G.; Sun, J.M.; Jiang, D.L.; Xu, R.S. Epidermal growth factor receptor (EGFR) as a therapeutic target in rheumatoid arthritis. Clin. Rheumatol. 2013, 32, 289–292. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.R.; Kim, K.W.; Kim, B.M.; Cho, M.L.; Lee, S.H. The effect of vascular endothelial growth factor on osteoclastogenesis in rheumatoid arthritis. PLoS ONE 2015, 10, e0124909. [Google Scholar] [CrossRef]
- Oikawa, T.; Kuroda, Y.; Matsuo, K. Regulation of osteoclasts by membrane-derived lipid mediators. Cell Mol. Life Sci. 2013, 70, 3341–3353. [Google Scholar] [CrossRef] [Green Version]
- Grandaunet, B.; Syversen, S.W.; Hoff, M.; Sundan, A.; Haugeberg, G.; van Der Heijde, D.; Kvien, T.K.; Standal, T. Association between high plasma levels of hepatocyte growth factor and progression of radiographic damage in the joints of patients with rheumatoid arthritis. Arthritis Rheum. 2011, 63, 662–669. [Google Scholar] [CrossRef] [PubMed]
- Pi, H.; Zhou, H.; Jin, H.; Ning, Y.; Wang, Y. Abnormal Glucose Metabolism in Rheumatoid Arthritis. Biomed. Res. Int. 2017, 2017, 9670434. [Google Scholar] [CrossRef]
- Arias de la Rosa, I.; Escudero-Contreras, A.; Rodríguez-Cuenca, S.; Ruiz-Ponce, M.; Jiménez-Gómez, Y.; Ruiz-Limón, P.; Pérez-Sánchez, C.; Ábalos-Aguilera, M.C.; Cecchi, I.; Ortega, R.; et al. Defective glucose and lipid metabolism in rheumatoid arthritis is determined by chronic inflammation in metabolic tissues. J. Intern. Med. 2018, 284, 61–77. [Google Scholar] [CrossRef] [Green Version]
- Stagakis, I.; Bertsias, G.; Karvounaris, S.; Kavousanaki, M.; Virla, D.; Raptopoulou, A.; Kardassis, D.; Boumpas, D.T.; Sidiropoulos, P.I. Anti-tumor necrosis factor therapy improves insulin resistance, beta cell function and insulin signaling in active rheumatoid arthritis patients with high insulin resistance. Arthritis Res. Ther. 2012, 14, R141. [Google Scholar] [CrossRef]
- Guo, X.; Higgs, B.W.; Bay-Jensen, A.C.; Wu, Y.; Karsdal, M.A.; Kuziora, M.; Godwood, A.; Close, D.; Ryan, P.C.; Roskos, L.K.; et al. Blockade of GM-CSF pathway induced sustained suppression of myeloid and T cell activities in rheumatoid arthritis. Rheumatology 2018, 57, 175–184. [Google Scholar] [CrossRef]
- Brühl, H.; Cihak, J.; Niedermeier, M.; Denzel, A.; Rodriguez Gomez, M.; Talke, Y.; Goebel, N.; Plachý, J.; Stangassinger, M.; Mack, M. Important role of interleukin-3 in the early phase of collagen-induced arthritis. Arthritis Rheum. 2009, 60, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Isomäki, P.; Alanärä, T.; Isohanni, P.; Lagerstedt, A.; Korpela, M.; Moilanen, T.; Visakorpi, T.; Silvennoinen, O. The expression of SOCS is altered in rheumatoid arthritis. Rheumatology 2007, 46, 1538–1546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Chen, H.; Zhang, Q.; Xu, J.; Shi, Q.; Wang, M. MiR-650 inhibits proliferation, migration and invasion of rheumatoid arthritis synovial fibroblasts by targeting AKT2. Biomed. Pharmacother. 2017, 88, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Kawamata, A.; Izu, Y.; Yokoyama, H.; Amagasa, T.; Wagner, E.F.; Nakashima, K.; Ezura, Y.; Hayata, T.; Noda, M. JunD suppresses bone formation and contributes to low bone mass induced by estrogen depletion. J. Cell Biochem. 2008, 103, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Makarov, S.S. NF-kappa B in rheumatoid arthritis: A pivotal regulator of inflammation, hyperplasia, and tissue destruction. Arthritis Res. 2001, 3, 200–206. [Google Scholar] [CrossRef] [PubMed]
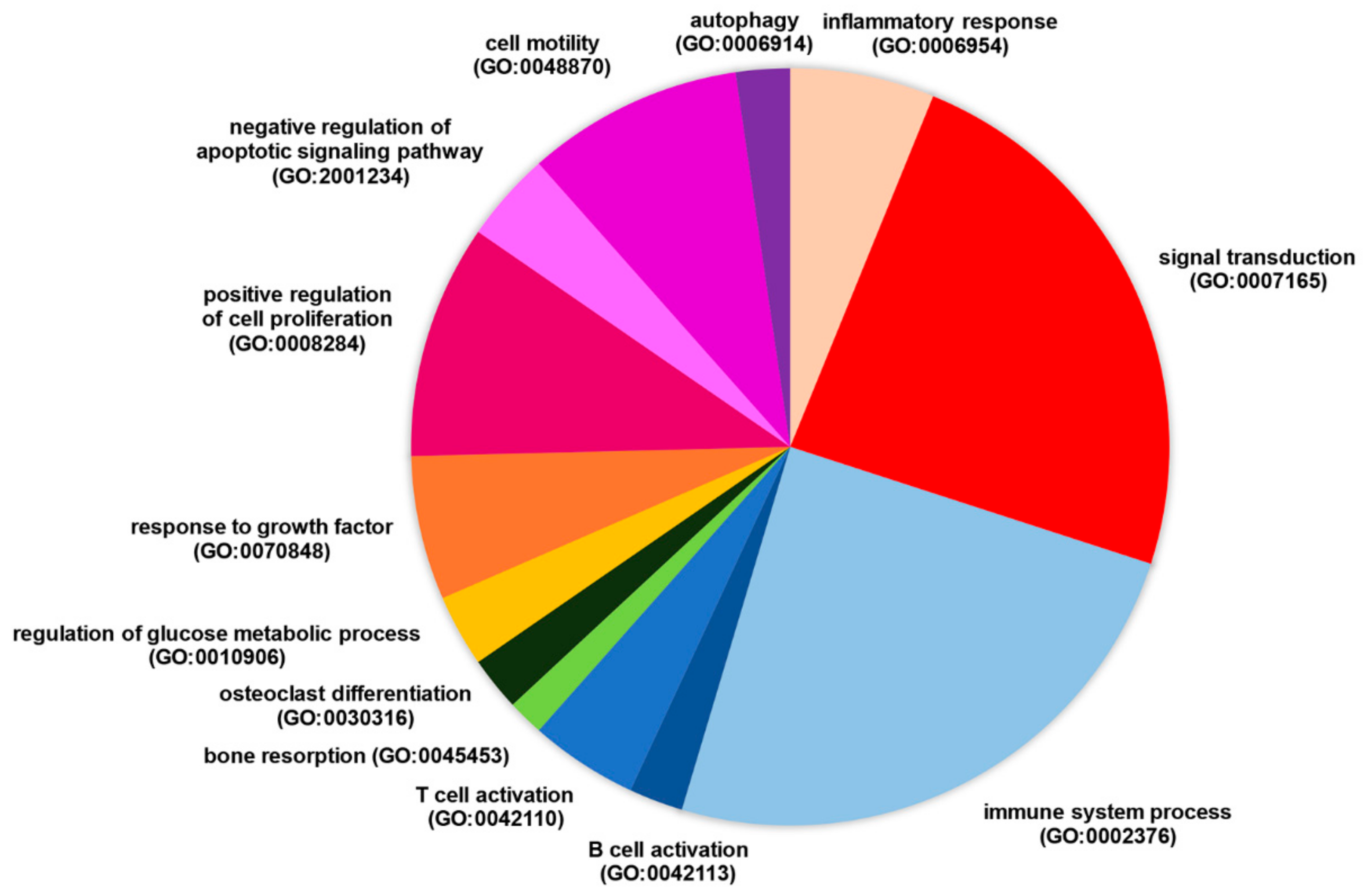

| ID | Fold Change | FDR p-Value | Gene Symbol | Description | mRNA Accession |
|---|---|---|---|---|---|
| Apoptosis | |||||
| TC0300009843.hg.1 | 2.12 | 0.050 | TP63 | tumour protein p63 | NM_001114978 |
| TC2200007783.hg.1 | 5.34 | 0.008 | PIM3 | Pim-3 proto-oncogene, serine/threonine kinase | NM_001001852 |
| TC0300010770.hg.1 | 12.5 | 0.008 | CSRNP1 | cysteine-serine-rich nuclear protein 1 | NM_033027 |
| TC1900009429.hg.1 | 2.3 | 0.018 | KHSRP | KH-type splicing regulatory protein | NM_003685 |
| TC0600011441.hg.1 | 3.47 | 0.008 | BAG6 | BCL2-associated athanogene 6 | NM_001199697 |
| TC1100013230.hg.1 | 2.08 | 0.023 | BCL9L | B-cell CLL/lymphoma 9-like | NM_182557 |
| TC0100007449.hg.1 | 2.2 | 0.049 | SH3BGRL3 | SH3 domain binding glutamate-rich protein like 3 | NM_031286 |
| TC1100006815.hg.1 | 3 | 0.037 | WEE1 | WEE1 G2 checkpoint kinase | NM_001143976 |
| Cell proliferation | |||||
| TC0300009702.hg.1 | 2.49 | 0.044 | EIF4G1 | eukaryotic translation initiation factor 4 gamma, 1 | NM_001194946 |
| TC0X00007132.hg.1 | 3.18 | 0.008 | CDK16 | cyclin-dependent kinase 16 | NM_001170460 |
| TC0200015764.hg.1 | 3.79 | 0.004 | CNPPD1 | cyclin Pas1/PHO80 domain containing 1 | NM_015680 |
| TC0200016494.hg.1 | 2.04 | 0.046 | CNNM4 | cyclin and CBS domain divalent metal cation transport mediator 4 | NM_020184 |
| TC1900010696.hg.1 | 5.13 | 0.007 | AKT2 | v-akt murine thymoma viral oncogene homolog 2 | NM_001243027 |
| Cell migration | |||||
| TC1500010018.hg.1 | 2.3 | 0.018 | SEMA7A | semaphorin 7A, GPI membrane anchor | NM_001146029 |
| TC1200012859.hg.1 | 3.65 | 0.006 | RHOF | ras homolog family member F (in filopodia) | NM_019034 |
| TC1100009864.hg.1 | 3.29 | 0.004 | RHOG | ras homolog family member G | NM_001665 |
| TC1700009528.hg.1 | 2.46 | 0.028 | CXCL16 | chemokine (C-X-C motif) ligand 16 | NM_001100812 |
| Inflammatory response | |||||
| TC1500006925.hg.1 | 24.36 | 0.018 | THBS1 | thrombospondin 1 | NM_003246 |
| TC1900006977.hg.1 | 11.9 | 0.050 | ICAM1 | intercellular adhesion molecule 1 | NM_000201 |
| TC0500007231.hg.1 | 3.28 | 0.022 | PTGER4 | prostaglandin E receptor 4 (subtype EP4) | NM_000958 |
| TC0100011384.hg.1 | 2.52 | 0.025 | MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 | NM_004759 |
| TC0100009364.hg.1 | 3.82 | 0.008 | CSF1 | colony stimulating factor 1 (macrophage) | NM_000757 |
| TC0300006985.hg.1 | 6.3 | 0.034 | CCR4 | chemokine (C-C motif) receptor 4 | NM_005508 |
| TC1900010743.hg.1 | 2.7 | 0.019 | TGFB1 | transforming growth factor beta 1 | NM_000660 |
| TC1000007990.hg.1 | 4.92 | 0.020 | DDIT4 | DNA damage inducible transcript 4 | NM_019058 |
| TC1600011060.hg.1 | 2.28 | 0.017 | COTL1 | coactosin-like F-actin binding protein 1 | NM_021149 |
| TC2000009401.hg.1 | 3.54 | 0.011 | SPATA2 | Spermatogenesis-associated 2 | NM_00113577 |
| TC1100011243.hg.1 | 4.25 | 0.007 | RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | NM_001145138 |
| TC0900011623.hg.1 | 2.16 | 0.030 | PTGES2 | prostaglandin E synthase 2 | NM_001256335 |
| TC0200014672.hg.1 | 7.65 | 0.042 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 | NM_006186 |
| TC1900007270.hg.1 | 6.2 | 0.007 | KLF2 | Kruppel-like factor 2 | NM_016270 |
| TC1700011903.hg.1 | 4.41 | 0.046 | SOCS3 | suppressor of cytokine signalling 3 | NM_003955 |
| TC1600009395.hg.1 | 3.76 | 0.010 | SOCS1 | suppressor of cytokine signalling 1 | NM_003745 |
| TC0100017107.hg.1 | 2.71 | 0.041 | IL10 | interleukin 10 | NM_000572 |
| TC1100009225.hg.1 | 7.04 | 0.006 | CXCR5 | chemokine (C-X-C motif) receptor 5 | NM_001716 |
| TC1100013178.hg.1 | 2.22 | 0.023 | MAP4K2 | mitogen-activated protein kinase kinase kinase kinase 2 | NM_001307990 |
| TC1700007262.hg.1 | 4.43 | 0.023 | MAP2K3 | mitogen-activated protein kinase kinase 3 | NM_002756 |
| TC1900009325.hg.1 | 3.56 | 0.038 | MAP2K2 | mitogen-activated protein kinase kinase 2 | NM_030662 |
| TC0600011438.hg.1 | 2.07 | 0.045 | LTB | lymphotoxin beta (TNF superfamily, member 3) | NM_002341 |
| TC1700011903.hg.1 | 4.41 | 0.046 | SOCS3 | suppressor of cytokine signalling 3 | NM_003955 |
| Immune response | |||||
| TC0200008452.hg.1 | 2.85 | 0.024 | MAL | mal, T-cell differentiation protein | NM_002371 |
| TC1100006576.hg.1 | 3.74 | 0.015 | CD81 | CD81 molecule | NM_001297649 |
| TC0600007495.hg.1 | 2.08 | 0.037 | HLA-A | major histocompatibility complex, class I, A | NM_001242758 |
| TC0100007291.hg.1 | 2.08 | 0.028 | C1QC | complement component 1, q subcomponent, C chain | NM_001114101 |
| TC1100007787.hg.1 | 3.29 | 0.035 | CD6 | CD6 molecule | NM_001254750 |
| TC1600011368.hg.1 | 3.14 | 0.008 | LAT | linker for activation of T-cells | NM_001014987 |
| TC1900008279.hg.1 | 5.58 | 0.006 | BCL3 | B-cell CLL/lymphoma 3 | NM_005178 |
| TC1900008166.hg.1 | 3.13 | 0.004 | CD79A | CD79a molecule, immunoglobulin-associated alpha | NM_001783 |
| TC1200010950.hg.1 | 2.47 | 0.034 | STAT6 | signal transducer and activator of transcription 6, interleukin-4 induced | NM_001178078 |
| TC1000008891.hg.1 | 10.97 | 0.008 | DUSP5 | dual specificity phosphatase 5 | NM_004419 |
| TC0600008972.hg.1 | 5.51 | 0.018 | PRDM1 | PR domain containing 1, with ZNF domain | NM_001198 |
| TC0100016000.hg.1 | 4.15 | 0.004 | MEF2D | myocyte enhancer factor 2D | NM_001271629 |
| TC0900008891.hg.1 | 2.21 | 0.023 | LRRC8A | leucine rich repeat containing 8 family, member A | NM_001127244 |
| TC1900009320.hg.1 | 2.71 | 0.045 | ZBTB7A | zinc finger and BTB domain containing 7A | NM_015898 |
| TC1900008505.hg.1 | 2.93 | 0.018 | BAX | BCL2-associated X protein | NM_001291428 |
| TC1800007805.hg.1 | 2.22 | 0.027 | NFATC1 | nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 | NM_001278669 |
| TC2200008637.hg.1 | 3.64 | 0.015 | IL2RB | interleukin 2 receptor, beta | NM_000878 |
| Angiogenesis | |||||
| TC1200010839.hg.1 | 2 | 0.029 | ITGA5 | integrin alpha 5 | NM_002205 |
| TC2000007336.hg.1 | 2.83 | 0.015 | PPP1R16B | protein phosphatase 1, regulatory subunit 16B | NM_001172735 |
| TC1600008971.hg.1 | 2.83 | 0.018 | JMJD8 | jumonji domain containing 8 | NM_001005920 |
| TC1700011818.hg.1 | 2.6 | 0.050 | JMJD6 | jumonji domain containing 6 | NM_001081461 |
| TC0100007832.hg.1 | 17.12 | 0.008 | ZC3H12A | zinc finger CCCH-type containing 12A | NM_025079 |
| TC0100018300.hg.1 | 2.44 | 0.032 | ADAM15 | ADAM metallopeptidase domain 15 | NM_001261464 |
| Bone resorption | |||||
| Positive regulation of bone resorption | |||||
| TC0X00008831.hg.1 | 2.62 | 0.021 | ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | NM_001183 |
| TC2000007283.hg.1 | 2.11 | 0.026 | SRC | SRC proto-oncogene, non-receptor tyrosine kinase | NM_005417 |
| TC0300009916.hg.1 | 4.17 | 0.030 | HES1 | hes family bHLH transcription factor 1 | NM_005524 |
| TC0100015891.hg.1 | 2.76 | 0.008 | CRTC2 | CREB regulated transcription coactivator 2 | NM_181715 |
| TC1900010009.hg.1 | 3.41 | 0.023 | JUND | jun D proto-oncogene | NM_001286968 |
| Positive regulation of osteoclast proliferation/differentiation | |||||
| TC0300007380.hg.1 | 2.4 | 0.027 | TCTA | T-cell leukaemia translocation altered | NM_022171 |
| TC1900009134.hg.1 | 9.56 | 0.002 | SBNO2 | strawberry notch homolog 2 | NM_014963 |
| TC0100009364.hg.1 | 3.82 | 0.008 | CSF1 | colony stimulating factor 1 (macrophage) | NM_000757 |
| TC0X00008831.hg.1 | 2.62 | 0.021 | ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | NM_001183 |
| TC0300009916.hg.1 | 4.17 | 0.030 | HES1 | hes family bHLH transcription factor 1 | NM_005524 |
| TC0500007231.hg.1 | 3.28 | 0.022 | PTGER4 | prostaglandin E receptor 4 (subtype EP4) | NM_000958 |
| TC1900007096.hg.1 | 5.16 | 0.032 | JUNB | jun B proto-oncogene | NM_002229 |
| Osteoblast differentiation | |||||
| TC2000009887.hg.1 | −2.8 | 0.0172 | PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) | NM_015192 |
| Extracellular matrix degradation | |||||
| TC2000007514.hg.1 | 3.14 | 0.041 | MMP9 | matrix metallopeptidase 9 | NM_004994 |
| TC0500012599.hg.1 | 1.9 | 0.040 | ADAM19 | ADAM metallopeptidase domain 19 | NM_033274 |
| TC0100018300.hg.1 | 2.44 | 0.032 | ADAM15 | ADAM metallopeptidase domain 15 | NM_001261464 |
| TC1900006470.hg.1 | 3.01 | 0.011 | BSG | basigin | NM_001728 |
| ID | Fold Change | FDR p-Value | Gene Symbol | Description | mRNA Accession |
|---|---|---|---|---|---|
| Wnt signalling pathway | |||||
| TC1900009272.hg.1 | 2.4 | 0.018 | AES | amino-terminal enhancer of split | NM_001130 |
| TC1100008181.hg.1 | 2.03 | 0.029 | LRP5 | LDL-receptor-related protein 5 | NM_001291902 |
| TC0200014672.hg.1 | 7.65 | 0.042 | NR4A2 | nuclear receptor subfamily 4, group A, member 2 | NM_006186 |
| TC1100007913.hg.1 | 2.69 | 0.048 | MARK2 | MAP/microtubule affinity-regulating kinase 2 | NM_001039469 |
| TC1200007595.hg.1 | 2.88 | 0.023 | SMARCD1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 1 | NM_003076 |
| TC1900011639.hg.1 | 2.1 | 0.032 | STK11 | serine/threonine kinase 11 | NM_000455 |
| TC1100008181.hg.1 | 2.03 | 0.029 | LRP5 | LDL-receptor-related protein 5 | NM_001291902 |
| TNF signalling pathway | |||||
| TC0900011385.hg.1 | −1.99 | 0.040 | PSMD5 | proteasome 26S subunit, non-ATPase 5 | NM_001270427 |
| TC0600011438.hg.1 | 2.07 | 0.045 | LTB | lymphotoxin beta (TNF superfamily, member 3) | NM_002341 |
| TC1700010879.hg.1 | 2.36 | 0.026 | MAP3K14 | mitogen-activated protein kinase kinase kinase 14 | NM_003954 |
| TC1600010616.hg.1 | 2.14 | 0.035 | TRADD | TNFRSF1A-associated via death domain | NM_003789 |
| TC1100011243.hg.1 | 4.25 | 0.007 | RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | NM_001145138 |
| Type I interferon signalling | |||||
| TC0600007495.hg.1 | 2.08 | 0.037 | HLA-A | major histocompatibility complex, class I, A | NM_001242758 |
| TC0200008452.hg.1 | 2.85 | 0.024 | MAL | mal, T-cell differentiation protein | NM_002371 |
| TC0100017107.hg.1 | 2.71 | 0.041 | IL10 | interleukin 10 | NM_000572 |
| TC1600009395.hg.1 | 3.76 | 0.010 | SOCS1 | suppressor of cytokine signalling 1 | NM_003745 |
| TC1100011243.hg.1 | 4.25 | 0.007 | RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | NM_001145138 |
| TC1900009627.hg.1 | 2.63 | 0.040 | TYK2 | tyrosine kinase 2 | NM_003331 |
| TC1000008727.hg.1 | 2.64 | 0.024 | NFKB2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) | NM_001077494 |
| p38 MAP kinase signalling | |||||
| TC1400009524.hg.1 | 2.91 | 0.029 | ZFP36L1 | ZFP36 ring finger protein-like 1 | NM_001244698 |
| TC1700007262.hg.1 | 4.43 | 0.023 | MAP2K3 | mitogen-activated protein kinase kinase 3 | NM_002756 |
| TC0100011384.hg.1 | 2.52 | 0.025 | MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 | NM_004759 |
| TC0X00009581.hg.1 | 2.99 | 0.013 | ELK1 | ELK1, member of ETS oncogene family | NM_001114123 |
| TC0100016000.hg.1 | 4.15 | 0.004 | MEF2D | myocyte enhancer factor 2D | NM_001271629 |
| NF-kB signalling pathway | |||||
| TC1900008300.hg.1 | 2.78 | 0.034 | RELB | v-rel avian reticuloendotheliosis viral oncogene homolog B | NM_006509 |
| TC1900008279.hg.1 | 5.58 | 0.006 | BCL3 | B-cell CLL/lymphoma 3 | NM_005178 |
| TC1600010616.hg.1 | 2.14 | 0.035 | TRADD | TNFRSF1A-associated via death domain | NM_003789 |
| TC1100011243.hg.1 | 4.25 | 0.007 | RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | NM_001145138 |
| TC1700010879.hg.1 | 2.36 | 0.026 | MAP3K14 | mitogen-activated protein kinase kinase kinase 14 | NM_003954 |
| TC0800012190.hg.1 | 3.65 | 0.018 | SHARPIN | SHANK-associated RH domain interactor | NM_030974 |
| TOLL-like receptors signalling pathways | |||||
| TC0200008452.hg.1 | 2.85 | 0.024 | MAL | mal, T-cell differentiation protein | NM_002371 |
| TC0X00009581.hg.1 | 2.99 | 0.013 | ELK1 | ELK1, member of ETS oncogene family | NM_001114123 |
| TC0100011384.hg.1 | 2.52 | 0.025 | MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 | NM_004759 |
| TC1000008727.hg.1 | 2.64 | 0.024 | NFKB2 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 | NM_001077494 |
| TC1700007262.hg.1 | 4.43 | 0.023 | MAP2K3 | mitogen-activated protein kinase kinase 3 | NM_002756 |
| TC1100011243.hg.1 | 4.25 | 0.007 | RELA | v-rel avian reticuloendotheliosis viral oncogene homolog A | NM_001145138 |
| Jak-Stat signalling pathway | |||||
| TC1600009395.hg.1 | 3.76 | 0.010 | SOCS1 | suppressor of cytokine signalling 1 | NM_003745 |
| TC1700011903.hg.1 | 4.41 | 0.046 | SOCS3 | suppressor of cytokine signalling 3 | NM_003955 |
| TC1900009991.hg.1 | 2.63 | 0.042 | JAK3 | Janus kinase 3 | NM_000215 |
| TC1200010950.hg.1 | 2.47 | 0.034 | STAT6 | signal transducer and activator of transcription 6, interleukin-4 induced | NM_001178078 |
| PI3K signalling pathway | |||||
| TC1900010696.hg.1 | 5.13 | 0.007 | AKT2 | v-akt murine thymoma viral oncogene homolog 2 | NM_001243027 |
| TC1100008136.hg.1 | 2.31 | 0.023 | RPS6KB2 | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | NM_003952 |
| TC1000007990.hg.1 | 4.92 | 0.020 | DDIT4 | DNA damage inducible transcript 4 | NM_019058 |
| TC1000008891.hg.1 | 10.97 | 0.008 | DUSP5 | dual specificity phosphatase 5 | NM_004419 |
| TC0700008560.hg.1 | 3.76 | 0.026 | GNB2 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | NM_005273 |
| TC1900010009.hg.1 | 3.41 | 0.023 | JUND | jun D proto-oncogene | NM_001286968 |
| TC1900007096.hg.1 | 5.16 | 0.032 | JUNB | jun B proto-oncogene | NM_002229 |
| TC0600008972.hg.1 | 5.51 | 0.018 | PRDM1 | PR domain containing 1, with ZNF domain | NM_001198 |
| mTOR signalling pathway | |||||
| TC0300013684.hg.1 | 2.21 | 0.043 | TFRC | transferrin receptor | NM_001128148 |
| TC1200007595.hg.1 | 2.88 | 0.023 | SMARCD1 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily d, member 1 | NM_003076 |
| TC1700011903.hg.1 | 4.41 | 0.046 | SOCS3 | suppressor of cytokine signalling 3 | NM_003955 |
| TC2000007283.hg.1 | 2.11 | 0.026 | SRC | SRC proto-oncogene, non-receptor tyrosine kinase | NM_005417 |
| TC0400009765.hg.1 | 3.06 | 0.018 | MXD4 | MAX dimerization protein 4 | NM_006454 |
| TC0100016000.hg.1 | 4.15 | 0.004 | MEF2D | myocyte enhancer factor 2D | NM_001271629 |
| Biological Pathway | Bonferroni Corrected p-Value |
|---|---|
| Proteoglycan syndecan-mediated signalling events | 0.006 |
| Alpha9 beta1 integrin signalling events | 0.020 |
| GMCSF-mediated signalling events | 0.025 |
| Beta1 integrin cell surface interactions | 0.026 |
| IL3-mediated signalling events | 0.027 |
| IFN-gamma pathway | 0.028 |
| PAR1-mediated thrombin signalling events | 0.031 |
| Thrombin/protease-activated receptor (PAR) pathway | 0.032 |
| Syndecan-1-mediated signalling events | 0.032 |
| Integrin family cell surface interactions | 0.032 |
| Plasma membrane oestrogen receptor signalling | 0.033 |
| Endothelins | 0.040 |
| Signalling events mediated by focal adhesion kinase | 0.040 |
| PDGFR-beta signalling pathway | 0.040 |
| Arf6 trafficking events | 0.040 |
| Class I PI3K signalling events mediated by Akt | 0.040 |
| mTOR signalling pathway | 0.040 |
| Internalization of ErbB1 | 0.040 |
| EGF receptor (ErbB1) signalling pathway | 0.040 |
| Class I PI3K signalling events | 0.040 |
| Arf6 signalling events | 0.040 |
| ErbB1 downstream signalling | 0.040 |
| Arf6 downstream pathway | 0.040 |
| Insulin Pathway | 0.040 |
| Urokinase-type plasminogen activator (uPA) and uPAR-mediated signalling | 0.040 |
| S1P1 pathway | 0.040 |
| EGFR-dependent Endothelin signalling events | 0.041 |
| IGF1 pathway | 0.044 |
| ErbB receptor signalling network | 0.045 |
| Sphingosine 1-phosphate (S1P) pathway | 0.045 |
| IL5-mediated signalling events | 0.045 |
| Signalling events mediated by hepatocyte growth factor receptor (c-Met) | 0.047 |
| PDGF receptor signalling network | 0.047 |
| Nectin adhesion pathway | 0.049 |
| Signalling events mediated by VEGFR1 and VEGFR2 | 0.051 |
| Glypican 1 network | 0.056 |
| Glypican pathway | 0.055 |
| VEGF and VEGFR signalling network | 0.055 |
| Integrin-linked kinase signalling | 0.053 |
| ID | Fold Change | FDR p-Value | Gene Symbol | mRNA Accession | miRNA Targets | Targeted Modulated Genes |
|---|---|---|---|---|---|---|
| TC0X00010064.hg.1 | −2.03 | 0.049 | FTX | NR_028379 | 64 | 96 |
| TC0100018570.hg.1 | −2.64 | 0.034 | HNRNPU-AS1 | NR_026778 | 55 | 161 |
| TC2200009240.hg.1 | −2 | 0.042 | MIATNB | NR_110543 | 16 | 11 |
| TC1700012077.hg.1 | 2.5 | 0.039 | RP11-498C9.15 | ENST00000582866.1 | 27 | 106 |
| TC0100009198.hg.1 | −3.23 | 0.008 | RP4-714D9.5 | ENST00000564623.1 | 6 | 27 |
| TC1400006883.hg.1 | 2.74 | 0.018 | RP11-73E17.2 | ENST00000557373.1 | 1 | 4 |
| Biological Pathway | Bonferroni Corrected p-Value |
|---|---|
| Beta1 integrin cell surface interactions | 0.004 |
| Integrin family cell surface interactions | 0.006 |
| IFN-gamma pathway | 0.008 |
| PAR1-mediated thrombin signalling events | 0.008 |
| Thrombin/protease-activated receptor (PAR) pathway | 0.009 |
| Plasma membrane oestrogen receptor signalling | 0.009 |
| Endothelins | 0.009 |
| Glypican pathway | 0.014 |
| Proteoglycan syndecan-mediated signalling events | 0.016 |
| Signalling events mediated by focal adhesion kinase | 0.030 |
| Class I PI3K signalling events mediated by Akt | 0.030 |
| Internalization of ErbB1 | 0.030 |
| Arf6 downstream pathway | 0.030 |
| Arf6 trafficking events | 0.030 |
| EGF receptor (ErbB1) signalling pathway | 0.030 |
| Urokinase-type plasminogen activator (uPA) and uPAR-mediated signalling | 0.030 |
| ErbB1 downstream signalling | 0.030 |
| S1P1 pathway | 0.030 |
| Arf6 signalling events | 0.030 |
| mTOR signalling pathway | 0.030 |
| Insulin pathway | 0.030 |
| PDGFR-beta signalling pathway | 0.030 |
| Class I PI3K signalling events | 0.030 |
| EGFR-dependent Endothelin signalling events | 0.030 |
| IGF1 pathway | 0.031 |
| GMCSF-mediated signalling events | 0.031 |
| IL5-mediated signalling events | 0.031 |
| Signalling events mediated by hepatocyte growth factor receptor (c-Met) | 0.032 |
| PDGF receptor signalling network | 0.032 |
| IL3-mediated signalling events | 0.032 |
| Nectin adhesion pathway | 0.032 |
| Signalling events mediated by VEGFR1 and VEGFR2 | 0.033 |
| Glypican 1 network | 0.034 |
| Syndecan-1-mediated signalling events | 0.035 |
| VEGF and VEGFR signalling network | 0.036 |
| Alpha9 beta1 integrin signalling events | 0.037 |
| Sphingosine 1-phosphate (S1P) pathway | 0.040 |
| ErbB receptor signalling network | 0.040 |
| Integrin-linked kinase signalling | 0.045 |
| Module | miRNAs | Gene |
|---|---|---|
| M1 | hsa-miR-23c (1.56 up) | CUL3 |
| hsa-miR-23b-3p (2.06 up) | CUL3 | |
| hsa-miR-23a-3p (1.74 down) | CUL3 | |
| hsa-miR-221-3p (2.34 up) | HNRNPA0 | |
| hsa-miR-302c-3p (2.48 down) | SOCS3 | |
| hsa-miR-221-3p (2.34 up) | SOCS3 | |
| M2 | hsa-miR-372-3p (1.63 up) | ANO6 |
| M3 | hsa-miR-101-3p (1.80 up) | ATP5G2 |
| hsa-miR-137 (2.54 up) | PTGES2 | |
| hsa-miR-101-3p (1.80 up) | TERF2 | |
| hsa-miR-613 (1.55 down) | TERF2 | |
| hsa-miR-221-3p (2.34 up) | TERF2 | |
| hsa-miR-206 (2.04 up) | TERF2 | |
| M4 | hsa-miR-4735-3p (2.53 up) | ATM |
| hsa-miR-101-3p (1.80 up) | NACA | |
| hsa-miR-137 (2.54 up) | PCGF5 | |
| hsa-miR-101-3p (1.80 up) | SEL1L | |
| hsa-miR-101-3p (1.80 up) | SURF4 | |
| hsa-miR-613 (1.55 down) | THBS1 | |
| hsa-miR-4735-3p (2.53 up) | THBS1 | |
| hsa-miR-221-3p (2.34 up) | THBS1 | |
| hsa-miR-206 (2.04 up) | THBS1 | |
| hsa-miR-18b-5p (1.67 down) | THBS1 | |
| hsa-miR-18a-5p (2.32 up) | THBS1 | |
| hsa-miR-613 (1.55 down) | VPS45 | |
| hsa-miR-206 (2.04 up) | VPS45 | |
| M5 | hsa-miR-137 (2.54 up) | AKT2 |
| hsa-miR-613 (1.55 down) | JUND | |
| hsa-miR-206 (2.04 up) | JUND | |
| hsa-miR-520e (1.83 down) | RELA | |
| hsa-miR-520d-3p (1.92 down) | RELA | |
| hsa-miR-520c-3p (1.52 down) | RELA | |
| hsa-miR-520b (1.70 down) | RELA | |
| hsa-miR-520a-3p (1.50 down) | RELA | |
| hsa-miR-372-3p (1.63 up) | RELA | |
| hsa-miR-302e (2.18 down) | RELA | |
| hsa-miR-302d-3p (1.84 down) | RELA | |
| hsa-miR-302c-3p (2.48 down) | RELA | |
| hsa-miR-302b-3p (1.66 down) | RELA | |
| hsa-miR-302a-3p (1.58 down) | RELA | |
| hsa-miR-137 (2.54 up) | SRC | |
| M6 | hsa-miR-23c (1.56 up) | STX12 |
| hsa-miR-23b-3p (2.06 up) | STX12 | |
| hsa-miR-23a-3p (1.74 down) | STX12 | |
| hsa-miR-206 (2.04 up) | STX12 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dolcino, M.; Tinazzi, E.; Puccetti, A.; Lunardi, C. Long Non-Coding RNAs Target Pathogenetically Relevant Genes and Pathways in Rheumatoid Arthritis. Cells 2019, 8, 816. https://doi.org/10.3390/cells8080816
Dolcino M, Tinazzi E, Puccetti A, Lunardi C. Long Non-Coding RNAs Target Pathogenetically Relevant Genes and Pathways in Rheumatoid Arthritis. Cells. 2019; 8(8):816. https://doi.org/10.3390/cells8080816
Chicago/Turabian StyleDolcino, Marzia, Elisa Tinazzi, Antonio Puccetti, and Claudio Lunardi. 2019. "Long Non-Coding RNAs Target Pathogenetically Relevant Genes and Pathways in Rheumatoid Arthritis" Cells 8, no. 8: 816. https://doi.org/10.3390/cells8080816
APA StyleDolcino, M., Tinazzi, E., Puccetti, A., & Lunardi, C. (2019). Long Non-Coding RNAs Target Pathogenetically Relevant Genes and Pathways in Rheumatoid Arthritis. Cells, 8(8), 816. https://doi.org/10.3390/cells8080816





