ERBB2 Regulates MED24 during Cancer Progression in Mice with Pten and Smad4 Deletion in the Pulmonary Epithelium
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Derivation, Culture, and siRNA Knockdown
2.3. Cell Proliferation
2.4. Histopathology and Immunohistochemistry
2.5. RNA Isolation, qRT-PCR, and Microarray Analysis
2.5.1. RNA Isolation
2.5.2. qRT-PCR
2.5.3. Experiment Processes of Microarray Analysis
2.5.4. Expression Array Analysis of Microarray Analysis
2.5.5. Functional Analysis of Differentially Changed Genes (DEGs)
2.5.6. Heatmap
2.6. Western Blot
2.7. Correlation Analysis and Survival Analysis
2.8. Quantification and Statistical Analysis
2.9. Data Availability
3. Results
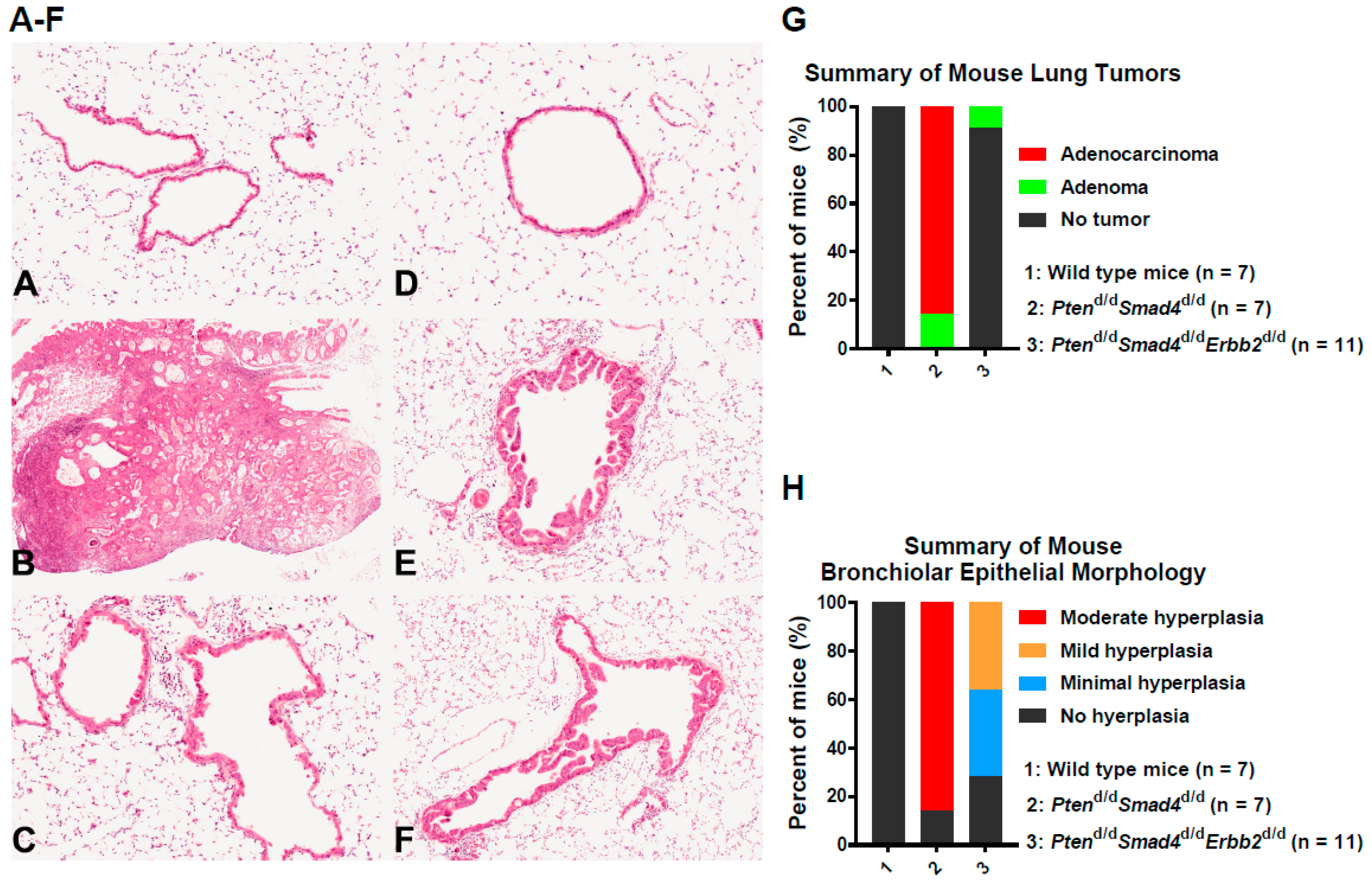
3.1. In Vivo Ablation of Erbb2 Prevents Pten−/−Smad4−/−-Induced Lung Tumor Development
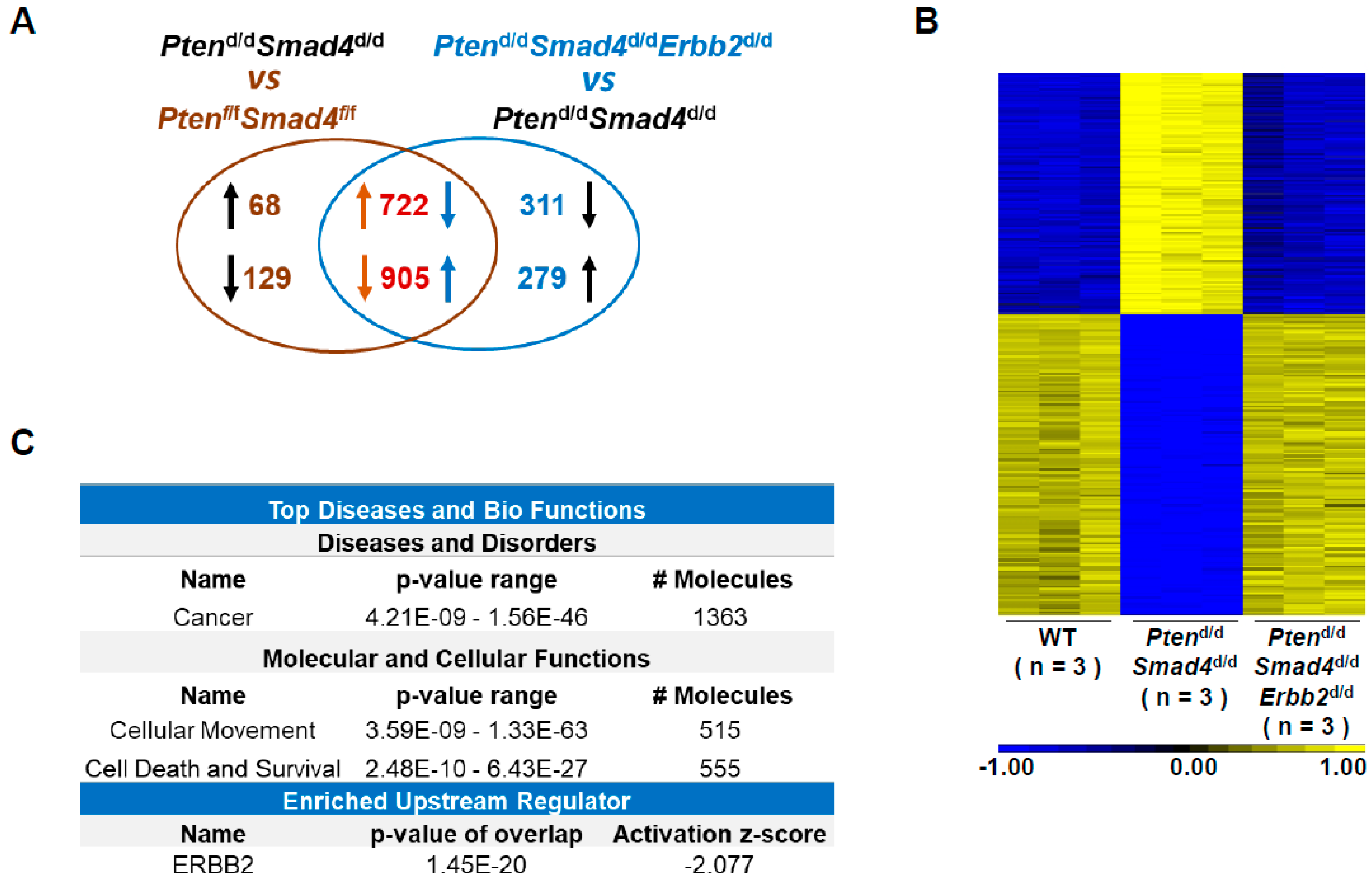
3.2. Transcriptome Analysis Identifies ERBB2-Regulated Cancer Genes
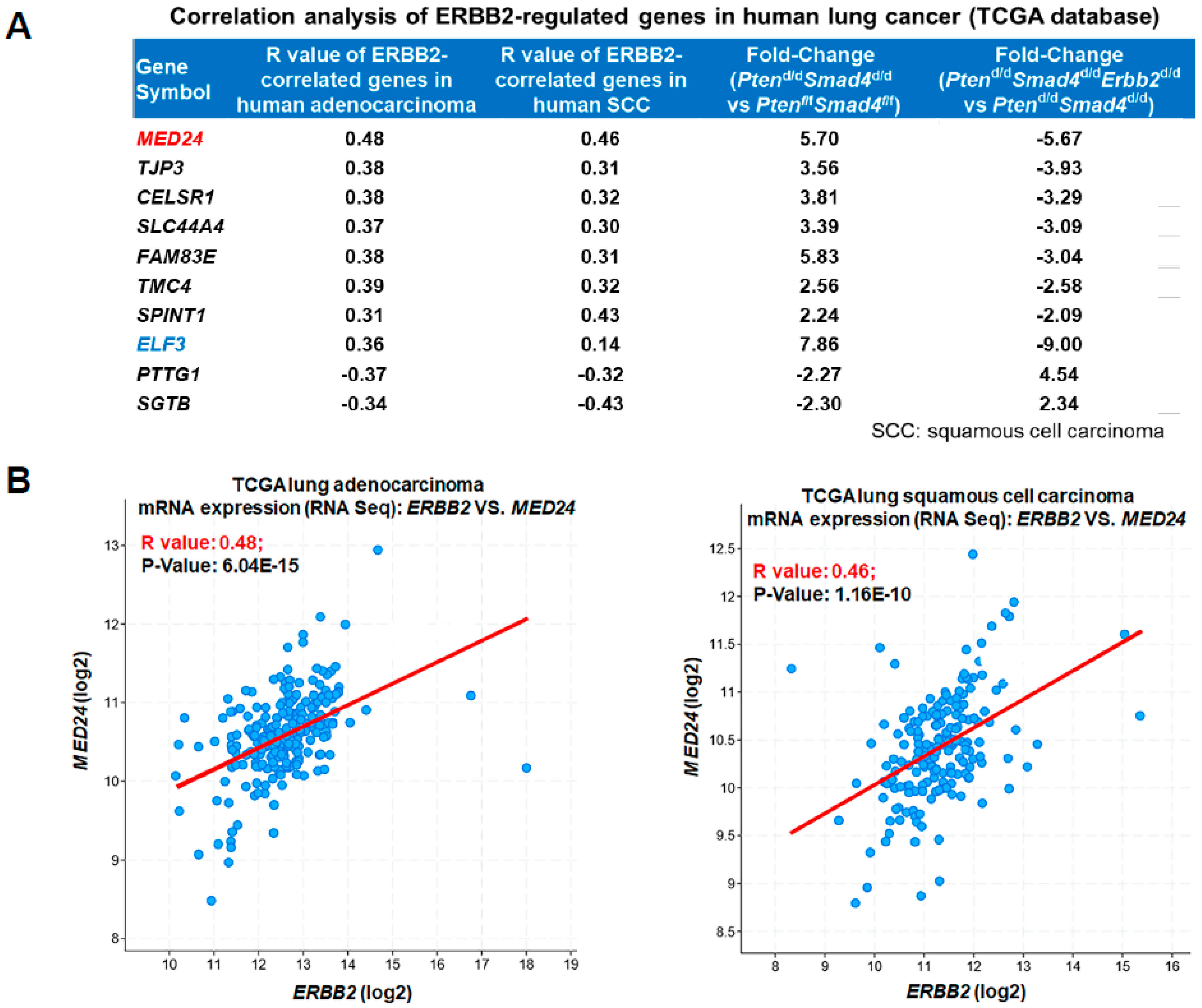
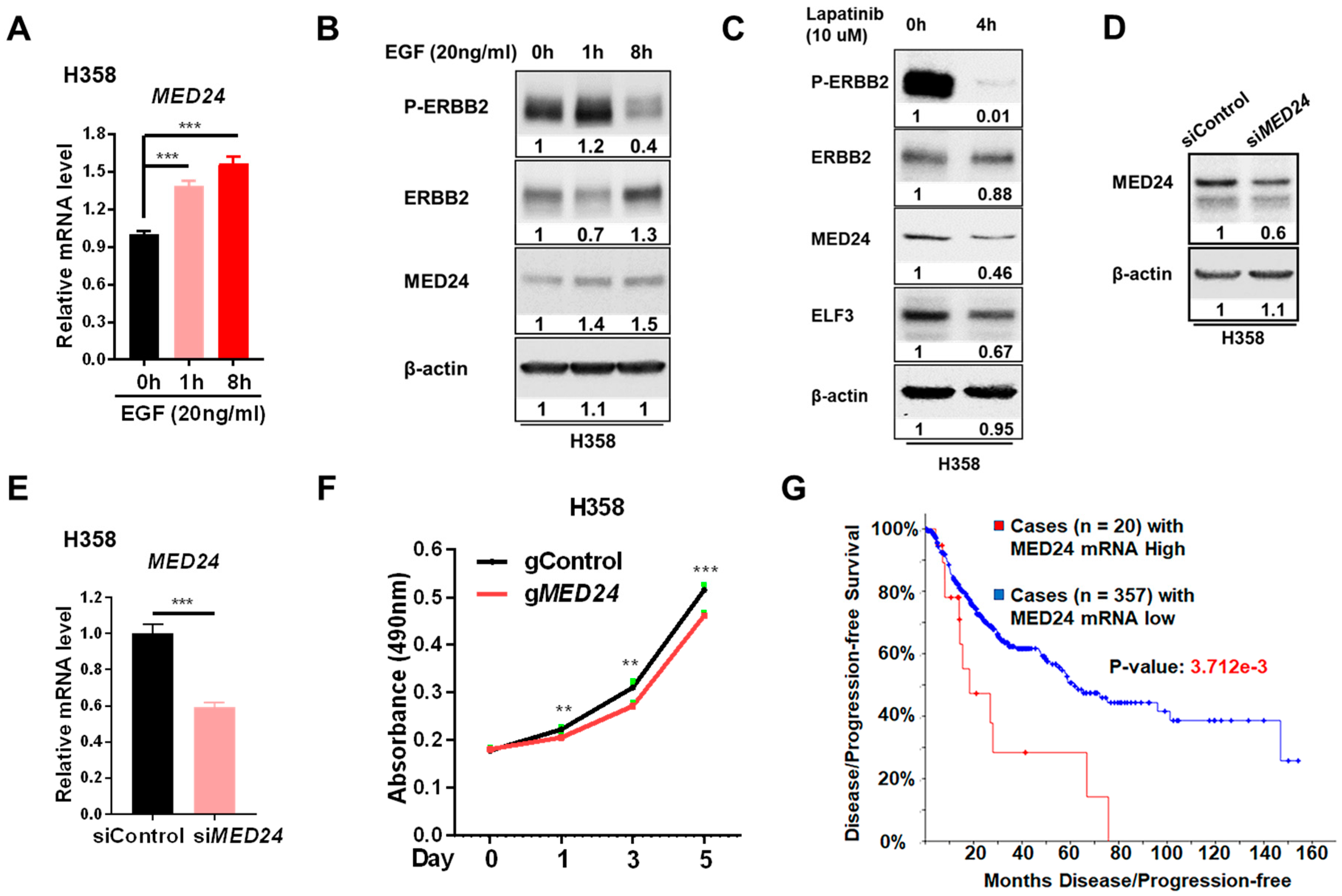
3.3. Conserved ERBB2-Downstream Genes (e.g., MED24) between Mouse and Human Lung Cancer
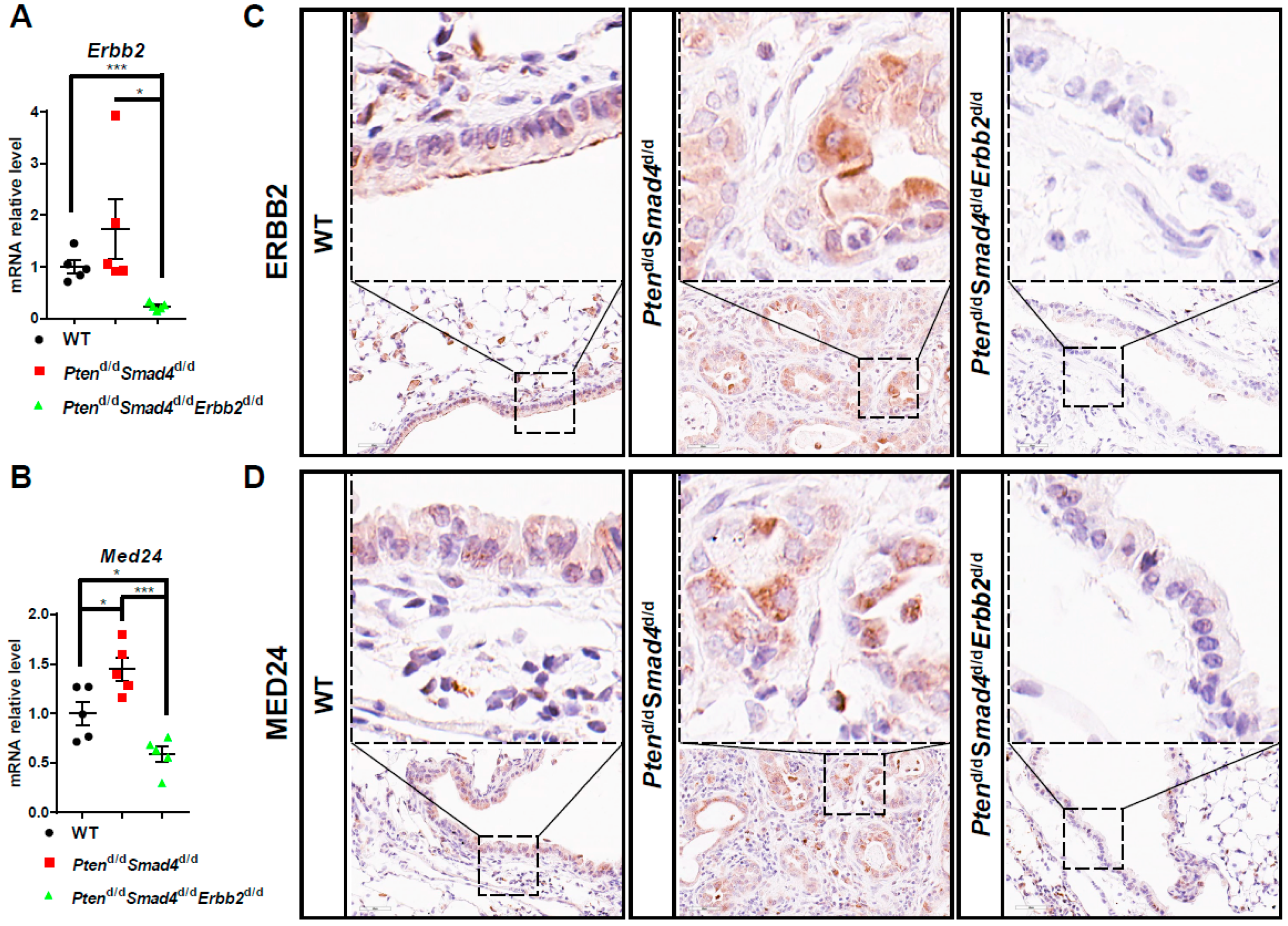
3.4. MED24 May Be an Oncogenic Player in ERBB2-Dependent Lung Cancer Development
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Siegel, R.L.; Jemal, A. Lung Cancer Statistics. Adv. Exp. Med. Biol. 2016, 893, 1–19. [Google Scholar] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Raparia, K.; Villa, C.; DeCamp, M.M.; Patel, J.D.; Mehta, M.P. Molecular profiling in non-small cell lung cancer: A step toward personalized medicine. Arch. Pathol. Lab. Med. 2013, 137, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Petersen, I. The morphological and molecular diagnosis of lung cancer. Dtsch. Arztebl. Int. 2011, 108, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Wee, P.; Wang, Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers 2017, 9, 52. [Google Scholar] [CrossRef]
- Liu, J.; Cho, S.N.; Wu, S.P.; Jin, N.; Moghaddam, S.J.; Gilbert, J.L.; Wistuba, I.; DeMayo, F.J. Mig-6 deficiency cooperates with oncogenic Kras to promote mouse lung tumorigenesis. Lung Cancer 2017, 112, 47–56. [Google Scholar] [CrossRef]
- Keshamouni, V.G.; Mattingly, R.R.; Reddy, K.B. Mechanism of 17-beta-estradiol-induced Erk1/2 activation in breast cancer cells. A role for HER2 AND PKC-delta. J. Biol. Chem. 2002, 277, 22558–22565. [Google Scholar] [CrossRef]
- Rusnak, D.W.; Affleck, K.; Cockerill, S.G.; Stubberfield, C.; Harris, R.; Page, M.; Smith, K.J.; Guntrip, S.B.; Carter, M.C.; Shaw, R.J.; et al. The characterization of novel, dual ErbB-2/EGFR, tyrosine kinase inhibitors: potential therapy for cancer. Cancer Res. 2001, 61, 7196–7203. [Google Scholar]
- Olayioye, M.A. Update on HER-2 as a target for cancer therapy: intracellular signaling pathways of ErbB2/HER-2 and family members. Breast Cancer Res. 2001, 3, 385–389. [Google Scholar] [CrossRef]
- Macdonald-Obermann, J.L.; Piwnica-Worms, D.; Pike, L.J. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc. Natl. Acad. Sci. USA 2012, 109, 137–142. [Google Scholar] [CrossRef]
- Pahuja, K.B.; Nguyen, T.T.; Jaiswal, B.S.; Prabhash, K.; Thaker, T.M.; Senger, K.; Chaudhuri, S.; Kljavin, N.M.; Antony, A.; Phalke, S.; et al. Actionable Activating Oncogenic ERBB2/HER2 Transmembrane and Juxtamembrane Domain Mutations. Cancer Cell 2018, 34, 792–806 e5. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cho, S.N.; Akkanti, B.; Jin, N.; Mao, J.; Long, W.; Chen, T.; Zhang, Y.; Tang, X.; Wistub, I.I.; et al. ErbB2 Pathway Activation upon Smad4 Loss Promotes Lung Tumor Growth and Metastasis. Cell Rep. 2015, 10, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Seshacharyulu, P.; Ponnusamy, M.P.; Haridas, D.; Jain, M.; Ganti, A.K.; Batra, S.K. Targeting the EGFR signaling pathway in cancer therapy. Expert. Opin. Ther. Targets 2012, 16, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Mates, M.; Fletcher, G.G.; Freedman, O.C.; Eisen, A.; Gandhi, S.; Trudeau, M.E.; Dent, S.F. Systemic targeted therapy for her2-positive early female breast cancer: a systematic review of the evidence for the 2014 Cancer Care Ontario systemic therapy guideline. Curr. Oncol. 2015, 22, S114–S122. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Herter-Sprie, G.S.; Greulich, H.; Wong, K.K. Activating Mutations in ERBB2 and Their Impact on Diagnostics and Treatment. Front. Oncol. 2013, 3, 86. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, S.; Hai, J.; Wang, X.; Chen, T.; Quinn, M.M.; Gao, P.; Zhang, Y.; Ji, H.; Cross, D.A.E.; et al. Targeting HER2 Aberrations in Non-Small Cell Lung Cancer with Osimertinib. Clin. Cancer Res. 2018, 24, 2594–2604. [Google Scholar] [CrossRef] [PubMed]
- Perera, S.A.; Li, D.; Shimamura, T.; Raso, M.G.; Ji, H.; Chen, L.; Borgman, C.L.; Zaghlul, S.; Brandstetter, K.A.; Kubo, S.; et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc. Natl. Acad. Sci. USA 2009, 106, 474–479. [Google Scholar] [CrossRef]
- Large, M.J.; Wetendorf, M.; Lanz, R.B.; Hartig, S.M.; Creighton, C.J.; Mancini, M.A.; Kovanci, E.; Lee, K.F.; Threadgill, D.W.; Lydon, J.P.; et al. The epidermal growth factor receptor critically regulates endometrial function during early pregnancy. PLoS Genet. 2014, 10, e1004451. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, T.; Creighton, C.J.; Wu, S.P.; Ray, M.; Janardhan, K.S.; Willson, C.J.; Cho, S.N.; Castro, P.D.; Ittmann, M.M.; et al. JNK(1/2) represses Lkb(1)-deficiency-induced lung squamous cell carcinoma progression. Nat. Commun. 2019, 10, 2148. [Google Scholar] [CrossRef]
- Liu, J.; Yu, G.; Zhao, Y.; Zhao, D.; Wang, Y.; Wang, L.; Liu, J.; Li, L.; Zeng, Y.; Dang, Y.; et al. REGgamma modulates p53 activity by regulating its cellular localization. J. Cell Sci. 2010, 123, 4076–4084. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Bartel, C.A.; Parameswaran, N.; Cipriano, R.; Jackson, M.W. FAM83 proteins: Fostering new interactions to drive oncogenic signaling and therapeutic resistance. Oncotarget 2016, 7, 52597–52612. [Google Scholar] [CrossRef] [PubMed]
- Cipriano, R.; Miskimen, K.L.; Bryson, B.L.; Foy, C.R.; Bartel, C.A.; Jackson, M.W. Conserved oncogenic behavior of the FAM83 family regulates MAPK signaling in human cancer. Mol. Cancer Res. 2014, 12, 1156–1165. [Google Scholar] [CrossRef] [PubMed]
- Diermeier, S.; Horvath, G.; Knuechel-Clarke, R.; Hofstaedter, F.; Szollosi, J.; Brockhoff, G. Epidermal growth factor receptor coexpression modulates susceptibility to Herceptin in HER2/neu overexpressing breast cancer cells via specific erbB-receptor interaction and activation. Exp. Cell Res. 2005, 304, 604–619. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Yin, J.W.; Wang, G. The Mediator complex: A master coordinator of transcription and cell lineage development. Development 2014, 141, 977–987. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, M.; Xia, M.; Liu, Y.; Yan, J.; Ji, H.; Wang, G. Selective requirement for Mediator MED23 in Ras-active lung cancer. Proc. Natl. Acad. Sci. USA 2012, 109, E2813–E2822. [Google Scholar] [CrossRef]
- Sun, M.; Jiang, R.; Li, J.D.; Luo, S.L.; Gao, H.W.; Jin, C.Y.; Shi, D.L.; Wang, C.G.; Wang, B.; Zhang, X.Y. MED19 promotes proliferation and tumorigenesis of lung cancer. Mol. Cell. Biochem. 2011, 355, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Yoon, N.K.; Maresh, E.L.; Elshimali, Y.; Li, A.; Horvath, S.; Seligson, D.B.; Chia, D.; Goodglick, L. Elevated MED28 expression predicts poor outcome in women with breast cancer. BMC Cancer 2010, 10, 335. [Google Scholar] [CrossRef]
- Hasegawa, N.; Sumitomo, A.; Fujita, A.; Aritome, N.; Mizuta, S.; Matsui, K.; Ishino, R.; Inoue, K.; Urahama, N.; Nose, J.; et al. Mediator subunits MED1 and MED24 cooperatively contribute to pubertal mammary gland development and growth of breast carcinoma cells. Mol. Cell. Biol. 2012, 32, 1483–1495. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Okano, H.J.; Darnell, R.B.; Roeder, R.G. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002, 21, 3464–3475. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Wang, T.; Willson, C.J.; Janardhan, K.S.; Wu, S.-P.; Li, J.-L.; DeMayo, F.J. ERBB2 Regulates MED24 during Cancer Progression in Mice with Pten and Smad4 Deletion in the Pulmonary Epithelium. Cells 2019, 8, 615. https://doi.org/10.3390/cells8060615
Liu J, Wang T, Willson CJ, Janardhan KS, Wu S-P, Li J-L, DeMayo FJ. ERBB2 Regulates MED24 during Cancer Progression in Mice with Pten and Smad4 Deletion in the Pulmonary Epithelium. Cells. 2019; 8(6):615. https://doi.org/10.3390/cells8060615
Chicago/Turabian StyleLiu, Jian, Tianyuan Wang, Cynthia J. Willson, Kyathanahalli S. Janardhan, San-Pin Wu, Jian-Liang Li, and Francesco J. DeMayo. 2019. "ERBB2 Regulates MED24 during Cancer Progression in Mice with Pten and Smad4 Deletion in the Pulmonary Epithelium" Cells 8, no. 6: 615. https://doi.org/10.3390/cells8060615
APA StyleLiu, J., Wang, T., Willson, C. J., Janardhan, K. S., Wu, S.-P., Li, J.-L., & DeMayo, F. J. (2019). ERBB2 Regulates MED24 during Cancer Progression in Mice with Pten and Smad4 Deletion in the Pulmonary Epithelium. Cells, 8(6), 615. https://doi.org/10.3390/cells8060615




