Evolutionary Divergence of Duplicated Hsf Genes in Populus
Abstract
1. Introduction
2. Materials and Methods
2.1. Characteristics of Populus Hsf Genes
2.2. Promoter Analysis
2.3. Gene Expression and Co-Expression Network
2.4. Splice Variants of PtHsfs
2.5. Natural Variation in PtHsfs
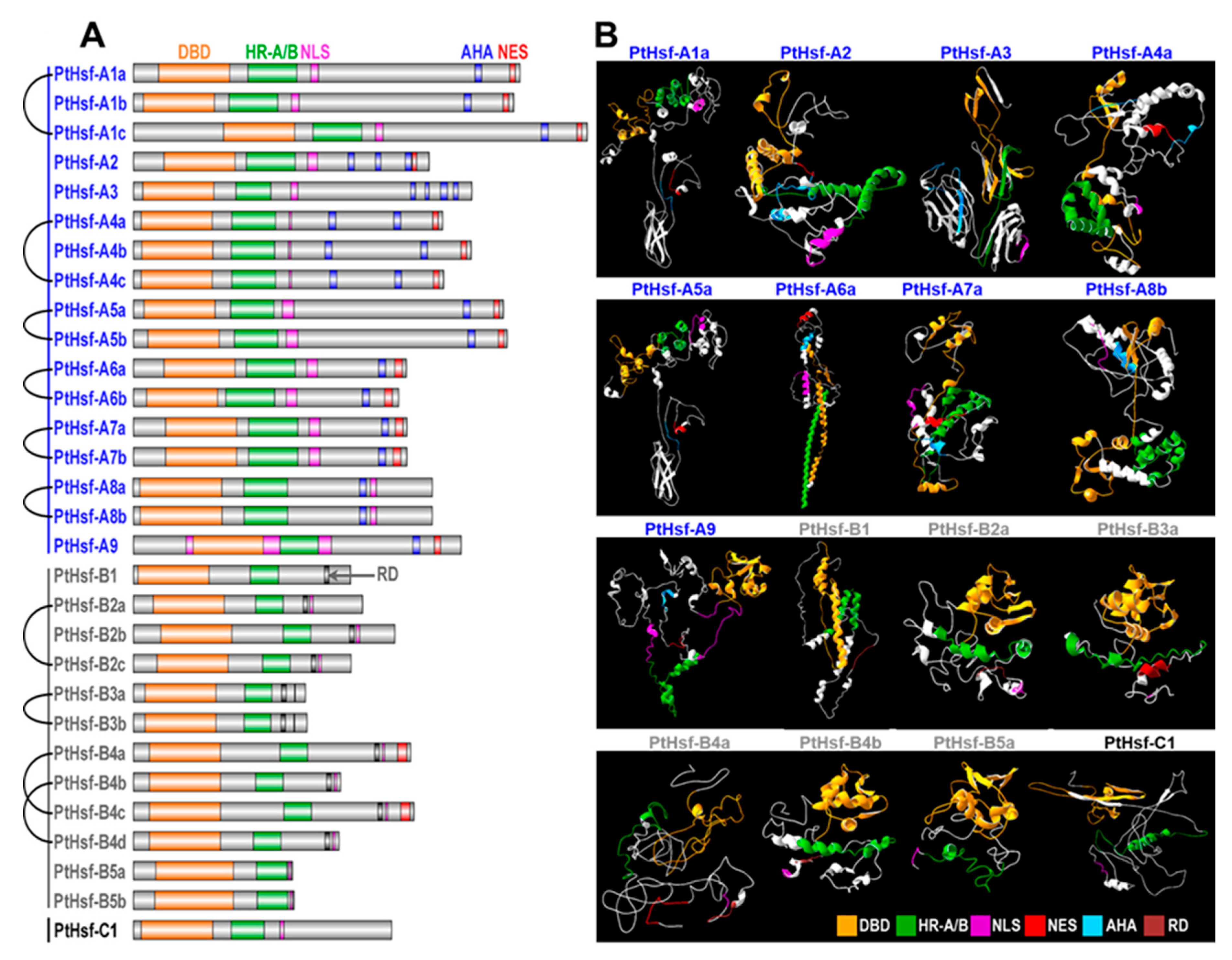
2.6. Protein Structural Modeling
3. Results
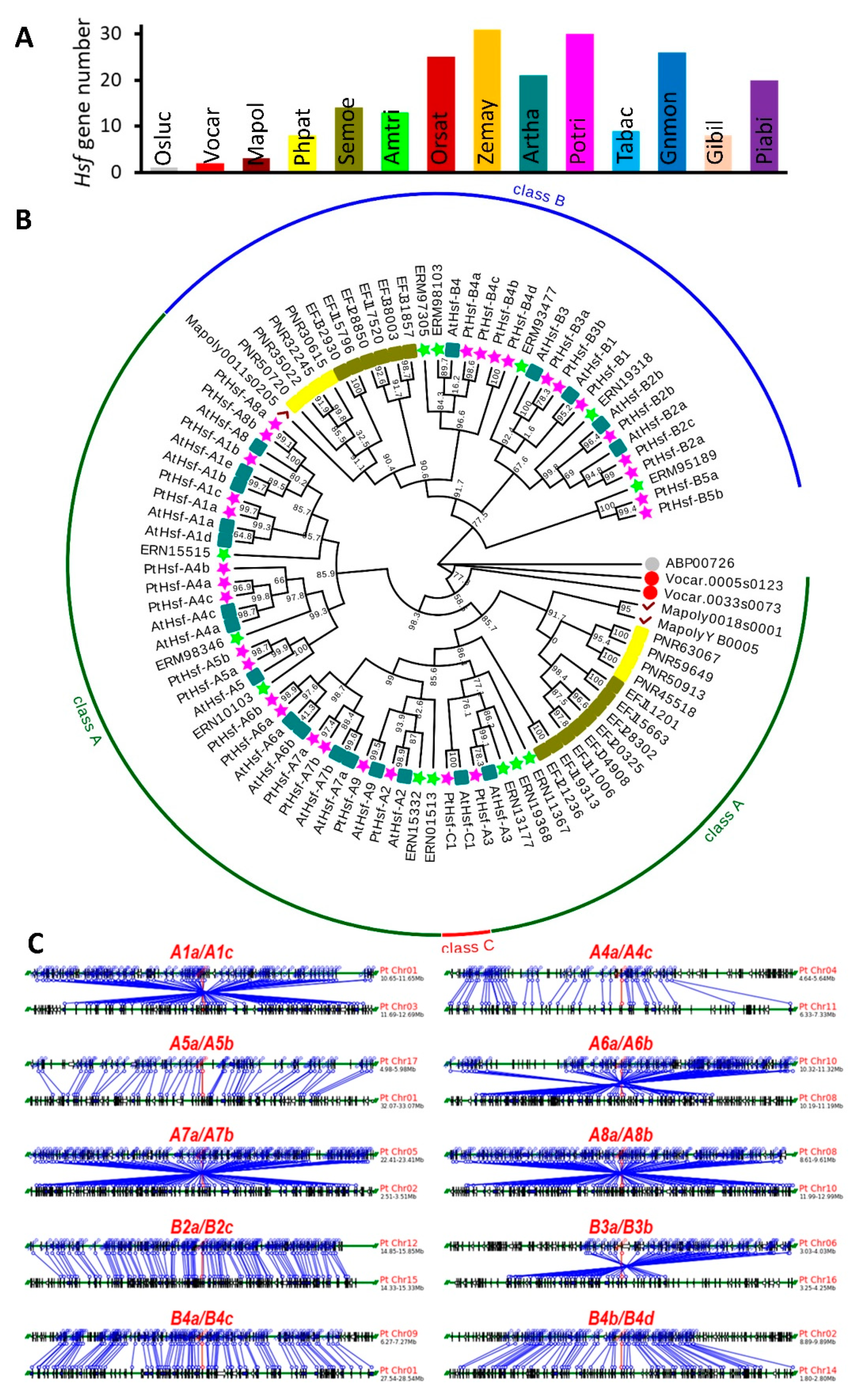
3.1. Genome-Wide Duplication of Hsf Genes in Populus
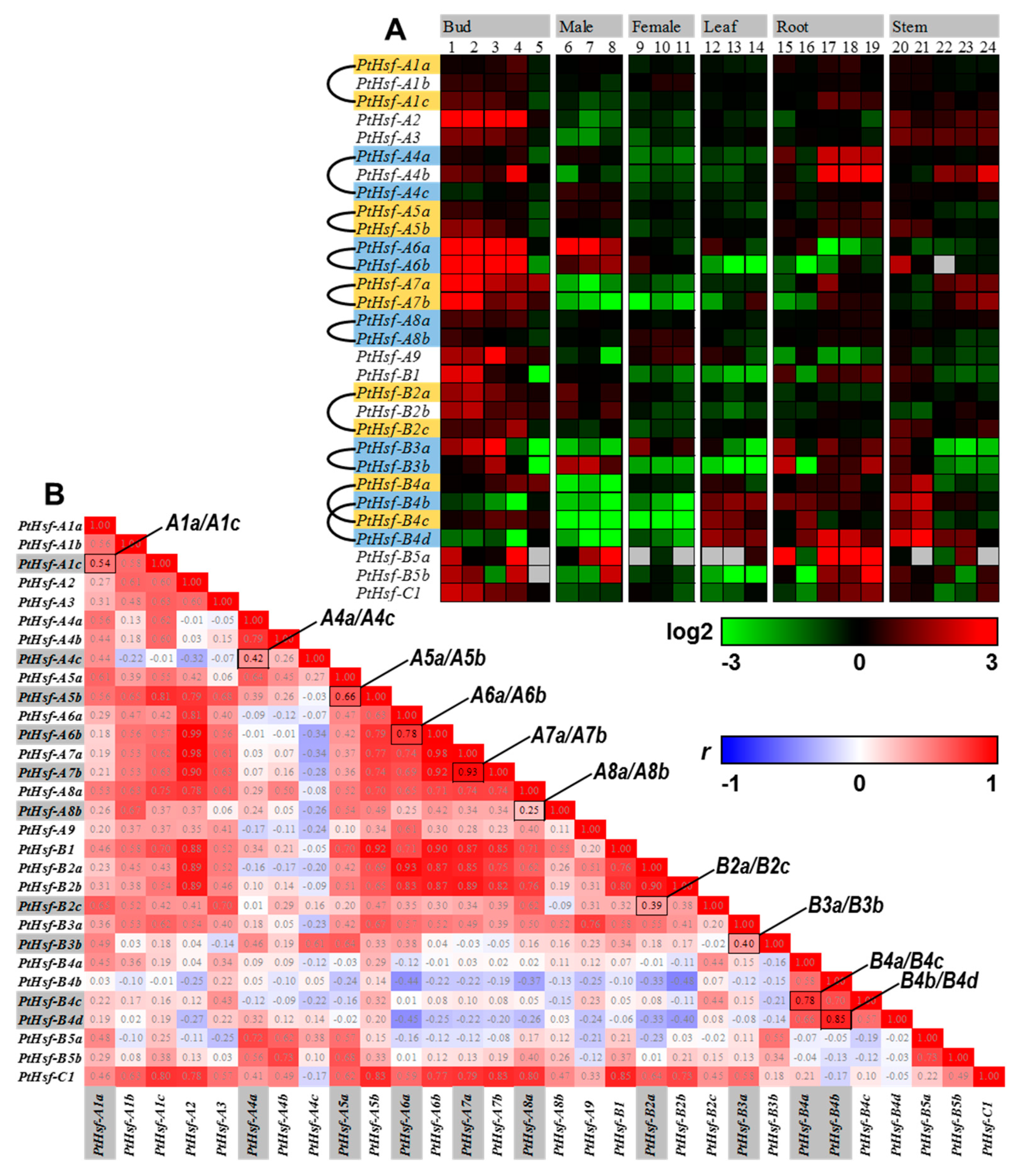
3.2. Expression Similarity and Divergence of PtHsf Genes
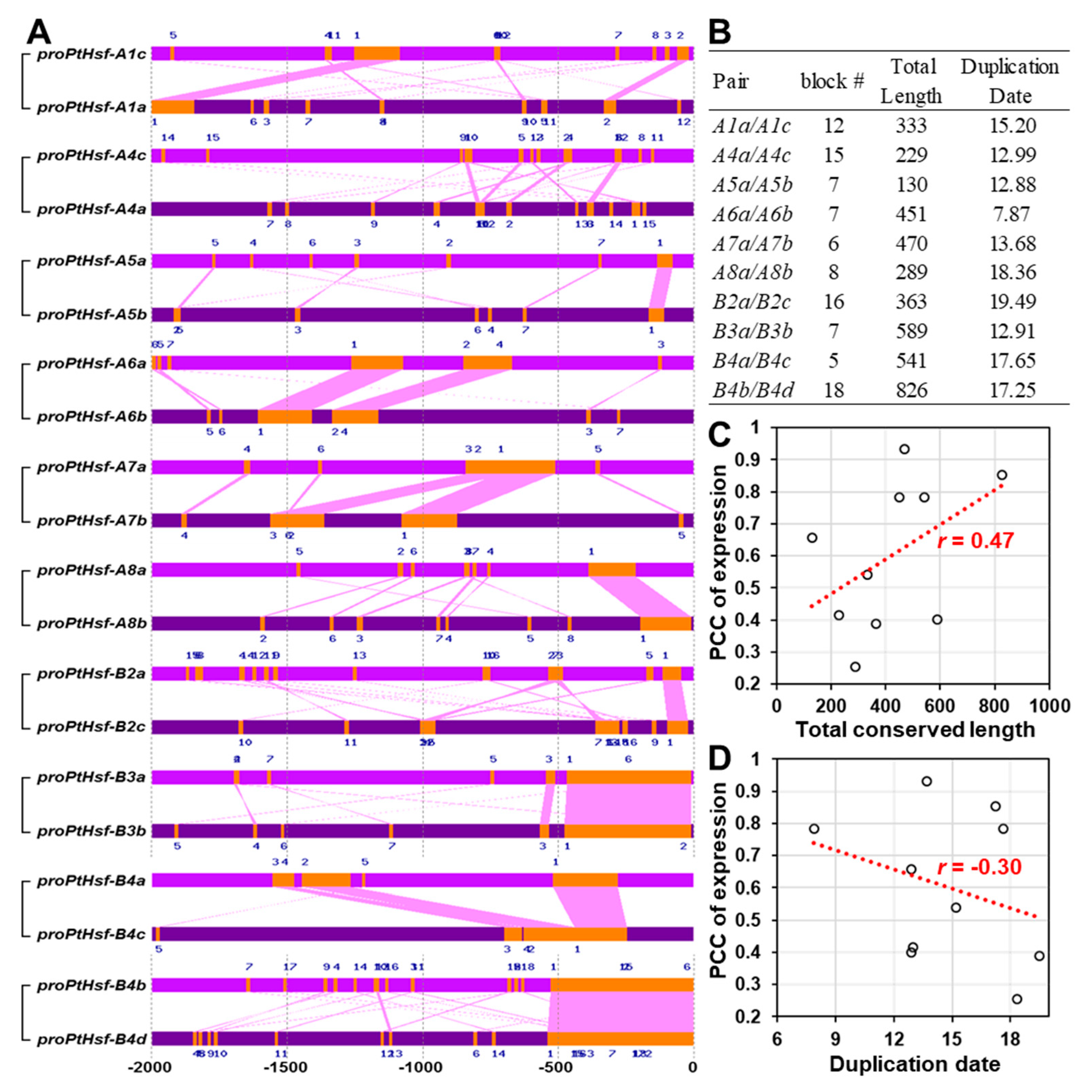
3.3. Promoter Similarity between the PtHsf Paralogous Pairs
3.4. Co-Expression Network of PtHsfs
3.5. Alternative Splicing of PtHsf
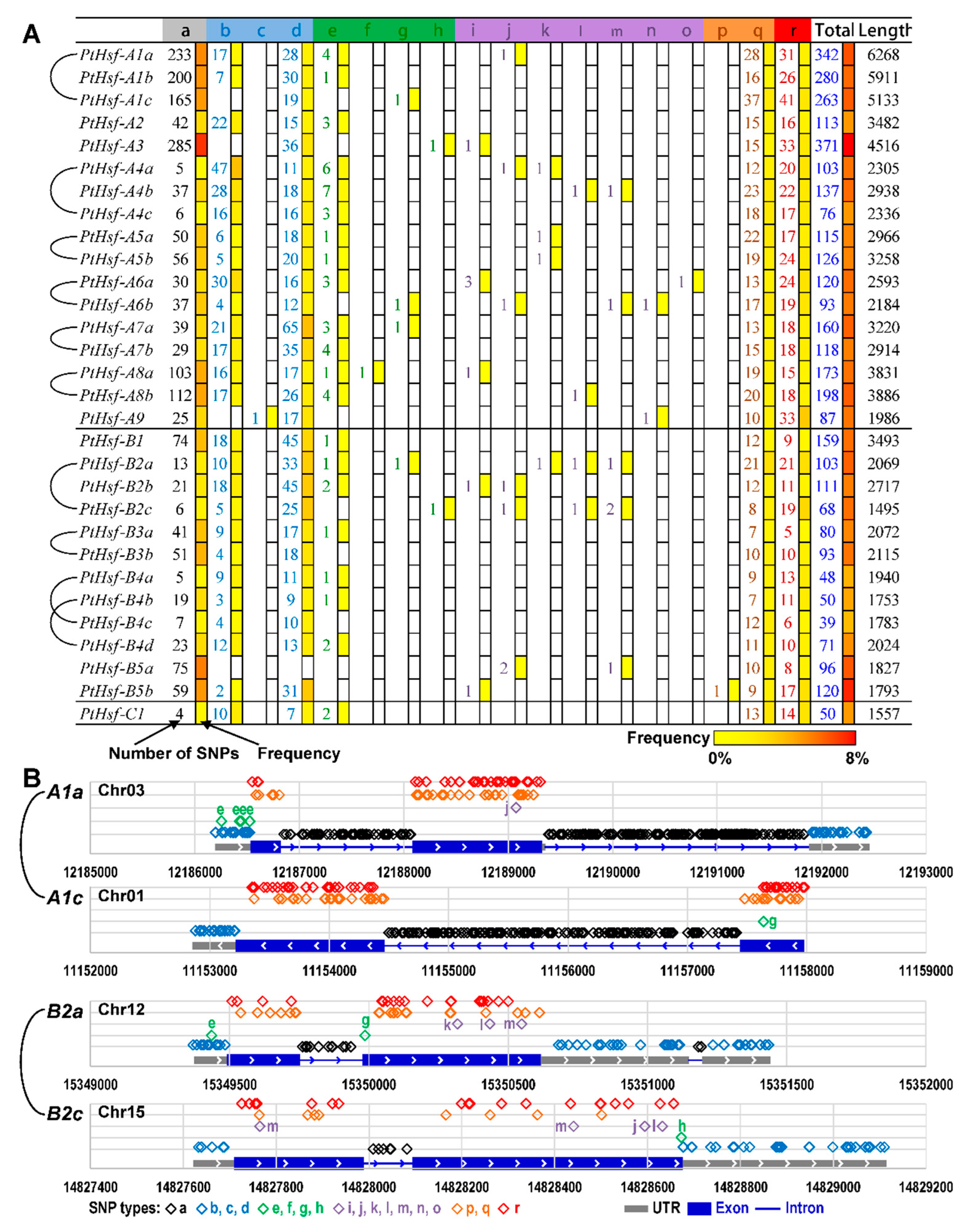
3.6. The SNP Differences between PtHsf Paralogous Pairs
3.7. Divergence of Protein 3D Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Akerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, S. The heat-shock response. Ann. Rev. Biochem. 1986, 55, 1151–1191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, J.; Liu, B.; Zhang, L.; Chen, J.; Lu, M. Genome-wide analysis of the populus hsp90 gene family reveals differential expression patterns, localization, and heat stress responses. BMC Genom. 2013, 14, 532. [Google Scholar] [CrossRef] [PubMed]
- Anckar, J.; Sistonen, L. Regulation of hsf1 function in the heat stress response: Implications in aging and disease. Ann. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef]
- Jedlicka, P.; Mortin, M.A.; Wu, C. Multiple functions of drosophila heat shock transcription factor In Vivo. EMBO J. 1997, 16, 2452–2462. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.C.; Charng, Y.Y. Common and distinct functions of arabidopsis class a1 and a2 heat shock factors in diverse abiotic stress responses and development. Plant Physiol. 2013, 163, 276–290. [Google Scholar] [CrossRef]
- Wu, C. Activating protein factor binds in vitro to upstream control sequences in heat shock gene chromatin. Nature 1984, 311, 81–84. [Google Scholar] [CrossRef]
- Gomez-Pastor, R.; Burchfiel, E.T.; Thiele, D.J. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 4–19. [Google Scholar] [CrossRef]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Jia, H.X.; Li, J.B.; Huang, J.; Lu, M.Z.; Hu, J.J. The heat shock factor gene family in salix suchowensis: A genome-wide survey and expression profiling during development and abiotic stresses. Front. Plant Sci. 2015, 6, 748. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, B.; Li, J.; Zhang, L.; Wang, Y.; Zheng, H.; Lu, M.; Chen, J. Hsf and hsp gene families in populus: Genome-wide identification, organization and correlated expression during development and in stress responses. BMC Genom. 2015, 16, 181. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, H.; Li, J.; Li, Y.; Lu, M.; Hu, J. Molecular evolution and expression divergence of the populus euphratica hsf genes provide insight into the stress acclimation of desert poplar. Sci. Rep. 2016, 6, 30050. [Google Scholar] [CrossRef]
- Wang, X.; Shi, X.; Chen, S.; Ma, C.; Xu, S. Evolutionary origin, gradual accumulation and functional divergence of heat shock factor gene family with plant evolution. Front. Plant Sci. 2018, 9, 71. [Google Scholar] [CrossRef]
- Taylor, G. Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 2002, 90, 681–689. [Google Scholar] [CrossRef]
- Gordon, J.C. Poplars: Trees of the people, trees of the future. For. Chron. 2001, 77, 217–219. [Google Scholar] [CrossRef]
- Demura, T.; Ye, Z.H. Regulation of plant biomass production. Curr. Opin. Plant Biol. 2010, 13, 298–303. [Google Scholar] [CrossRef]
- Sannigrahi, P.; Ragauskas, A.J.; Tuskan, G.A. Poplar as a feedstock for biofuels: A review of compositional characteristics. Biofuels Bioprod. Biorefining 2010, 4, 209–226. [Google Scholar] [CrossRef]
- Wilkinson, A. Poplars and willows for soil erosion control in new zealand. Biomass Bioenergy 1999, 16, 263–274. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, populus trichocarpa (torr. & gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Jansson, S.; Douglas, C.J. Populus: A model system for plant biology. Ann. Rev. Plant Biol. 2007, 58, 435–458. [Google Scholar] [CrossRef]
- Evans, L.M.; Slavov, G.T.; Rodgers-Melnick, E.; Martin, J.; Ranjan, P.; Muchero, W.; Brunner, A.M.; Schackwitz, W.; Gunter, L.; Chen, J.G.; et al. Population genomics of populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 2014, 46, 1089–1096. [Google Scholar] [CrossRef]
- Oubida, R.W.; Gantulga, D.; Zhang, M.; Zhou, L.; Bawa, R.; Holliday, J.A. Partitioning of multivariate phenotypes using regression trees reveals complex patterns of adaptation to climate across the range of black cottonwood (populus trichocarpa). Front. Plant Sci. 2015, 6, 181. [Google Scholar] [CrossRef]
- McKown, A.D.; Guy, R.D.; Klapste, J.; Geraldes, A.; Friedmann, M.; Cronk, Q.C.; El-Kassaby, Y.A.; Mansfield, S.D.; Douglas, C.J. Geographical and environmental gradients shape phenotypic trait variation and genetic structure in populus trichocarpa. New Phytol. 2014, 201, 1263–1276. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Von Koskull-Döring, P. Characterization of c-terminal domains of arabidopsis heat stress transcription factors (hsfs) and identification of a new signature combination of plant class a hsfs with aha and nes motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the nbs-lrr gene family in the cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; Minh, B.Q.; von Haeseler, A. W-iq-tree: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. Modelfinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. Ufboot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Chow, C.N.; Lee, T.Y.; Hung, Y.C.; Li, G.Z.; Tseng, K.C.; Liu, Y.H.; Kuo, P.L.; Zheng, H.Q.; Chang, W.C. Plantpan3. 0: A new and updated resource for reconstructing transcriptional regulatory networks from chip-seq experiments in plants. Nucleic Acids Res. 2018, 47, D1155–D1163. [Google Scholar] [CrossRef]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2010, 27, 431–432. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The i-tasser suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7. [Google Scholar] [CrossRef]
- Project, A.G. The amborella genome and the evolution of flowering plants. Science 2013, 342, 1241089. [Google Scholar] [CrossRef]
- Cooke, J.E.; Eriksson, M.E.; Junttila, O. The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant Cell Environ. 2012, 35, 1707–1728. [Google Scholar] [CrossRef]
- Rinne, P.L.; Kaikuranta, P.M.; van der Schoot, C. The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. Plant J. 2001, 26, 249–264. [Google Scholar] [CrossRef]
- Ruprecht, C.; Vaid, N.; Proost, S.; Persson, S.; Mutwil, M. Beyond genomics: Studying evolution with gene coexpression networks. Trends Plant Sci. 2017, 22, 298–307. [Google Scholar] [CrossRef]
- Reddy, T.V.; Kaur, J.; Agashe, B.; Sundaresan, V.; Siddiqi, I. The duet gene is necessary for chromosome organization and progression during male meiosis in arabidopsis and encodes a phd finger protein. Development 2003, 130, 5975–5987. [Google Scholar] [CrossRef]
- Volkenburgh, E.V. Leaf expansion–an integrating plant behaviour. Plant Cell Environ. 1999, 22, 1463–1473. [Google Scholar] [CrossRef]
- Iniguez, L.P.; Hernandez, G. The evolutionary relationship between alternative splicing and gene duplication. Front. Genet. 2017, 8, 14. [Google Scholar] [CrossRef]
- Su, Z.; Wang, J.; Yu, J.; Huang, X.; Gu, X. Evolution of alternative splicing after gene duplication. Genome Res. 2006, 16, 182–189. [Google Scholar] [CrossRef]
- Shastry, B.S. Snps: Impact on gene function and phenotype. Methods Mol. Biol. 2009, 578, 3–22. [Google Scholar]
- Shastry, B.S. Snp alleles in human disease and evolution. J. Hum. Genet. 2002, 47, 561–566. [Google Scholar] [CrossRef]
- Jaeger, A.M.; Pemble, C.W.T.; Sistonen, L.; Thiele, D.J. Structures of hsf2 reveal mechanisms for differential regulation of human heat-shock factors. Nat. Struct. Mol. Biol. 2016, 23, 147–154. [Google Scholar] [CrossRef]
- Dunn, M.J.; Kinney, G.M.; Washington, P.M.; Berman, J.; Anderson, M.Z. Functional diversification accompanies gene family expansion of med2 homologs in candida albicans. PLoS Genet. 2018, 14, e1007326. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.H.; Ma, X.; Luo, D.X.; Gong, Z.H.; Lu, M.H. The plant heat stress transcription factors (hsfs): Structure, regulation, and function in response to abiotic stresses. Front. Plant Sci. 2016, 7, 114. [Google Scholar] [CrossRef]
- McConnell, J.R.; Emery, J.; Eshed, Y.; Bao, N.; Bowman, J.; Barton, M.K. Role of phabulosa and phavoluta in determining radial patterning in shoots. Nature 2001, 411, 709–713. [Google Scholar] [CrossRef]
- Aguilar-Martinez, J.A.; Sinha, N. Analysis of the role of arabidopsis class i tcp genes attcp7, attcp8, attcp22, and attcp23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar] [CrossRef]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. Tcp14 and tcp15 affect internode length and leaf shape in arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef]
- Cramer, P.; Pesce, C.G.; Baralle, F.E.; Kornblihtt, A.R. Functional association between promoter structure and transcript alternative splicing. Proc. Natl. Acad. Sci. USA 1997, 94, 11456–11460. [Google Scholar] [CrossRef]
- Pecci, A.; Viegas, L.R.; Baranao, J.L.; Beato, M. Promoter choice influences alternative splicing and determines the balance of isoforms expressed from the mouse bcl-x gene. J. Biol. Chem. 2001, 276, 21062–21069. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.B.; Tchourbanov, A.Y.; Fan, H.; Danilov, S.M.; Wang, T.; Garcia, J.G. Mechanical stress and single nucleotide variants regulate alternative splicing of the mylk gene. Am. J. Resp. Cell Mol. 2017, 56, 29–37. [Google Scholar] [CrossRef]
- Guyon-Debast, A.; Lécureuil, A.; Bonhomme, S.; Guerche, P.; Gallois, J.L. A snp associated with alternative splicing of rpt5b causes unequal redundancy between rpt5a and rpt5b among arabidopsis thaliana natural variation. BMC Plant Biol. 2010, 10, 158. [Google Scholar] [CrossRef]
- Takii, R.; Fujimoto, M. Structure and function of the hsf family members. In Heat Shock Factor; Springer: Berlin, Germany, 2016; pp. 31–50. [Google Scholar]
- Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 1999, 12, 85–94. [Google Scholar] [CrossRef]
- Yang, W.Z.; Ko, T.P.; Corselli, L.; Johnson, R.C.; Yuan, H.S. Conversion of a beta-strand to an alpha-helix induced by a single-site mutation observed in the crystal structure of fis mutant pro26ala. Protein Sci. 1998, 7, 1875–1883. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, N.; Biswas, P. Helical ambivalency induced by point mutations. BMC Struct. Biol. 2013, 13, 9. [Google Scholar] [CrossRef] [PubMed]
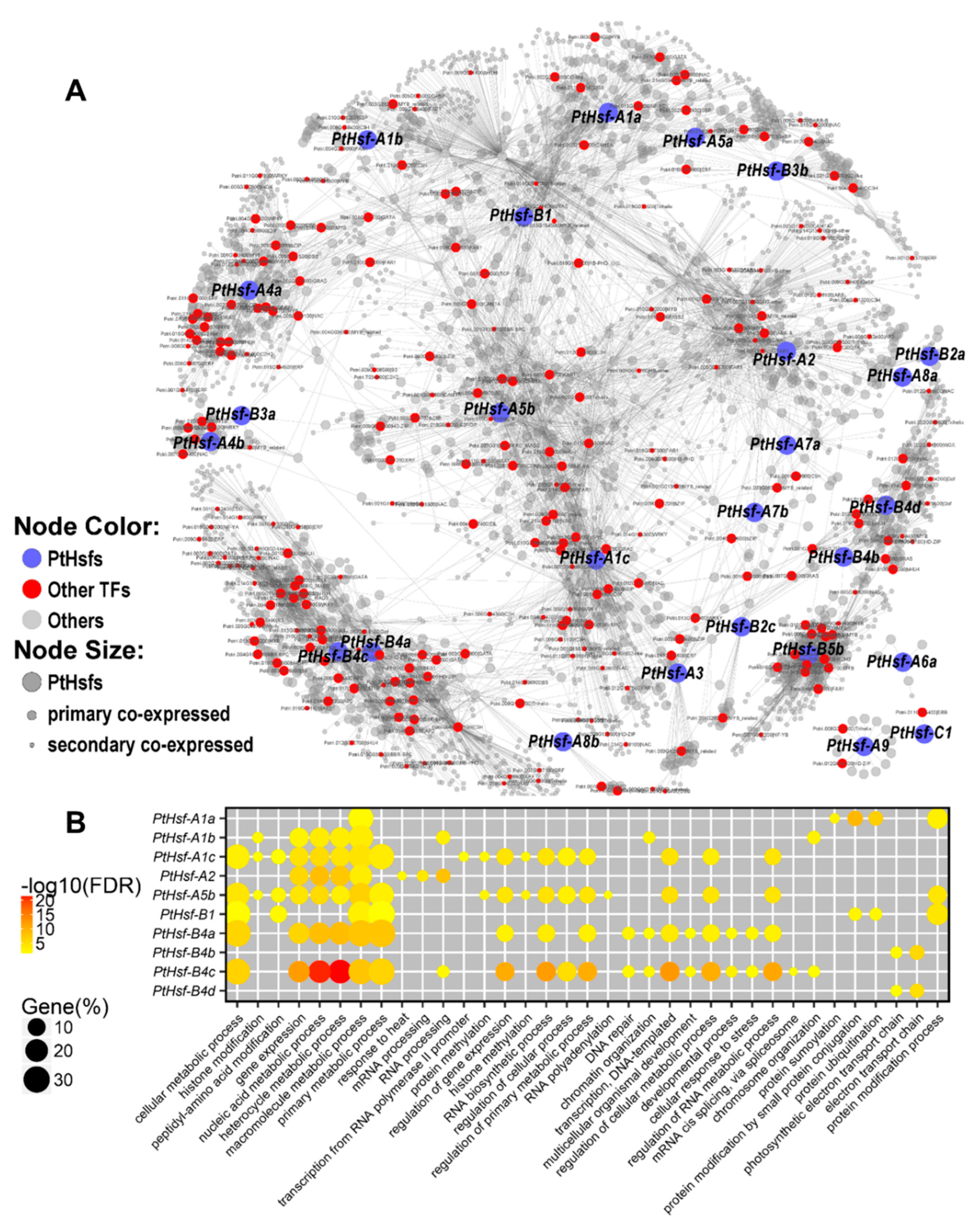
| Gene Name | Co-Expressed Gene # | Co-Expressed TF # | Enriched GO Term | Enriched Protein Domain | Enriched Pathway |
|---|---|---|---|---|---|
| PtHsf-A1a | 268 | 8 | 9 | 9 | 1 |
| PtHsf-A1b | 281 | 2 | 17 | 20 | 1 |
| PtHsf-A1c | 458 | 21 | 57 | 2 | 1 |
| PtHsf-A2 | 439 | 6 | 16 | 41 | 2 |
| PtHsf-A3 | 84 | 3 | n.a. | 2 | n.a. |
| PtHsf-A4a | 275 | 24 | n.a. | 5 | n.a. |
| PtHsf-A4b | 17 | 4 | n.a. | n.a. | n.a. |
| PtHsf-A4c | 0 | 0 | n.a. | n.a. | n.a. |
| PtHsf-A5a | 137 | 4 | n.a. | n.a. | n.a. |
| PtHsf-A5b | 626 | 21 | 56 | 28 | 1 |
| PtHsf-A6a | 6 | 0 | n.a. | n.a. | n.a. |
| PtHsf-A6b | 0 | 0 | n.a. | n.a. | n.a. |
| PtHsf-A7a | 91 | 4 | n.a. | 13 | 1 |
| PtHsf-A7b | 54 | 3 | n.a. | n.a. | n.a. |
| PtHsf-A8a | 8 | 1 | n.a. | n.a. | n.a. |
| PtHsf-A8b | 17 | 2 | n.a. | n.a. | n.a. |
| PtHsf-A9 | 15 | 2 | n.a. | n.a. | n.a. |
| PtHsf-B1 | 269 | 11 | 19 | 1 | 1 |
| PtHsf-B2a | 18 | 0 | n.a. | 6 | 1 |
| PtHsf-B2b | 0 | 0 | n.a. | n.a. | n.a. |
| PtHsf-B2c | 39 | 3 | n.a. | n.a. | n.a. |
| PtHsf-B3a | 27 | 1 | n.a. | n.a. | n.a. |
| PtHsf-B3b | 103 | 5 | n.a. | 2 | n.a. |
| PtHsf-B4a | 315 | 10 | 53 | n.a. | 1 |
| PtHsf-B4b | 176 | 6 | 7 | 11 | 3 |
| PtHsf-B4c | 391 | 29 | 59 | 5 | n.a. |
| PtHsf-B4d | 200 | 8 | 6 | 11 | 3 |
| PtHsf-B5a | 0 | 0 | n.a. | n.a. | n.a. |
| PtHsf-B5b | 154 | 12 | n.a. | 2 | n.a. |
| PtHsf-C1 | 2 | 1 | n.a. | n.a. | n.a. |
| Class | Gene Name | Gene ID | Transcript ID | AS type a | Resultant |
|---|---|---|---|---|---|
| Class A | PtHsf-A1a | Potri.003G095000 | Potri.003G095000.1 | (primary transcript) | |
| Potri.003G095000.2 | A5SS | 3′ UTR change | |||
| PtHsf-A1b | Potri.013G079800 | Potri.013G079800.1 | (primary transcript) | ||
| PtHsf-A1c | Potri.001G138900 | Potri.001G138900.3 | (primary transcript) | ||
| Potri.001G138900.2 | A3SS | loss partial of DBD | |||
| PtHsf-A2 | Potri.006G226800 | Potri.006G226800.4 | (primary transcript) | ||
| Potri.006G226800.2 | A5SS, SE | loss partial of C-terminal and 3′ UTR change | |||
| Potri.006G226800.3 | A5SS | loss partial of C-terminal and 3′ UTR change | |||
| PtHsf-A3 | Potri.006G115700 | Potri.006G115700.2 | (primary transcript) | ||
| PtHsf-A4a | Potri.011G071700 | Potri.011G071700.1 | (primary transcript) | ||
| PtHsf-A4b | Potri.014G141400 | Potri.014G141400.1 | (primary transcript) | ||
| Potri.014G141400.2 | RI | loss DBD | |||
| PtHsf-A4c | Potri.004G062300 | Potri.004G062300.1 | (primary transcript) | ||
| Potri.004G062300.2 | A3SS | loss partial DBD | |||
| PtHsf-A5a | Potri.017G059600 | Potri.017G059600.1 | (primary transcript) | ||
| PtHsf-A5b | Potri.001G320900 | Potri.001G320900.1 | (primary transcript) | ||
| PtHsf-A6a | Potri.010G082000 | Potri.010G082000.2 | (primary transcript) | ||
| Potri.010G082000.1 | SE | 3′ UTR change | |||
| PtHsf-A6b | Potri.008G157600 | Potri.008G157600.1 | (primary transcript) | ||
| PtHsf-A7a | Potri.005G214800 | Potri.005G214800.2 | (primary transcript) | ||
| Potri.005G214800.4 | SE, SE | 3′ UTR change | |||
| Potri.005G214800.1 | SE | 3′ UTR change | |||
| Potri.005G214800.3 | SE, SE, SE | 3′ UTR change | |||
| Potri.005G214800.5 | SE, SE | 3′ UTR change | |||
| Potri.005G214800.6 | RI | N-terminal | |||
| PtHsf-A7b | Potri.002G048200 | Potri.002G048200.1 | (primary transcript) | ||
| Potri.002G048200.2 | RI | 3′ UTR change | |||
| Potri.002G048200.3 | SE | 3′ UTR change | |||
| Potri.002G048200.4 | RI, SE | 3′ UTR change | |||
| PtHsf-A8a | Potri.008G136800 | Potri.008G136800.2 | (primary transcript) | ||
| Potri.008G136800.6 | A5SS | 3′ UTR change | |||
| PtHsf-A8b | Potri.010G104300 | Potri.010G104300.2 | (primary transcript) | ||
| Potri.010G104300.1 | SE | 3′ UTR change | |||
| PtHsf-A9 | Potri.006G148200 | Potri.006G148200.2 | (primary transcript) | ||
| Class B | PtHsf-B1 | Potri.007G043800 | Potri.007G043800.1 | (primary transcript) | |
| PtHsf-B2a | Potri.012G138900 | Potri.012G138900.1 | (primary transcript) | ||
| Potri.012G138900.2 | SE, RI | 3′ UTR change | |||
| Potri.012G138900.3 | RI | 3′ UTR change | |||
| PtHsf-B2b | Potri.001G108100 | Potri.001G108100.1 | (primary transcript) | ||
| Potri.001G108100.3 | A3SS | 5′ UTR change | |||
| Potri.001G108100.2 | A5SS | 5′ UTR change | |||
| PtHsf-B2c | Potri.015G141100 | Potri.015G141100.1 | (primary transcript) | ||
| Potri.015G141100.2 | SE | loss partial of internal sequence | |||
| PtHsf-B3a | Potri.006G049200 | Potri.006G049200.1 | (primary transcript) | ||
| PtHsf-B3b | Potri.016G056500 | Potri.016G056500.1 | (primary transcript) | ||
| PtHsf-B4a | Potri.002G124800 | Potri.002G124800.1 | (primary transcript) | ||
| PtHsf-B4b | Potri.009G068000 | Potri.009G068000.1 | (primary transcript) | ||
| PtHsf-B4c | Potri.014G027100 | Potri.014G027100.3 | (primary transcript) | ||
| Potri.014G027100.1 | RI | loss partial of C-terminal | |||
| PtHsf-B4d | Potri.001G273700 | Potri.001G273700.1 | (primary transcript) | ||
| PtHsf-B5a | Potri.004G042600 | Potri.004G042600.1 | (primary transcript) | ||
| PtHsf-B5b | Potri.011G051600 | Potri.011G051600.1 | (primary transcript) | ||
| Class C | PtHsf-C1 | Potri.T137400 | Potri.T137400.1 | (primary transcript) |
| Gene Name | DBD | HR-A/B | AHA | Others (NLS, NES, or RD) |
|---|---|---|---|---|
| PtHsf-A1a | Chr03:12188111 G->T (W105C) | Chr03:12188394 A->G (M200V) | ||
| Chr03:12188143 A->T (Q116L) | ||||
| PtHsf-A1b | Chr13:6946101 T->C (K107E) | Chr13:6946028 G->A (S131F) | Chr13:6945777 T->G (K215Q) <NLS> | |
| Chr13:6945966 C->A (V152F) | ||||
| Chr13:6945909 G->C (R171G) | ||||
| PtHsf-A1c | Chr01:11154369 C->T (R211Q) | Chr01:11154258 T->C (E248G) | Chr01:11153373 G->A (A543V) | |
| Chr01:11154213 C->T (R263K) | ||||
| Chr01:11154182 G->T (S273R) | ||||
| PtHsf-A2 | Chr06:23831182 C->A (S94I) | Chr06:23830387 C->G (M171I) | ||
| Chr06:23830564 C->A (W112C) | Chr06:23830281 T->C (T207A) | |||
| PtHsf-A3 | Chr06:9090513 G->T (P15H) | Chr06:9088646 C->T (V367I) | ||
| Chr06:9090444 A->G (I38T) | Chr06:9088592 G->A (P385S) | |||
| Chr06:9089476 G->A (S90F) | ||||
| Chr06:9089434 C->A (R104M) | ||||
| PtHsf-A4a | Chr11:6830049 T->A (N37Y) | Chr11:6829640 C->T (D132N) | Chr11:6828857 G->A (L393F) <NES> | |
| Chr11:6830025 G->T (P45T) | Chr11:6829620 T->A (K138N) | |||
| Chr11:6829607 C->A (A143S) | ||||
| Chr11:6829480 C->G (R185P) | ||||
| Chr11:6829475 A->T (L187M) | ||||
| PtHsf-A4b | Chr14:10767449 A->C (S6R) | Chr14:10766450 T->C (K131R) | Chr14:10765533 T->C (I437V) <NES> | |
| Chr14:10766596 C->A (Q82H) | Chr14:10766449 C->A (K131N) | |||
| Chr14:10766553 T->G (I97L) | Chr14:10766399 T->C (Q148R) | |||
| Chr14:10766333 C->G (S170T) | ||||
| Chr14:10766295 C->A (G183C) | ||||
| Chr14:10766294 C->G (G183A) | ||||
| PtHsf-A4c | Chr04:5146215 G->A (P25L) | Chr04:5145571 T->A (M187L) | Chr04:5145100 C->G (V344L) | |
| Chr04:5146204 G->A (P29S) | ||||
| Chr04:5146203 G->T (P29Q) | ||||
| Chr04:5146185 T->G (Q35P) | ||||
| Chr04:5146182 C->G (S36T) | ||||
| PtHsf-A5a | Chr17:5478669 C->T (S43N) | Chr17:5477318 G->C (H138D) | Chr17:5476292 T->C (M480V) <NES> | |
| Chr17:5477446 T->C (K95R) | Chr17:5477236 T->A (Q165L) | |||
| Chr17:5477207 C->T (E175K) | ||||
| PtHsf-A5b | Chr01:32571330 G->T (F23L) | Chr01:32569699 T->C (K144R) | ||
| Chr01:32571193 T->C (N69S) | Chr01:32569688 T->C (K148E) | |||
| Chr01:32569818 G->T (H104Q) | Chr01:32569582 A->C (L183R) | |||
| PtHsf-A6a | Chr10:10826222 C->A (S64I) | Chr10:10825317 G->C (L156V) | Chr10:10825077 T->A (I236F) <NLS> | |
| Chr10:10826111 G->T (T101K) | Chr10:10825283 T->C (K167R) | Chr10:10824747 A->C (L346V) <NES> | ||
| Chr10:10825416 T->C (K123E) | Chr10:10825281 G->T (Q168K) | Chr10:10824724 C->G (L353F) <NES> | ||
| Chr10:10825251 T->C (R178G) | ||||
| Chr10:10825200 C->T (V195I) | ||||
| Chr10:10825148 G->A (A212V) | ||||
| PtHsf-A6b | Chr08:10686052 C->T (P18S) | Chr08:10686988 T->A (L129Q) | Chr08:10687618 A->C (Y339S) <NES> | |
| Chr08:10687036 T->C (V145A) | ||||
| Chr08:10687083 A->C (I161L) | ||||
| Chr08:10687099 G->A (R166Q) | ||||
| Chr08:10687140 A->G (S180G) | ||||
| PtHsf-A7a | Chr05:22774061 G->T (A74S) | Chr05:22774833 G->T (R156L) | Chr05:22775403 C->T (T346I) <NES> | |
| Chr05:22774724 G->A (G120R) | Chr05:22774866 G->A (R167K) | |||
| Chr05:22774951 C->G (D195E) | ||||
| Chr05:22774954 A->C (Q196H) | ||||
| PtHsf-A7b | Chr02:3141391 T->C (D56G) | Chr02:3140451 G->C (H171Q) | Chr02:3140246 T->A (T240S) <NLS> | |
| Chr02:3141341 C->T (V73I) | Chr02:3140405 C->T (A187T) | |||
| Chr02:3141326 A->G (Y78H) | Chr02:3140318 T->A (M216L) | |||
| Chr02:3141309 A->T (N83K) | ||||
| Chr02:3140561 T->C (R135G) | ||||
| PtHsf-A8a | Chr08:9112715 G->A (E251K) <NLS> | |||
| PtHsf-A8b | Chr10:12489151 T->G (M19L) | |||
| Chr10:12489048 T->C (K53R) | ||||
| Chr10:12487083 T->A (D78V) | ||||
| Chr10:12487082 A->T (D78E) | ||||
| Chr10:12487060 C->T (G86R) | ||||
| Chr10:12486987 C->T (R110Q) | ||||
| PtHsf-A9 | Chr06:12750969 T->C (K103E) | Chr06:12750442 C->G (V206L) | Chr06:12750381 C->T (S226N) <NLS> | |
| Chr06:12750927 A->G (S117P) | Chr06:12750409 T->G (K217Q) | |||
| Chr06:12750921 T->C (N119D) | ||||
| Chr06:12750920 T->A (N119I) | ||||
| Chr06:12750618 G->A (P147L) | ||||
| PtHsf-B1 | Chr07:3779417 T->A (E174V) | |||
| Chr07:3779389 C->A (L183F) | ||||
| PtHsf-B2a | Chr12:15349619 G->A (D44N) | Chr12:15350210 G->T (E166D) | Chr12:15350412 G->A (G234R) <NLS> | |
| Chr12:15349721 T->C (F78L) | Chr12:15350293 C->G (S194W) | Chr12:15350397 G->T (V229F) <RD> | ||
| Chr12:15350046 T->A (L112M) | ||||
| Chr12:15350051 A->T (L113F) | ||||
| Chr12:15350067 A->G (R119G) | ||||
| PtHsf-B2b | Chr01:8582316 C->G (D55E) | Chr01:8582880 C->T (S209L) | Chr01:8583138 T->C (V295A) <NLS> | |
| Chr01:8582335 G->A (D62N) | ||||
| Chr01:8582651 A->C (T133P) | ||||
| PtHsf-B2c | Chr15:14827877 A->T (E56D) | Chr15:14828338 A->G (E175G) | Chr15:14828531 G->T (E239D) <RD> | |
| Chr15:14827921 G->T (R71I) | ||||
| Chr15:14827935 A->C (K76Q) | ||||
| PtHsf-B3a | Chr06:3535947 A->G (K152R) | |||
| Chr06:3535970 G->T (V160F) | ||||
| Chr06:3535988 A->T (T166S) | ||||
| PtHsf-B3b | Chr16:3748780 T->A (I36L) | Chr16:3747556 G->T (T162N) | ||
| Chr16:3748759 T->C (T43A) | Chr16:3747537 G->C (N168K) | |||
| Chr16:3748734 G->T (A51E) | Chr16:3747526 T->C (K172R) | |||
| Chr16:3747757 C->A (R95L) | ||||
| PtHsf-B4a | Chr02:9388712 C->G (T49S) | Chr02:9389317 T->C (S209P) | Chr02:9389755 G->C (G355R) <NES> | |
| Chr02:9388795 G->T (V77F) | Chr02:9389350 G->A (D220N) | Chr02:9389656 T->C (S322P) <RD> | ||
| PtHsf-B4b | Chr09:6774240 C->A (L80I) | Chr09:6774986 A->C (K167Q) | Chr09:6775257 G->A (G257E) <RD> | |
| Chr09:6775259 G->T (V258F) <RD> | ||||
| PtHsf-B4c | Chr14:2300725 A->C (M211L) | Chr14:2301145 A->T (M351L) <NES> | ||
| Chr14:2301156 C->A (D354E) <NES> | ||||
| PtHsf-B4d | Chr01:28036766 T->G (N81K) | Chr01:28037553 A->C (N169H) | ||
| Chr01:28037311 T->C (V88A) | ||||
| Chr01:28037358 G->A (A104T) | ||||
| PtHsf-B5a | Chr04:3233937 G->T (P53H) | Chr04:3232326 G->C (T195S) | ||
| Chr04:3232591 A->T (S107T) | ||||
| Chr04:3232540 A->T (L124M) | ||||
| PtHsf-B5b | Chr11:4423922 C->A (A45S) | Chr11:4422794 G->A (T162M) | ||
| Chr11:4423915 T->C (D47G) | Chr11:4422692 G->T (T196N) | |||
| Chr11:4423904 C->A (D51Y) | Chr11:4422679 T->C (I200M) | |||
| Chr11:4423865 C->T (E64K) | ||||
| Chr11:4423834 G->T (A74D) | ||||
| Chr11:4423825 G->A (S77L) | ||||
| Chr11:4422971 T->C (K103R) | ||||
| Chr11:4422954 G->T (Q109K) | ||||
| Chr11:4422936 C->T (E115K) | ||||
| Chr11:4422931 C->A (K116N) | ||||
| PtHsf-C1 | scaffold_294:25641 C->G (G149A) | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, B.; Hu, J.; Zhang, J. Evolutionary Divergence of Duplicated Hsf Genes in Populus. Cells 2019, 8, 438. https://doi.org/10.3390/cells8050438
Liu B, Hu J, Zhang J. Evolutionary Divergence of Duplicated Hsf Genes in Populus. Cells. 2019; 8(5):438. https://doi.org/10.3390/cells8050438
Chicago/Turabian StyleLiu, Bobin, Jianjun Hu, and Jin Zhang. 2019. "Evolutionary Divergence of Duplicated Hsf Genes in Populus" Cells 8, no. 5: 438. https://doi.org/10.3390/cells8050438
APA StyleLiu, B., Hu, J., & Zhang, J. (2019). Evolutionary Divergence of Duplicated Hsf Genes in Populus. Cells, 8(5), 438. https://doi.org/10.3390/cells8050438





