Nuclear Phosphoinositides—Versatile Regulators of Genome Functions
Abstract
1. Introduction
2. Transport of Phosphoinositides into the Nucleus
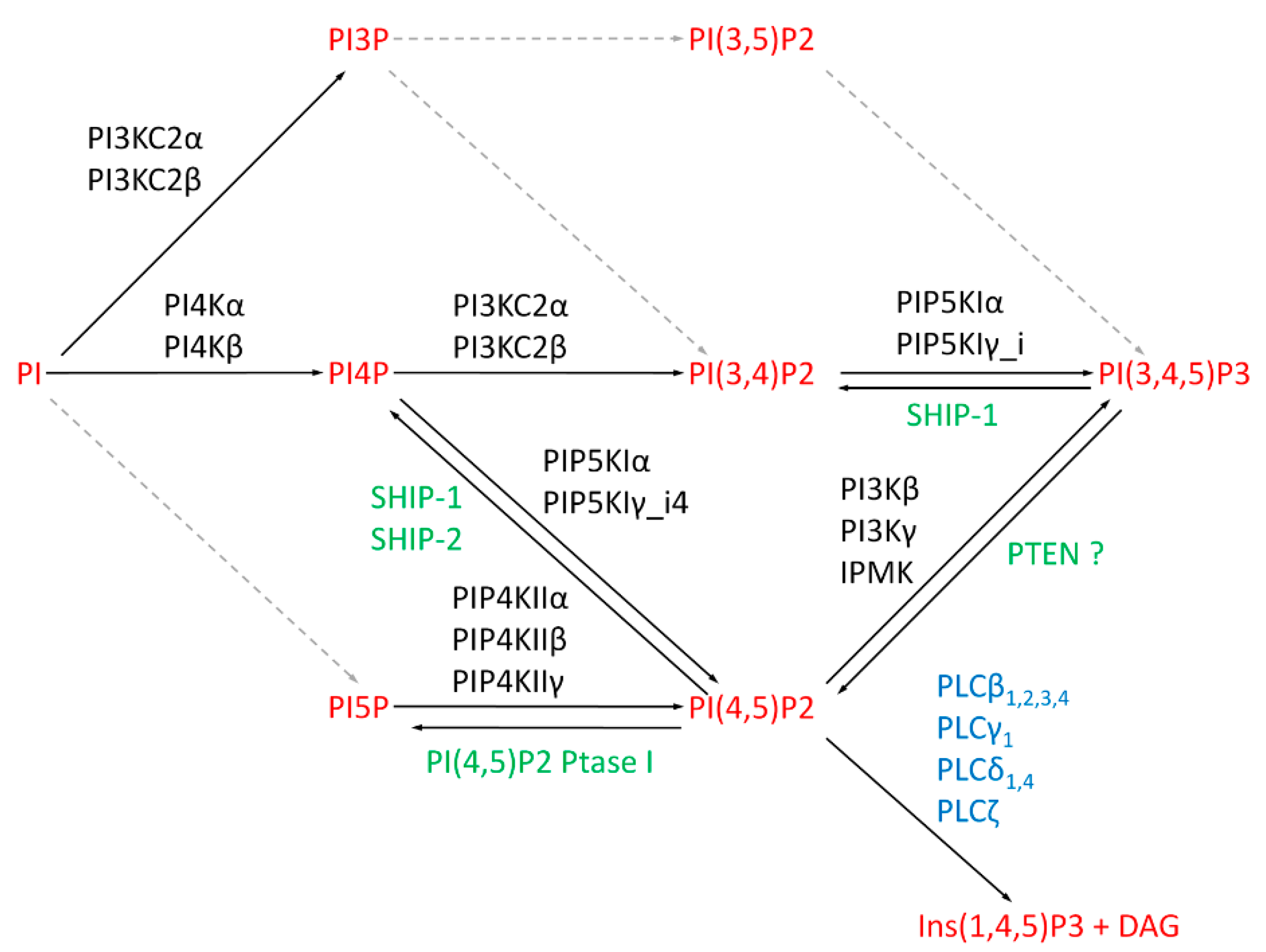
3. Metabolic Pathways for Nuclear Phosphoinositides
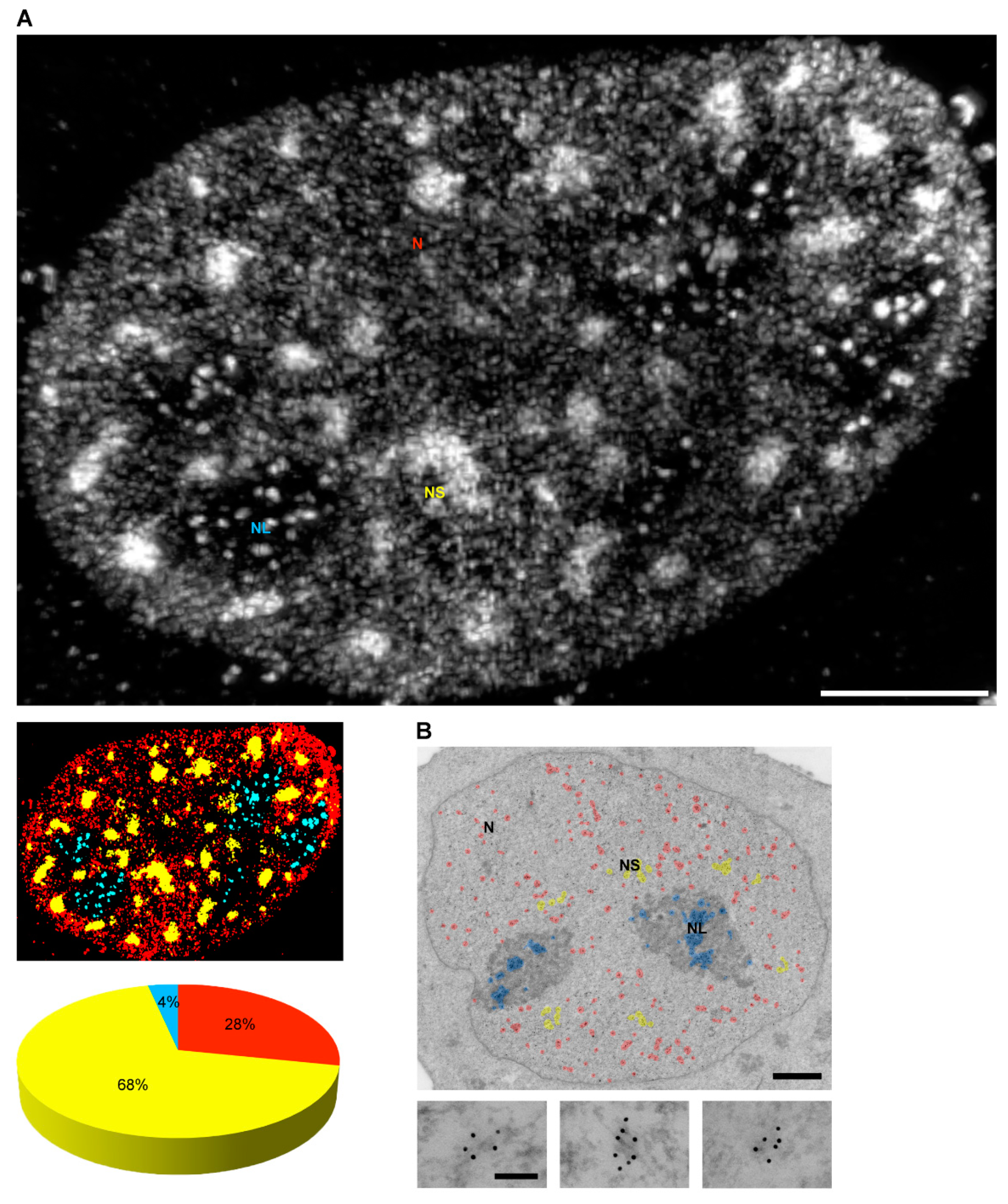
4. Localization of Nuclear Phosphoinositides
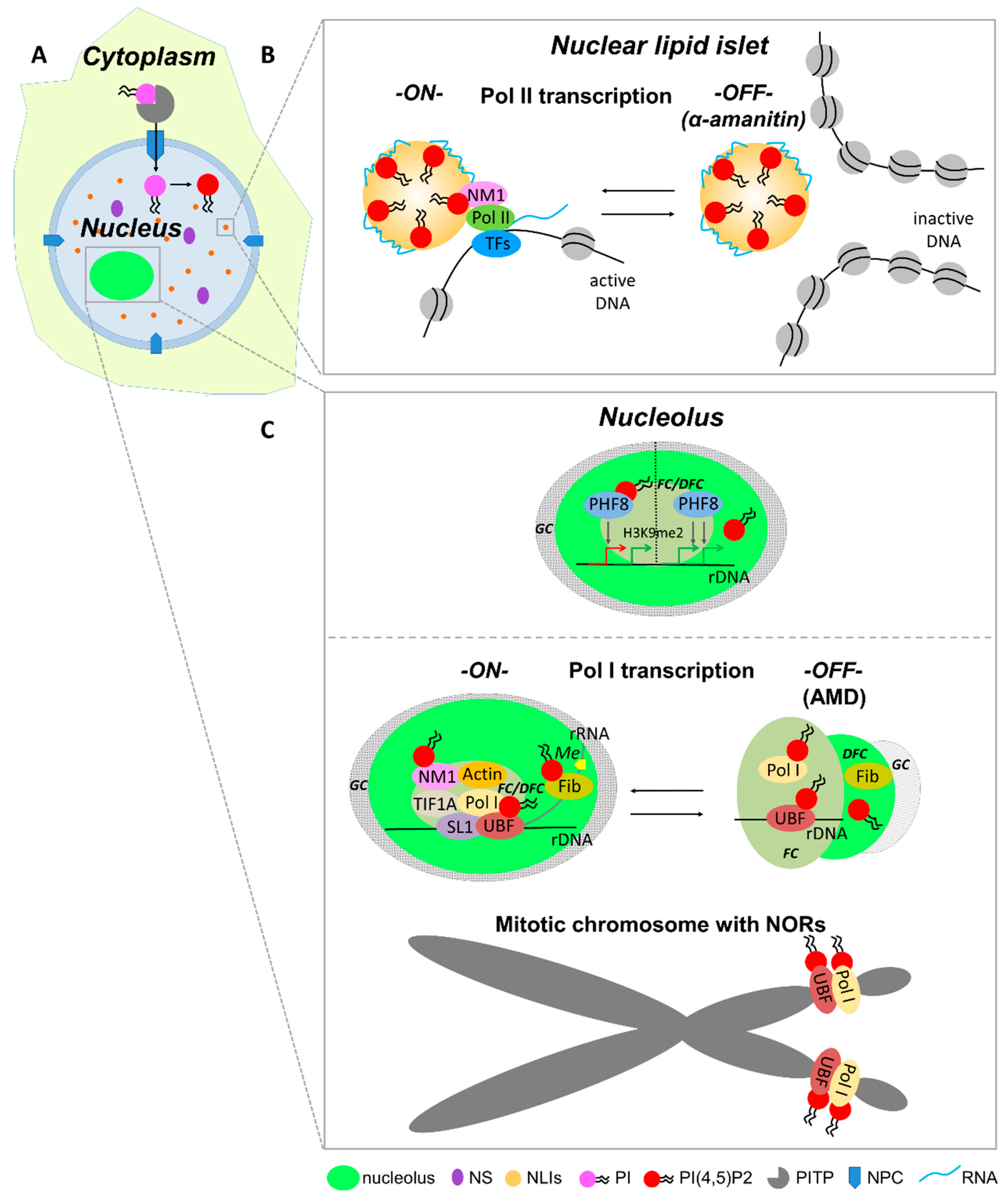
5. Nuclear Processes Regulated by Phosphoinositides
5.1. Anti-Apoptotic Signaling
5.2. Chromatin Remodeling
5.3. Epigenetics
5.4. Gene Expression
5.5. RNA Pol II-Dependent Transcription Initiation
5.6. Pre-mRNA Processing
5.7. rDNA Modification and RNA Pol I Transcription
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Warwick, N.; Somerville, C.R.; Slack, C.R. Fluxes through the prokaryotic and eukaryotic pathways of lipid synthesis in the ‘16:3’ plant Arabidopsis thaliana. Biochem. J. 1986, 235, 25–31. [Google Scholar]
- Thompson, W.; Macdonald, G. Cytidine diphosphate diglyceride of bovine brain. Positional distribution of fatty acids and analysis of major molecular species. Eur. J. Biochem. 1976, 65, 107–111. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, G.; De Camilli, P. Phosphoinositides in cell regulation and membrane dynamics. Nature 2006, 443, 651–657. [Google Scholar] [CrossRef] [PubMed]
- Balla, T.; Szentpetery, Z.; Kim, Y.J. Phosphoinositide signaling: New tools and insights. Physiology 2009, 24, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Saarikangas, J.; Zhao, H.; Lappalainen, P. Regulation of the actin cytoskeleton-plasma membrane interplay by phosphoinositides. Physiol. Rev. 2010, 90, 259–289. [Google Scholar] [CrossRef] [PubMed]
- Shewan, A.; Eastburn, D.J.; Mostov, K. Phosphoinositides in cell architecture. Cold Spring Harb Perspect. Biol. 2011, 3, a004796. [Google Scholar] [CrossRef] [PubMed]
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Thapa, N.; Hedman, A.C.; Anderson, R.A. Phosphatidylinositol 4,5-bisphosphate: Targeted production and signaling. Bioessays 2013, 35, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.K.; Williams, S.A.; Bindra, G.K.; Lay, F.T.; Poon, I.K.; Hulett, M.D. Phosphoinositides: Multipurpose cellular lipids with emerging roles in cell death. Cell Death Differ. 2019, 26, 781–793. [Google Scholar] [CrossRef] [PubMed]
- Dickson, E.J.; Hille, B. Understanding phosphoinositides: Rare, dynamic, and essential membrane phospholipids. Biochem. J. 2019, 476, 1–23. [Google Scholar] [CrossRef]
- Ratti, S.; Ramazzotti, G.; Faenza, I.; Fiume, R.; Mongiorgi, S.; Billi, A.M.; McCubrey, J.A.; Suh, P.G.; Manzoli, L.; Cocco, L.; et al. Nuclear inositide signaling and cell cycle. Adv. Biol. Regul. 2018, 67, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hamann, B.L.; Blind, R.D. Nuclear phosphoinositide regulation of chromatin. J. Cell Physiol. 2018, 233, 107–123. [Google Scholar] [CrossRef] [PubMed]
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marášek, P.; Castaño, E.; Hozák, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Osborne, S.L.; Thomas, C.L.; Gschmeissner, S.; Schiavo, G. Nuclear PtdIns (4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 2001, 14, 2501–2511. [Google Scholar]
- Prasanth, K.V.; Sacco-Bubulya, P.A.; Prasanth, S.G.; Spector, D.L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell. 2003, 14, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- de Vries, K.J.; Heinrichs, A.A.J.; Cunningham, E.; Brunink, F.; Westerman, J.; Somerharju, P.J.; Cockcroft, S.; Wirtz, K.W.A.; Snoek, G.T. An isoform of the phosphatidylinositol-transfer protein transfers sphingomyelin and is associated with the Golgi system. Biochem. J. 1995, 310, 643–649. [Google Scholar] [CrossRef] [PubMed]
- De Vries, K.J.; Westerman, J.; Bastiaens, P.I.; Jovin, T.M.; Wirtz, K.W.; Snoek, G.T. Fluorescently labeled phosphatidylinositol transfer protein isoforms (a and b), microinjected into fetal bovine heart endothelial cells, are targeted to distinct intracellular sites. Exp. Cell Res. 1996, 227, 33–39. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rubbini, S.; Cocco, L.; Manzoli, L.; Lutterman, J.; Billi, A.M.; Matteucci, A.; Wirtz, K.W. Phosphoinositide signalling in nuclei of friend cells: DMSO-induced differentiation reduces the association of phosphatidylinositol-transfer protein with the nucleus. Biochem. Biophys. Res. Commun. 1997, 230, 30230–30235. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.D.; Wells, W.W. Phosphorylation of rat liver nuclear envelopes. I. Characterization of in vitro protein phosphorylation. J. Biol. Chem. 1983, 258, 9360–9367. [Google Scholar]
- Cocco, L.; Gilmour, R.S.; Ognibene, A.; Letcher, A.J.; Manzoli, F.A.; Irvine, R.F. Synthesis of polyphosphoinositides in nuclei of Friend cells. Evidence for polyphosphoinositide metabolism inside the nucleus which changes with cell differentiation. Biochem. J. 1987, 248, 765–770. [Google Scholar] [CrossRef] [PubMed]
- Vann, R.L.; Wooding, P.F.; Irvine, F.R.; Divecha, N. Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 1997, 327, 569–576. [Google Scholar] [CrossRef] [PubMed]
- Payrastre, B.; Nievers, M.; Boonstra, J.; Breton, M.; Verkleij, A.J.; en Henegouwen, P.V.B. A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 1992, 267, 5078–5084. [Google Scholar] [PubMed]
- York, J.D.; Majerus, P.W. Nuclear phosphatidylinositols decrease during S-phase of the cell cycle in HeLa cells. J. Biol. Chem. 1994, 269, 7847–7850. [Google Scholar] [PubMed]
- Clarke, J.H.; Letcher, A.J.; D’Santos, C.S.; Halstead, J.R.; Irvine, R.F.; Divecha, N. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem. J. 2001, 357, 905–910. [Google Scholar] [CrossRef] [PubMed]
- Višnjić, D.; Ćurić, J.; Crljen, V.; Batinić, D.; Volinia, S.; Banfić, H. Nuclear phosphoinositide 3-kinase C2beta activation during G2/M phase of the cell cycle in HL-60 cells. Biochim. Biophys. Acta 2003, 1631, 61–71. [Google Scholar] [CrossRef]
- Stallings, J.D.; Tall, E.G.; Pentyala, S.; Rebecchi, M.J. Nuclear translocation of phospholipase C-delta1 is linked to the cell cycle and nuclear phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 2005, 280, 22060–22069. [Google Scholar] [CrossRef]
- Zheng, L.; Lee, W.H. The retinoblastoma gene: A prototypic and multifunctional tumor suppressor. Exp. Cell Res. 2001, 264, 2–18. [Google Scholar] [CrossRef]
- Divecha, N.; Roefs, M.; Los, A.; Halstead, J.; Bannister, A.; D’Santos, C. Type I PIPkinases interact with and are regulated by the retinoblastoma susceptibility gene product-pRB. Curr. Biol. 2002, 12, 582–587. [Google Scholar] [CrossRef]
- Didichenko, S.A.; Thelen, M. Phosphatidylinositol 3-kinase c2alpha contains a nuclear localization sequence and associates with nuclear speckles. J. Biol. Chem. 2001, 276, 48135–48142. [Google Scholar] [CrossRef]
- Sinđić, A.; Aleksandrova, A.; Fields, A.P.; Volinia, S.; Banfić, H. Presence and activation of nuclear phosphoinositide 3-kinase C2beta during compensatory liver growth. J. Biol. Chem. 2001, 276, 17754–17761. [Google Scholar] [CrossRef]
- De Graaf, P.; Klapisz, E.E.; Schulz, T.K.; Cremers, A.F.; Verkleij, A.J.; en Henegouwen, P.M.V.B. Nuclear localization of phosphatidylinositol 4-kinase beta. J. Cell Sci. 2002, 115, 1769–1775. [Google Scholar] [PubMed]
- Strahl, T.; Hama, H.; DeWald, D.B.; Thorner, J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J. Cell Biol. 2005, 171, 967–979. [Google Scholar] [CrossRef] [PubMed]
- Kakuk, A.; Friedländer, E.; Vereb Jr, G.; Kása, A.; Balla, A.; Balla, T.; Heilmeyer Jr, L.M.; Gergely, P.; Vereb, G. Nucleolar localization of phosphatidylinositol 4-kinase PI4K230 in various mammalian cells. Cytom. A 2006, 69, 1174–1183. [Google Scholar] [CrossRef] [PubMed]
- Damen, J.E.; Liu, L.; Rosten, P.; Humphries, R.K.; Jefferson, A.B.; Majerus, P.W.; Krystal, G. The 145-kDa protein induced to associate with Shc by multiple cytokines is an inositol tetraphosphate and phosphatidylinositol 3,4,5-triphosphate 5-phosphatase. Proc. Natl. Acad. Sci. USA 1996, 93, 1689–1693. [Google Scholar] [CrossRef] [PubMed]
- Lioubin, M.N.; Algate, P.A.; Tsai, S.; Carlberg, K.; Aebersold, A.; Rohrschneider, L.R. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes. Dev. 1996, 10, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Derua, R.; Janssens, V.; Nakamura, T.; Vanderwinden, J.M.; Waelkens, E.; Erneux, C. Evidence of SHIP2 Ser132 phosphorylation, its nuclear localization and stability. Biochem. J. 2011, 439, 391–401. [Google Scholar]
- Boronenkov, I.V.; Loijens, J.C.; Umeda, M.; Anderson, R.A. Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 1998, 9, 3547–3560. [Google Scholar] [CrossRef]
- Mellman, D.L.; Gonzales, M.L.; Song, C.; Barlow, C.A.; Wang, P.; Kendziorski, C.; Anderson, R.A. A PtdIns4,5P2-regulated nuclear poly (A) polymerase controls expression of select mRNAs. Nature 2008, 451, 1013–1017. [Google Scholar] [CrossRef]
- Szivak, I.; Lamb, N.; Heilmeyer, L.M. Subcellular localization and structural function of endogenous phosphorylated phosphatidylinositol 4-kinase (PI4K92). J. Biol. Chem. 2006, 281, 16740–16749. [Google Scholar] [CrossRef]
- Richardson, J.P.; Wang, M.; Clarke, J.H.; Patel, K.J.; Irvine, R.F. Genomic tagging of endogenous type IIbeta phosphatidylinositol 5-phosphate 4-kinase in DT40 cells reveals a nuclear localisation. Cell Signal. 2007, 19, 1309–1314. [Google Scholar] [CrossRef]
- Jones, D.R.; Bultsma, Y.; Keune, W.J.; Halstead, J.R.; Elouarrat, D.; Mohammed, S.; Heck, A.J.; D’Santos, C.S.; Divecha, N. Nuclear PtdIns5P as a transducer of stress signaling: An in vivo role for PIP4Kbeta. Mol. Cell 2006, 23, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Marjanovic, J.; Kisseleva, M.V.; Wilson, M.; Majerus, P.W. Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. USA 2007, 104, 16834–16839. [Google Scholar] [CrossRef] [PubMed]
- Schill, N.J.; Anderson, R.A. Two novel phosphatidylinositol-4-phosphate 5-kinase type Igamma splice variants expressed in human cells display distinctive cellular targeting. Biochem. J. 2009, 422, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Bond, N.J.; Letcher, A.J.; Richardson, J.P.; Lilley, K.S.; Irvine, R.F.; Clarke, J.H. Genomic tagging reveals a random association of endogenous PtdIns5P 4-kinases IIalpha and IIbeta and a partial nuclear localization of the IIalpha isoform. Biochem. J. 2010, 430, 215–221. [Google Scholar] [CrossRef]
- Clarke, J.H.; Irvine, R.F. The activity, evolution and association of phosphatidylinositol 5-phosphate 4-kinases. Adv. Biol. Regul. 2012, 52, 40–45. [Google Scholar] [CrossRef]
- Doughman, R.L.; Firestone, A.J.; Wojtasiak, M.L.; Bunce, M.W.; Anderson, R.A. Membrane ruffling requires coordination between type Ialpha phosphatidylinositol phosphate kinase and Rac signaling. J. Biol. Chem. 2003, 278, 23036–23045. [Google Scholar] [CrossRef]
- Neri, L.M.; Milani, D.; Bertolaso, L.; Stroscio, M.; Bertagnolo, V.; Capitani, S. Nuclear translocation of phosphatidylinositol 3-kinase in rat pheochromocytoma PC 12 cells after treatment with nerve growth factor. Cell Mol. Biol. 1994, 40, 619–626. [Google Scholar]
- Zini, N.; Ognibene, A.; Bavelloni, A.; Santi, S.; Sabatelli, P.; Baldini, N.; Scotlandi, K.; Serra, M.; Maraldi, N.M. Cytoplasmic and nuclear localization sites of phosphatidylinositol 3-kinase in human osteosarcoma sensitive and multidrug-resistant Saos-2 cells. Histochem. Cell Biol. 1996, 106, 457–464. [Google Scholar] [CrossRef]
- Metjian, A.; Roll, R.L.; Ma, A.D.; Abrams, C.S. Agonists cause nuclear translocation of phosphatidylinositol 3-kinase gamma. A Gbetagamma-dependent pathway that requires the p110gamma amino terminus. J. Biol. Chem. 1999, 274, 27943–27947. [Google Scholar] [CrossRef]
- Bacqueville, D.; Déléris, P.; Mendre, C.; Pieraggi, M.T.; Chap, H.; Guillon, G.; Perret, B.; Breton-Douillon, M. Characterization of a G protein-activated phosphoinositide 3-kinase in vascular smooth muscle cell nuclei. J. Biol. Chem. 2001, 276, 22170–22176. [Google Scholar] [CrossRef]
- Resnick, A.C.; Snowman, A.M.; Kang, B.N.; Hurt, K.J.; Snyder, S.H.; Saiardi, A. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. USA 2005, 102, 12783–12788. [Google Scholar] [CrossRef] [PubMed]
- Maag, D.; Maxwell, M.J.; Hardesty, D.A.; Boucher, K.L.; Choudhari, N.; Hanno, A.G.; Ma, J.F.; Snowman, A.S.; Pietropaoli, J.W.; Xu, R.; et al. Inositol polyphosphate multikinase is a physiologic PI3-kinase that activates Akt/PKB. Proc. Natl. Acad. Sci. USA 2011, 108, 1391–1396. [Google Scholar] [CrossRef] [PubMed]
- Vanhaesebroeck, B.; Leevers, S.J.; Ahmadi, K.; Timms, J.; Katso, R.; Driscoll, P.C.; Woscholski, R.; Parker, P.J.; Waterfield, M.D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001, 70, 535–602. [Google Scholar] [CrossRef] [PubMed]
- Lachyankar, M.B.; Sultana, N.; Schonhoff, C.M.; Mitra, P.; Poluha, W.; Lambert, S.; Quesenberry, P.J.; Litofsky, N.S.; Recht, L.D.; Nabi, R.; et al. A role for nuclear PTEN in neuronal differentiation. J. Neurosci. 2000, 20, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Gimm, O.; Perren, A.; Weng, L.P.; Marsh, D.J.; Yeh, J.J.; Ziebold, U.; Gil, E.; Hinze, R.; Delbridge, L.; Lees, J.A.; et al. Differential nuclear and cytoplasmic expression of PTEN in normal thyroid tissue, and benign and malignant epithelial thyroid tumors. Am. J. Pathol. 2000, 156, 1693–1700. [Google Scholar] [CrossRef]
- Déléris, P.; Bacqueville, D.; Gayral, S.; Carrez, L.; Salles, J.P.; Perret, B.; Breton-Douillon, M. SHIP-2 and PTEN are expressed and active in vascular smooth muscle cell nuclei, but only SHIP-2 is associated with nuclear speckles. J. Biol. Chem. 2003, 278, 38884–38891. [Google Scholar] [CrossRef] [PubMed]
- Lindsay, Y.; McCoull, D.; Davidson, L.; Leslie, N.R.; Fairservice, A.; Gray, A.; Lucocq, J.; Downes, C.P. Localization of agonist-sensitive PtdIns (3,4,5)P3 reveals a nuclear pool that is insensitive to PTEN expression. J. Cell Sci. 2006, 119, 5160–5168. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.; Gascard, P.; Berthon, B.; Fukami, K.; Takenawa, T.; Giraud, F.; Claret, M. Cellular distribution of polyphosphoinositides in rat hepatocytes. Cell Signal. 1993, 5, 565–581. [Google Scholar] [CrossRef]
- Kim, C.G.; Park, D.; Rhee, S.G. The role of carboxyl-terminal basic amino acids in Gqalpha-dependent activation, particulate association, and nuclear localization of phospholipase C-beta1. J. Biol. Chem. 1996, 271, 21187–21192. [Google Scholar] [CrossRef]
- Zini, N.; Martelli, A.M.; Cocco, L.; Manzoli, F.A.; Maraldi, N.M. Phosphoinositidase C isoforms are specifically localized in the nuclear matrix and cytoskeleton of Swiss 3T3 cells. Exp. Cell Res. 1993, 208, 257–269. [Google Scholar] [CrossRef]
- Tabellini, G.; Bortul, R.; Santi, S.; Riccio, M.; Baldini, G.; Cappellini, A.; Billi, A.M.; Berezney, R.; Ruggeri, A.; Cocco, L.; et al. Diacylglycerol kinase-theta is localized in the speckle domains of the nucleus. Exp. Cell Res. 2003, 287, 143–154. [Google Scholar] [CrossRef]
- Bertagnolo, V.; Mazzoni, M.; Ricci, D.; Carini, C.; Neri, L.M.; Previati, M.; Capitani, S. Identification of PI-PLC beta 1, gamma 1, and delta 1 in rat liver: Subcellular distribution and relationship to inositol lipid nuclear signalling. Cell Signal. 1995, 7, 669–678. [Google Scholar] [CrossRef]
- Liu, N.; Fukami, K.; Yu, H.; Takenawa, T. A new phospholipase C delta 4 is induced at S-phase of the cell cycle and appears in the nucleus. J. Biol. Chem. 1996, 271, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Bertagnolo, V.; Marchisio, M.; Capitani, S.; Neri, L.M. Intranuclear translocation of phospholipase C beta2 during HL-60 myeloid differentiation. Biochem. Biophys. Res. Commun. 1997, 235, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Yamaga, M.; Fujii, M.; Kamata, H.; Hirata, H.; Yagisawa, H. Phospholipase C-delta1 contains a functional nuclear export signal sequence. J. Biol. Chem. 1999, 274, 28537–28541. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Aghdasi, B.; Luo, H.R.; Moriarity, J.L.; Wu, F.Y.; Hong, J.J.; Hurt, K.J.; Bae, S.S.; Suh, P.G.; Snyder, S.H. Phospholipase C gamma 1 is a physiological guanine nucleotide exchange factor for the nuclear GTPase PIKE. Nature 2002, 415, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Yoda, A.; Oda, S.; Shikano, T.; Kouchi, Z.; Awaji, T.; Shirakawa, H.; Kinoshita, K.; Miyazaki, S. Ca2+ oscillation-inducing phospholipase C zeta expressed in mouse eggs is accumulated to the pronucleus during egg activation. Dev. Biol. 2004, 268, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Larman, M.G.; Saunders, C.M.; Carroll, J.; Lai, F.A.; Swann, K. Cell cycle-dependent Ca2+ oscillations in mouse embryos are regulated by nuclear targeting of PLCzeta. J. Cell Sci. 2004, 117, 2513–2521. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.M.; Andrews, J.B.; Bannan, B.A.; Nazario-Toole, A.E.; Jenkins, T.C.; Christensen, K.D.; Oprisan, S.A.; Meyer-Bernstein, E.L. Phospholipase C beta 4 in mouse hepatocytes: Rhythmic expression and cellular distribution. Comp. Hepatol. 2008, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Cooney, M.A.; Malcuit, C.; Cheon, B.; Holland, M.K.; Fissore, R.A.; D’Cruz, N.T. Species-specific differences in the activity and nuclear localization of murine and bovine phospholipase C zeta 1. Biol. Reprod. 2010, 83, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Fiume, R.; Keune, W.J.; Faenza, I.; Bultsma, Y.; Ramazzotti, G.; Jones, D.R.; Martelli, A.M.; Somner, L.; Follo, M.Y.; Divecha, N.; et al. Nuclear phosphoinositides: Location, regulation and function. Subcell. Biochem. 2012, 59, 335–361. [Google Scholar] [PubMed]
- Kalasova, I.; Fáberová, V.; Kalendová, A.; Yildirim, S.; Uličná, L.; Venit, T.; Hozák, P. Tools for visualization of phosphoinositides in the cell nucleus. Histochem. Cell Biol. 2016, 145, 485–496. [Google Scholar] [CrossRef]
- Tsuji, T.; Takatori, S.; Fujimoto, T. Definition of phosphoinositide distribution in the nanoscale. Curr. Opin. Cell Biol. 2019, 57, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef] [PubMed]
- Yokogawa, T.; Nagata, S.; Nishio, Y.; Tsutsumi, T.; Ihara, S.; Shirai, R.; Morita, K.; Umeda, M.; Shirai, Y.; Saitoh, N.; et al. Evidence that 3’-phosphorylated polyphosphoinositides are generated at the nuclear surface: Use of immunostaining technique with monoclonal antibodies specific for PI 3,4-P (2). FEBS Lett. 2000, 473, 222–226. [Google Scholar] [CrossRef]
- Mazzotti, G.; Zini, N.; Rizzi, E.; Rizzoli, R.; Galanzi, A.; Ognibene, A.; Santi, S.; Matteucci, A.; Martelli, A.M.; Maraldi, N.M. Immunocytochemical detection of phosphatidylinositol 4,5-bisphosphate localization sites within the nucleus. J. Histochem. Cytochem. 1995, 43, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Kular, G.; Fleming, I.N.; Downes, C.P.; Lucocq, J.M. Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 2002, 363, 657–666. [Google Scholar]
- Yildirim, S.; Castano, E.; Sobol, M.; Philimonenko, V.V.; Dzijak, R.; Venit, T.; Hozák, P. Involvement of phosphatidylinositol 4,5-bisphosphate in RNA polymerase I transcription. J. Cell Sci. 2013, 126, 2730–2739. [Google Scholar] [CrossRef]
- Ulicna, L.; Kalendova, A.; Kalasova, I.; Vacik, T.; Hozák, P. PIP2 epigenetically represses rRNA genes transcription interacting with PHF8. Biochim. Biophys. Acta 2018, 1863, 266–275. [Google Scholar] [CrossRef]
- Sobol, M.; Krausová, A.; Yildirim, S.; Kalasová, I.; Fáberová, V.; Vrkoslav, V.; Philimonenko, V.; Marášek, P.; Pastorek, L.; Čapek, M.; et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018, 131, jcs211094. [Google Scholar] [CrossRef]
- Maraldi, N.M.; Zini, N.; Santi, S.; Manzoli, F.A. Topology of inositol lipid signal transduction in the nucleus. J. Cell. Physiol. 1999, 181, 203–217. [Google Scholar] [CrossRef]
- Sztacho, M.; Sobol, M.; Balaban, C.; Lopes, S.E.E.; Hozák, P. Nuclear phosphoinositides and phase separation: Important players in nuclear compartmentalization. Adv. Biol. Regul. 2019, 71, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Toska, E.; Campbell, H.A.; Shandilya, J.; Goodfellow, S.J.; Shore, P.; Medler, K.F.; Roberts, S.G. Repression of transcription by WT1-BASP1 requires the myristoylation of BASP1 and the PIP2-dependent recruitment of histone deacetylase. Cell Rep. 2012, 2, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Blind, R.D.; Suzawa, M.; Ingraham, H.A. Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Sci. Signal. 2012, 5, ra44. [Google Scholar] [CrossRef] [PubMed]
- Lewis, A.E.; Sommer, L.; Arntzen, M.Ø.; Strahm, Y.; Morrice, N.A.; Divecha, N.; D’Santos, C.S. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol. Cell Proteomics 2011, 10, M110-003376. [Google Scholar] [CrossRef] [PubMed]
- Gelato, K.A.; Tauber, M.; Ong, M.S.; Winter, S.; Hiragami-Hamada, K.; Sindlinger, J.; Lemak, A.; Bultsma, Y.; Houliston, S.; Schwarzer, D.; et al. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol. Cell 2014, 54, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Gozani, O.; Karuman, P.; Jones, D.R.; Ivanov, D.; Cha, J.; Lugovskoy, A.A.; Baird, C.L.; Zhu, H.; Field, S.J.; Lessnick, S.L.; et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 2003, 114, 99–111. [Google Scholar] [CrossRef]
- Ndamukong, I.; Jones, D.R.; Lapko, H.; Divecha, N.; Avramova, Z. Phosphatidylinositol 5-phosphate links dehydration stress to the activity of ARABIDOPSIS TRITHORAX-LIKE factor ATX1. PLoS ONE 2010, 5, e13396. [Google Scholar] [CrossRef]
- Jungmichel, S.; Sylvestersen, K.B.; Choudhary, C.; Nguyen, S.; Mann, M.; Nielsen, M.L. Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep. 2014, 6, 578–591. [Google Scholar] [CrossRef]
- Bertagnolo, V.; Neri, L.M.; Marchisio, M.; Mischiati, C.; Capitani, S. Phosphoinositide 3-kinase activity is essential for all-trans-retinoic acid-induced granulocytic differentiation of HL-60 cells. Cancer Res. 1999, 59, 542–546. [Google Scholar]
- Ahn, J.Y.; Rong, R.; Liu, X.; Ye, K. PIKE/nuclear PI 3-kinase signaling mediates the antiapoptotic actions of NGF in the nucleus. EMBO J. 2004, 23, 3995–4006. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.Y.; Liu, X.; Cheng, D.; Peng, J.; Chan, P.K.; Wade, P.A.; Ye, K. Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol. Cell 2005, 18, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, P.; Martelli, A.M.; Tabellini, G.; Bellacosa, A.; Capitani, S.; Neri, L.M. Threonine 308 phosphorylated form of Akt translocates to the nucleus of PC12 cells under nerve growth factor stimulation and associates with the nuclear matrix protein nucleolin. J. Cell Physiol. 2003, 196, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.L.X.; Choi, J.W.; Lee, S.B.; Ye, K.; Woo, S.D.; Lee, K.H.; Ahn, J.Y. Akt phosphorylation is essential for nuclear translocation and retention in NGF-stimulated PC12 cells. Biochem. Biophys. Res. Commun. 2006, 349, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Nguyen, T.L.X.; Choi, J.W.; Lee, K.H.; Cho, S.W.; Liu, Z.; Ye, K.; Bae, S.S.; Ahn, J.Y. Nuclear akt interacts with B23/NPM and protects it from proteolytic cleavage, enhancing cell survival. Proc. Natl. Acad. Sci. USA 2008, 105, 16584–165689. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns (3,4,5) P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, T.; Altankhuyag, A.; Dobrovolska, O.; Turcu, D.C.; Lewis, A.E. polybasic motif in ErbB3-binding protein 1 (EBP1) has key functions in nucleolar localization and polyphosphoinositide interaction. Biochem. J. 2016, 473, 2033–2047. [Google Scholar] [CrossRef]
- Bunce, M.W.; Boronenkov, I.V.; Anderson, R.A. Coordinated activation of the nuclear ubiquitin ligase Cul3-SPOP by the generation of phosphatidylinositol 5-phosphate. J. Biol. Chem. 2008, 283, 8678–8686. [Google Scholar] [CrossRef]
- Yu, H.; Fukami, K.; Watanabe, Y.; Ozaki, C.; Takenawa, T. Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur. J. Biochem. 1998, 251, 281–287. [Google Scholar] [CrossRef]
- Croston, G.E.; Kerrigan, L.A.; Lira, L.M.; Marshak, D.R.; Kadonaga, J.T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science 1991, 251, 643–649. [Google Scholar] [CrossRef]
- Johnson, C.A.; Goddard, J.P.; Adams, R.L. The effect of histone H1 and DNA methylation on transcription. Biochem. J. 1995, 305, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.; Yeivin, A.; Ben-Asher, E.; Aloni, Y.; Razin, A. Histone H1-mediated inhibition of transcription initiation of methylated templates in vitro. J. Biol. Chem. 1993, 268, 21754–21759. [Google Scholar] [PubMed]
- Rando, O.J.; Zhao, K.; Janmey, P.; Crabtree, G.R. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2002, 99, 2824–2829. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Wang, W.; Rando, O.J.; Xue, Y.; Swiderek, K.; Kuo, A.; Crabtree, G.R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 1998, 95, 625–636. [Google Scholar] [CrossRef]
- Viiri, K.M.; Jänis, J.; Siggers, T.; Heinonen, T.Y.; Valjakka, J.; Bulyk, M.L.; Mäki, M.; Lohi, O. DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell Biol. 2009, 29, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ramirez, M.; Rocchini, C.; Ausio, J. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 1995, 270, 17923–17928. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Hayes, J.J.; Pruss, D.; Wolffe, A.P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell 1993, 72, 73–84. [Google Scholar] [CrossRef]
- Nagashima, M.; Shiseki, M.; Miura, K.; Hagiwara, K.; Linke, S.P.; Pedeux, R.; Wang, X.W.; Yokota, J.; Riabowol, K.; Harris, C.C. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc. Natl. Acad. Sci. USA 2001, 98, 9671–9676. [Google Scholar] [CrossRef]
- Choi, S.; Chen, M.; Cryns, V.L.; Anderson, R.A. A nuclear phosphoinositide kinase complex regulates p53. Nature 2019, 21, 462–475. [Google Scholar] [CrossRef]
- Alvarez-Venegas, R.; Sadder, M.; Hlavacka, A.; Baluška, F.; Xia, Y.; Lu, G.; Firsov, A.; Sarath, G.; Moriyama, H.; Dubrovsky, J.G.; et al. The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. USA 2006, 103, 6049–6054. [Google Scholar] [CrossRef]
- Stijf-Bultsma, Y.; Sommer, L.; Tauber, M.; Baalbaki, M.; Giardoglou, P.; Jones, D.R.; Gelato, K.A.; van Pelt, J.; Shah, Z.; Rahnamoun, H.; et al. The basal transcription complex component TAF3 transduces changes in nuclear phosphoinositides into transcriptional output. Mol. Cell 2015, 58, 453–467. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Horiguchi, K.; Yoshida, T.; Takeda, M.; Fujisawa, H.; Takeuchi, K.; Umeda, M.; Kato, S.; Ihara, S.; Nagata, S.; et al. Evidence that a phosphatidylinositol 3,4,5-trisphosphate-binding protein can function in nucleus. J. Biol. Chem. 1999, 274, 3919–3922. [Google Scholar] [CrossRef] [PubMed]
- Blind, R.D.; Sablin, E.P.; Kuchenbecker, K.M.; Chiu, H.J.; Deacon, A.M.; Das, D.; Fletterick, R.J.; Ingraham, H.A. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc. Natl. Acad. Sci. USA 2014, 111, 15054–15059. [Google Scholar] [CrossRef] [PubMed]
- Lalli, E.; Doghman, M.; de Late, P.L.; El Wakil, A.; Mus-Veteau, I. Beyond steroidogenesis: Novel target genes for SF-1 discovered by genomics. Mol. Cell Endocrinol. 2013, 371, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Ulicna, L.; Rohozkova, J.; Hozak, P. Multiple aspects of PIP2 involvement in C. elegans gametogenesis. Int. J. Mol. Sci. 2018, 19, 2679. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Hirose, T.; Kimura, H.; Hagiwara, M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 2007, 282, 15645–15651. [Google Scholar] [CrossRef] [PubMed]
- Fuke, H.; Ohno, M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucl. Acids Res. 2008, 36, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Okada, M.; Jang, S.W.; Ye, K. Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. USA 2008, 105, 8649–8654. [Google Scholar] [CrossRef]
- Wickramasinghe, V.O.; Savill, J.M.; Chavali, S.; Jonsdottir, A.B.; Rajendra, E.; Grüner, T.; Laskey, R.A.; Babu, M.M.; Venkitaraman, A.R. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol. Cell 2013, 51, 737–750. [Google Scholar] [CrossRef]
- Grummt, I. Wisely chosen paths—Regulation of rRNA synthesis. FEBS J. 2010, 277, 4626–4639. [Google Scholar] [CrossRef]
- Hozák, P.; Cook, P.R.; Schofer, C.; Mosgoller, W.; Wachtler, F. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 1994, 107, 639–648. [Google Scholar] [PubMed]
- Bell, S.P.; Learned, R.M.; Jantzen, H.M.; Tjian, R. Functional cooperativity between transcription factor-Ubf1 and factor-Sl1 mediates human ribosomal-Rna synthesis. Science 1988, 241, 1192–1197. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, B.; Read, C.; Moss, T. Recognition of the Xenopus ribosomal core promoter by the transcription factor xUBF involves multiple HMG box domains and leads to an xUBF interdomain interaction. EMBO J. 1993, 12, 513–525. [Google Scholar] [CrossRef]
- Mais, C.; Wright, J.E.; Prieto, J.L.; Raggett, S.L.; McStay, B. UBF-binding site arrays form pseudo-NORs and sequester the RNA polymerase I transcription machinery. Genes Dev. 2005, 19, 50–64. [Google Scholar] [CrossRef] [PubMed]
- Mougey, E.B.; Pape, L.K.; Sollner-Webb, B. Virtually the entire Xenopus laevis rDNA multikilobase intergenic spacer serves to stimulate polymerase I transcription. J. Biol. Chem. 1996, 271, 27138–27145. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, A.C.; Sullivan, G.J.; McStay, B. UBF binding in vivo is not restricted to regulatory sequences within the vertebrate ribosomal DNA repeat. Mol. Cell Biol. 2002, 22, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Pikaard, C.S.; McStay, B.; Schultz, M.C.; Bell, S.P.; Reeder, R.H. The Xenopus ribosomal gene enhancers bind an essential polymerase I transcription factor, xUBF. Genes Dev. 1989, 3, 1779–1788. [Google Scholar] [CrossRef]
- Putnam, C.D.; Pikaard, C.S. Cooperative binding of the Xenopus RNA polymerase I transcription factor xUBF to repetitive ribosomal gene enhancers. Mol. Cell Biol. 1992, 12, 4970–4980. [Google Scholar] [CrossRef]
- Loza-Muller, L.; Rodríguez-Corona, U.; Sobol, M.; Rodríguez-Zapata, L.C.; Hozak, P.; Castano, E. Fibrillarin methylates H2A in RNA polymerase I trans-active promoters in Brassica oleracea. Front. Plant Sci. 2015, 6, 976. [Google Scholar] [CrossRef]
- Tollervey, D.; Lehtonen, H.; Jansen, R.; Kern, H.; Hurt, E.C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell 1993, 72, 443–457. [Google Scholar] [CrossRef]
- Newton, K.; Petfalski, E.; Tollervey, D.; Cáceres, J.F. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol. Cell Biol. 2003, 23, 8519–8527. [Google Scholar] [CrossRef] [PubMed]
- Uličná, L.; Paprčková, D.; Fáberová, V.; Hozák, P. Phospholipids and inositol phosphates linked to the epigenome. Histochem. Cell Biol. 2018, 150, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Tribble, E.K.; Ivanova, P.T.; Grabon, A.; Alb, J.G.; Faenza, I.; Cocco, L.; Brown, H.A.; Bankaitis, V.A. Quantitative profiling of the endonuclear glycerophospholipidome of murine embryonic fibroblasts. J. Lipid Res. 2016, 57, 1492–1506. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, G.O.; Renner, M.L.; Gil, G.A.; Rodríguez-Berdini, L.; Caputto, B.L. c-Fos-activated synthesis of nuclear phosphatidylinositol 4,5-bisphosphate PtdIns (4,5)P(2) promotes global transcriptional changes. Biochem. J. 2014, 461, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, R.; Sanyal, S.; Ghosh, A.; Bhar, K.; Das, C.; Siddhanta, A. Phosphatidylinositol-4-phosphate 5-kinase 1alpha modulates ribosomal RNA gene silencing through its interaction with histone H3 lysine 9 trimethylation and heterochromatin protein HP1-alpha. J. Biol. Chem. 2015, 290, 20893–20903. [Google Scholar] [CrossRef] [PubMed]
- Bua, D.J.; Martin, G.M.; Binda, O.; Gozani, O. Nuclear phosphatidylinositol-5-phosphate regulates ING2 stability at discrete chromatin targets in response to DNA damage. Sci. Rep. 2013, 3, 2137. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Laishram, R.S.; Ji, Z.; Barlow, C.A.; Tian, B.; Anderson, R.A. Star-PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIalpha and PKCdelta signaling. Mol. Cell 2012, 45, 25–37. [Google Scholar] [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Hyman, Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4334–4339. [Google Scholar] [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016, 165, 1686–1697. [Google Scholar] [CrossRef]
- Jost, D.; Carrivain, P.; Cavalli, G.; Vaillant, C. Modeling epigenome folding: Formation and dynamics of topologically associated chromatin domains. Nucl. Acids Res. 2014, 42, 9553–9561. [Google Scholar] [CrossRef]
- Strom, A.R.; Emelyanov, A.V.; Mir, M.; Fyodorov, D.V.; Darzacq, X.; Karpen, G.H. Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxyterminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Souquere, S.; Chujo, T.; Kobelke, S.; Chong, Y.S.; Fox, A.H.; Bond, C.S.; Nakagawa, S.; Pierron, G.; Hirose, T. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 2018, 70, 1038–1053. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Han, K.Y.; Khanna, N.; Ha, T.; Belmont, A.S. Nuclear speckle fusion via long-range directional motion regulates the number and size of speckles. J. Cell Sci. 2019, 132, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Keenen, M.M.; Larson, A.G.; Narlikar, G.J. Visualization and quantitation of phase-separated droplet Formation By Human HP1alpha. Methods Enzymol. 2018, 611, 51–66. [Google Scholar] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castano, E.; Yildirim, S.; Fáberová, V.; Krausová, A.; Uličná, L.; Paprčková, D.; Sztacho, M.; Hozák, P. Nuclear Phosphoinositides—Versatile Regulators of Genome Functions. Cells 2019, 8, 649. https://doi.org/10.3390/cells8070649
Castano E, Yildirim S, Fáberová V, Krausová A, Uličná L, Paprčková D, Sztacho M, Hozák P. Nuclear Phosphoinositides—Versatile Regulators of Genome Functions. Cells. 2019; 8(7):649. https://doi.org/10.3390/cells8070649
Chicago/Turabian StyleCastano, Enrique, Sukriye Yildirim, Veronika Fáberová, Alžběta Krausová, Lívia Uličná, Darina Paprčková, Martin Sztacho, and Pavel Hozák. 2019. "Nuclear Phosphoinositides—Versatile Regulators of Genome Functions" Cells 8, no. 7: 649. https://doi.org/10.3390/cells8070649
APA StyleCastano, E., Yildirim, S., Fáberová, V., Krausová, A., Uličná, L., Paprčková, D., Sztacho, M., & Hozák, P. (2019). Nuclear Phosphoinositides—Versatile Regulators of Genome Functions. Cells, 8(7), 649. https://doi.org/10.3390/cells8070649





