Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Lines
2.2. Adenoviral Infection of Cells
2.3. Cell Signaling Assay
2.4. Scratch Assay
2.5. Growth Curve
2.6. Immunoblot
3. Results
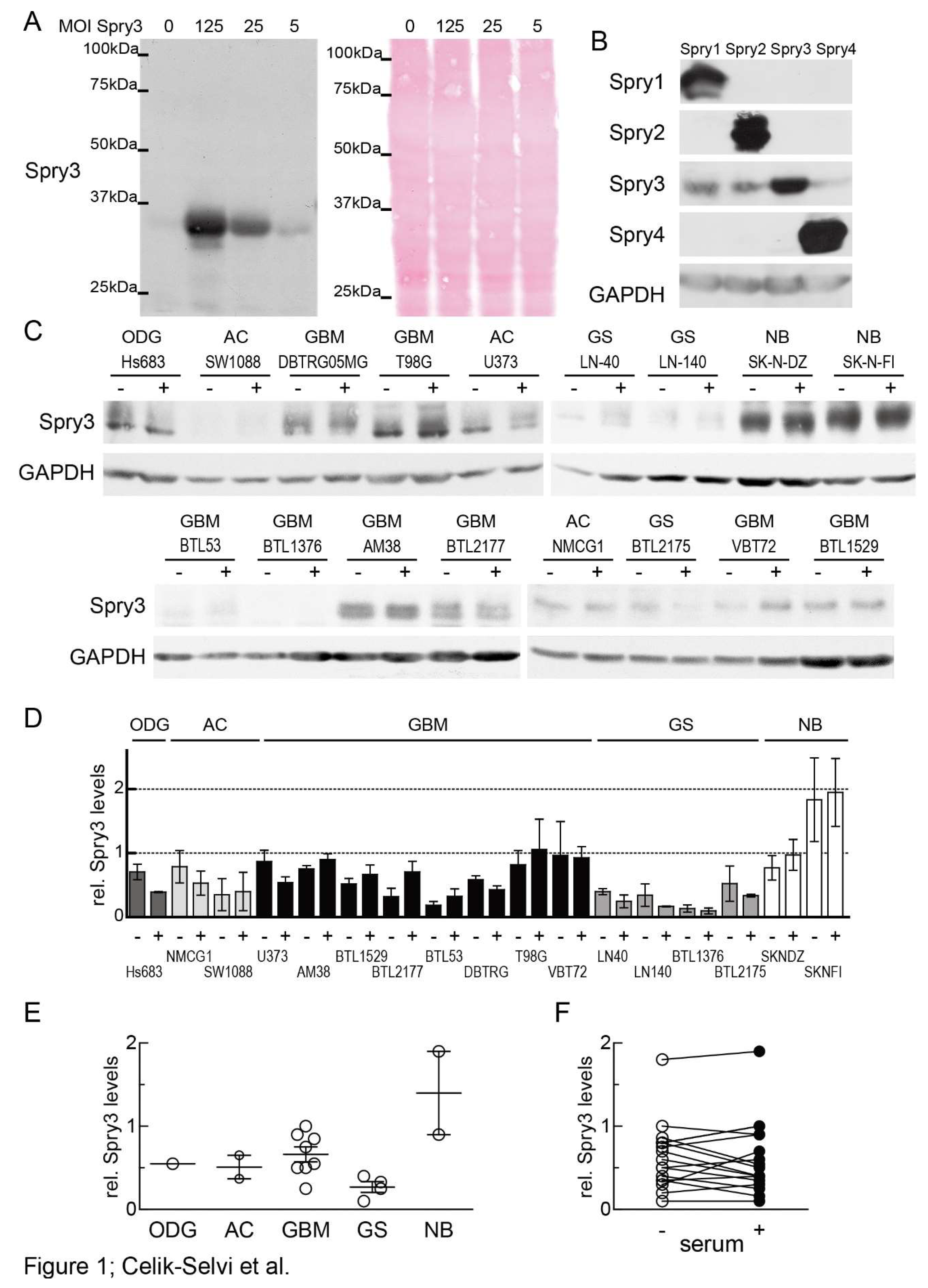
3.1. In Brain Cancer-Derived Cell Lines Spry3 Protein is Commonly Expressed Independent of Mitogen Availability
3.2. Spry4 Protein Expression is Repressed in Cell Lines Derived from More Malignant Brain Tumors, but Usually Still Serum-Dependent
3.3. MAPK Activation in Response to FGF and Serum is Effectively Inhibited by Spry4 While Spry3 Failed to Fulfill This Function in the Presence of Serum
3.4. In Brain Cancer-Derived Cell Lines, Spry3 and Spry4 Expressions Have an Opposing Effect on Cell Proliferation
3.5. Spry3 and Spry4 Exert a Contrary Effect on the Migratory Capabilities of Brain Cells
3.6. Repressed Expression of Spry3 Inhibits Cell Proliferation and Migration
4. Discussion
5. Conclusion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furnari, F.B.; Fenton, T.; Bachoo, R.M.; Mukasa, A.; Stommel, J.M.; Stegh, A.; Hahn, W.C.; Ligon, K.L.; Louis, D.N.; Brennan, C.; et al. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007, 21, 2683–2710. [Google Scholar] [CrossRef] [PubMed]
- Kruchko, C.; Ostrom, Q.T.; Boscia, A.; Truitt, G.; Gittleman, H.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro-Oncol. 2018, 20, iv1–iv86. [Google Scholar] [CrossRef]
- Schilling, F.H.; Spix, C.; Berthold, F.; Erttmann, R.; Fehse, N.; Hero, B.; Klein, G.; Sander, J.; Schwarz, K.; Treuner, J.; et al. Neuroblastoma screening at one year of age. N. Engl. J. Med. 2002, 346, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Hacohen, N.; Kramer, S.; Sutherland, D.; Hiromi, Y.; Krasnow, M.A. Sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell 1998, 92, 253–263. [Google Scholar] [CrossRef]
- Minowada, G.; Jarvis, L.A.; Chi, C.L.; Neubuser, A.; Sun, X.; Hacohen, N.; Krasnow, M.A.; Martin, G.R. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development 1999, 126, 4465–4475. [Google Scholar] [PubMed]
- Ning, Z.; McLellan, A.S.; Ball, M.; Wynne, F.; O’Neill, C.; Mills, W.; Quinn, J.P.; Kleinjan, D.A.; Anney, R.J.; Carmody, R.J.; et al. Regulation of SPRY3 by X chromosome and PAR2-linked promoters in an autism susceptibility region. Hum. Mol. Genet. 2015, 24, 7450. [Google Scholar] [CrossRef]
- Sasaki, A.; Taketomi, T.; Kato, R.; Saeki, K.; Nonami, A.; Sasaki, M.; Kuriyama, M.; Saito, N.; Shibuya, M.; Yoshimura, A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat. Cell Biol. 2003, 5, 427–432. [Google Scholar] [CrossRef]
- Hanafusa, H.; Torii, S.; Yasunaga, T.; Nishida, E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat. Cell Biol. 2002, 4, 850–858. [Google Scholar] [CrossRef]
- Gross, I.; Morrison, D.J.; Hyink, D.P.; Georgas, K.; English, M.A.; Mericskay, M.; Hosono, S.; Sassoon, D.; Wilson, P.D.; Little, M.; et al. The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. J. Biol. Chem. 2003, 278, 41420–41430. [Google Scholar] [CrossRef]
- Edwin, F.; Singh, R.; Endersby, R.; Baker, S.J.; Patel, T.B. The tumor suppressor PTEN is necessary for human Sprouty 2-mediated inhibition of cell proliferation. J. Biol. Chem. 2006, 281, 4816–4822. [Google Scholar] [CrossRef] [PubMed]
- Akbulut, S.; Reddi, A.L.; Aggarwal, P.; Ambardekar, C.; Canciani, B.; Kim, M.K.; Hix, L.; Vilimas, T.; Mason, J.; Basson, M.A.; et al. Sprouty proteins inhibit receptor-mediated activation of phosphatidylinositol-specific phospholipase C. Mol. Biol. Cell 2010, 21, 3487–3496. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.; Lim, J.; Low, B.C.; Chen, Q.; Guy, G.R. Evidence for direct interaction between Sprouty and Cbl. J. Biol. Chem. 2001, 276, 5866–5875. [Google Scholar] [CrossRef] [PubMed]
- Tennis, M.A.; Van Scoyk, M.M.; Freeman, S.V.; Vandervest, K.M.; Nemenoff, R.A.; Winn, R.A. Sprouty-4 inhibits transformed cell growth, migration and invasion, and epithelial-mesenchymal transition, and is regulated by Wnt7A through PPARgamma in non-small cell lung cancer. Mol. Cancer Res. 2010, 8, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Sutterluty, H.; Mayer, C.E.; Setinek, U.; Attems, J.; Ovtcharov, S.; Mikula, M.; Mikulits, W.; Micksche, M.; Berger, W. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol. Cancer Res. 2007, 5, 509–520. [Google Scholar] [CrossRef]
- Shaw, A.T.; Meissner, A.; Dowdle, J.A.; Crowley, D.; Magendantz, M.; Ouyang, C.; Parisi, T.; Rajagopal, J.; Blank, L.J.; Bronson, R.T.; et al. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007, 21, 694–707. [Google Scholar] [CrossRef] [Green Version]
- Fong, C.W.; Chua, M.S.; McKie, A.B.; Ling, S.H.; Mason, V.; Li, R.; Yusoff, P.; Lo, T.L.; Leung, H.Y.; So, S.K.; et al. Sprouty 2, an inhibitor of mitogen-activated protein kinase signaling, is down-regulated in hepatocellular carcinoma. Cancer Res. 2006, 66, 2048–2058. [Google Scholar] [CrossRef]
- Vanas, V.; Muhlbacher, E.; Kral, R.; Sutterluty-Fall, H. Sprouty4 interferes with cell proliferation and migration of breast cancer-derived cell lines. Tumour Biol. 2014, 35, 4447–4456. [Google Scholar] [CrossRef]
- Lo, T.L.; Yusoff, P.; Fong, C.W.; Guo, K.; McCaw, B.J.; Phillips, W.A.; Yang, H.; Wong, E.S.; Leong, H.F.; Zeng, Q.; et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res. 2004, 64, 6127–6136. [Google Scholar] [CrossRef]
- Kwabi-Addo, B.; Wang, J.; Erdem, H.; Vaid, A.; Castro, P.; Ayala, G.; Ittmann, M. The expression of Sprouty1, an inhibitor of fibroblast growth factor signal transduction, is decreased in human prostate cancer. Cancer Res. 2004, 64, 4728–4735. [Google Scholar] [CrossRef]
- Rathmanner, N.; Haigl, B.; Vanas, V.; Doriguzzi, A.; Gsur, A.; Sutterluty-Fall, H. Sprouty2 but not Sprouty4 is a potent inhibitor of cell proliferation and migration of osteosarcoma cells. FEBS Lett. 2013, 587, 2597–2605. [Google Scholar] [CrossRef] [Green Version]
- Barbachano, A.; Ordonez-Moran, P.; Garcia, J.M.; Sanchez, A.; Pereira, F.; Larriba, M.J.; Martinez, N.; Hernandez, J.; Landolfi, S.; Bonilla, F.; et al. SPROUTY-2 and E-cadherin regulate reciprocally and dictate colon cancer cell tumourigenicity. Oncogene 2010, 29, 4800–4813. [Google Scholar] [CrossRef] [Green Version]
- Holgren, C.; Dougherty, U.; Edwin, F.; Cerasi, D.; Taylor, I.; Fichera, A.; Joseph, L.; Bissonnette, M.; Khare, S. Sprouty-2 controls c-Met expression and metastatic potential of colon cancer cells: Sprouty/c-Met upregulation in human colonic adenocarcinomas. Oncogene 2010, 29, 5241–5253. [Google Scholar] [CrossRef]
- Schaaf, G.; Hamdi, M.; Zwijnenburg, D.; Lakeman, A.; Geerts, D.; Versteeg, R.; Kool, M. Silencing of SPRY1 triggers complete regression of rhabdomyosarcoma tumors carrying a mutated RAS gene. Cancer Res. 2010, 70, 762–771. [Google Scholar] [CrossRef]
- Park, J.W.; Wollmann, G.; Urbiola, C.; Fogli, B.; Florio, T.; Geley, S.; Klimaschewski, L. Sprouty2 enhances the tumorigenic potential of glioblastoma cells. Neuro-Oncol. 2018, 20, 1044–1054. [Google Scholar] [CrossRef] [Green Version]
- Walsh, A.M.; Kapoor, G.S.; Buonato, J.M.; Mathew, L.K.; Bi, Y.; Davuluri, R.V.; Martinez-Lage, M.; Simon, M.C.; O’Rourke, D.M.; Lazzara, M.J. Sprouty2 Drives Drug Resistance and Proliferation in Glioblastoma. Mol. Cancer Res. 2015, 13, 1227–1237. [Google Scholar] [CrossRef] [Green Version]
- Hausott, B.; Vallant, N.; Schlick, B.; Auer, M.; Nimmervoll, B.; Obermair, G.J.; Schwarzer, C.; Dai, F.; Brand-Saberi, B.; Klimaschewski, L. Sprouty2 and -4 regulate axon outgrowth by hippocampal neurons. Hippocampus 2012, 22, 434–441. [Google Scholar] [CrossRef]
- Hausott, B.; Vallant, N.; Auer, M.; Yang, L.; Dai, F.; Brand-Saberi, B.; Klimaschewski, L. Sprouty2 down-regulation promotes axon growth by adult sensory neurons. Mol. Cell. Neurosci. 2009, 42, 328–340. [Google Scholar] [CrossRef]
- Yu, T.; Yaguchi, Y.; Echevarria, D.; Martinez, S.; Basson, M.A. Sprouty genes prevent excessive FGF signalling in multiple cell types throughout development of the cerebellum. Development 2011, 138, 2957–2968. [Google Scholar] [CrossRef] [Green Version]
- Panagiotaki, N.; Dajas-Bailador, F.; Amaya, E.; Papalopulu, N.; Dorey, K. Characterisation of a new regulator of BDNF signalling, Sprouty3, involved in axonal morphogenesis in vivo. Development 2010, 137, 4005–4015. [Google Scholar] [CrossRef] [Green Version]
- Benke, S.; Agerer, B.; Haas, L.; Stoger, M.; Lercher, A.; Gabler, L.; Kiss, I.; Scinicariello, S.; Berger, W.; Bergthaler, A.; et al. Human tripartite motif protein 52 is required for cell context-dependent proliferation. Oncotarget 2018, 9, 13565–13581. [Google Scholar] [CrossRef]
- Sutterluty, H.; Chatelain, E.; Marti, A.; Wirbelauer, C.; Senften, M.; Muller, U.; Krek, W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1999, 1, 207–214. [Google Scholar] [CrossRef]
- Vanas, V.; Haigl, B.; Stockhammer, V.; Sutterluty-Fall, H. MicroRNA-21 Increases Proliferation and Cisplatin Sensitivity of Osteosarcoma-Derived Cells. PLoS ONE 2016, 11, e0161023. [Google Scholar] [CrossRef]
- Kral, R.M.; Mayer, C.E.; Vanas, V.; Gsur, A.; Sutterluty-Fall, H. In non-small cell lung cancer mitogenic signaling leaves Sprouty1 protein levels unaffected. Cell. Biochem. Funct. 2014, 32, 96–100. [Google Scholar] [CrossRef]
- Mayer, C.E.; Haigl, B.; Jantscher, F.; Siegwart, G.; Grusch, M.; Berger, W.; Sutterluty, H. Bimodal expression of Sprouty2 during the cell cycle is mediated by phase-specific Ras/MAPK and c-Cbl activities. Cell. Mol. Life Sci. 2010, 67, 3299–3311. [Google Scholar] [CrossRef]
- Impagnatiello, M.A.; Weitzer, S.; Gannon, G.; Compagni, A.; Cotten, M.; Christofori, G. Mammalian sprouty-1 and -2 are membrane-anchored phosphoprotein inhibitors of growth factor signaling in endothelial cells. J. Cell Biol. 2001, 152, 1087–1098. [Google Scholar] [CrossRef]
- Zhang, W.; Lv, Y.; Xue, Y.; Wu, C.; Yao, K.; Zhang, C.; Jin, Q.; Huang, R.; Li, J.; Sun, Y.; et al. Co-expression modules of NF1, PTEN and sprouty enable distinction of adult diffuse gliomas according to pathway activities of receptor tyrosine kinases. Oncotarget 2016, 7, 59098–59114. [Google Scholar] [CrossRef] [Green Version]
- Doriguzzi, A.; Salhi, J.; Sutterluty-Fall, H. Sprouty4 mRNA variants and protein expressions in breast and lung-derived cells. Oncol. Lett. 2016, 12, 4161–4166. [Google Scholar] [CrossRef] [Green Version]
- Faratian, D.; Sims, A.H.; Mullen, P.; Kay, C.; Um, I.; Langdon, S.P.; Harrison, D.J. Sprouty 2 is an independent prognostic factor in breast cancer and may be useful in stratifying patients for trastuzumab therapy. PLoS ONE 2011, 6, e23772. [Google Scholar] [CrossRef]
- Sirivatanauksorn, Y.; Sirivatanauksorn, V.; Srisawat, C.; Khongmanee, A.; Tongkham, C. Differential expression of sprouty genes in hepatocellular carcinoma. J. Surg. Oncol. 2012, 105, 273–276. [Google Scholar] [CrossRef]
- Kwak, H.J.; Kim, Y.J.; Chun, K.R.; Woo, Y.M.; Park, S.J.; Jeong, J.A.; Jo, S.H.; Kim, T.H.; Min, H.S.; Chae, J.S.; et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 2011, 30, 2433–2442. [Google Scholar] [CrossRef] [Green Version]
- Doriguzzi, A.; Haigl, B.; Gsur, A.; Sutterluty-Fall, H. The increased Sprouty4 expression in response to serum is transcriptionally controlled by Specific protein 1. Int. J. Biochem. Cell Biol. 2015, 64, 220–228. [Google Scholar] [CrossRef]
- Jiang, Z.; Price, C.A. Differential actions of fibroblast growth factors on intracellular pathways and target gene expression in bovine ovarian granulosa cells. Reproduction 2012, 144, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Masoumi-Moghaddam, S.; Amini, A.; Morris, D.L. The developing story of Sprouty and cancer. Cancer Metastasis Rev. 2014, 33, 695–720. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Xie, S.; Yuan, C.; Jiang, L.; Huang, X.; Li, L.; Chen, Y.; Luo, L.; Zhang, J.; Wang, D.; et al. Lower Expression of SPRY4 Predicts a Poor Prognosis and Regulates Cell Proliferation in Colorectal Cancer. Cell. Physiol. Biochem. 2016, 40, 1433–1442. [Google Scholar] [CrossRef]
- Zhang, Q.; Wei, T.; Shim, K.; Wright, K.; Xu, K.; Palka-Hamblin, H.L.; Jurkevich, A.; Khare, S. Atypical role of sprouty in colorectal cancer: Sprouty repression inhibits epithelial-mesenchymal transition. Oncogene 2016, 35, 3151–3162. [Google Scholar] [CrossRef]
- Masoumi-Moghaddam, S.; Amini, A.; Wei, A.Q.; Robertson, G.; Morris, D.L. Sprouty 2 protein, but not Sprouty 4, is an independent prognostic biomarker for human epithelial ovarian cancer. Int. J. Cancer 2015, 137, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Thompson, B.; Ren, C.; Ittmann, M.; Kwabi-Addo, B. Sprouty4, a suppressor of tumor cell motility, is down regulated by DNA methylation in human prostate cancer. Prostate 2006, 66, 613–624. [Google Scholar] [CrossRef]
- Jaggi, F.; Cabrita, M.A.; Perl, A.K.; Christofori, G. Modulation of endocrine pancreas development but not beta-cell carcinogenesis by Sprouty4. Mol. Cancer Res. 2008, 6, 468–482. [Google Scholar] [CrossRef]
- Gong, Y.; Yang, X.; He, Q.; Gower, L.; Prudovsky, I.; Vary, C.P.; Brooks, P.C.; Friesel, R.E. Sprouty4 regulates endothelial cell migration via modulating integrin beta3 stability through c-Src. Angiogenesis 2013, 16, 861–875. [Google Scholar] [CrossRef]
- Taniguchi, K.; Ishizaki, T.; Ayada, T.; Sugiyama, Y.; Wakabayashi, Y.; Sekiya, T.; Nakagawa, R.; Yoshimura, A. Sprouty4 deficiency potentiates Ras-independent angiogenic signals and tumor growth. Cancer Sci. 2009, 100, 1648–1654. [Google Scholar] [CrossRef] [PubMed]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celik-Selvi, B.E.; Stütz, A.; Mayer, C.-E.; Salhi, J.; Siegwart, G.; Sutterlüty, H. Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells. Cells 2019, 8, 808. https://doi.org/10.3390/cells8080808
Celik-Selvi BE, Stütz A, Mayer C-E, Salhi J, Siegwart G, Sutterlüty H. Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells. Cells. 2019; 8(8):808. https://doi.org/10.3390/cells8080808
Chicago/Turabian StyleCelik-Selvi, Burcu Emine, Astrid Stütz, Christoph-Erik Mayer, Jihen Salhi, Gerald Siegwart, and Hedwig Sutterlüty. 2019. "Sprouty3 and Sprouty4, Two Members of a Family Known to Inhibit FGF-Mediated Signaling, Exert Opposing Roles on Proliferation and Migration of Glioblastoma-Derived Cells" Cells 8, no. 8: 808. https://doi.org/10.3390/cells8080808




